Clinical Implications of Serum Hepatitis B Virus Pregenomic RNA Kinetics in Chronic Hepatitis B Patients Receiving Antiviral Treatment and Those Achieving HBsAg Loss
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Enrollment
2.2. Patient Monitoring
2.3. Quantification of Serum HBsAg
2.4. Quantification of Serum HBV DNA
2.5. Extraction and Reverse Transcription of HBV pgRNA
2.6. Quantification of Serum HBV pgRNA
2.7. HBV Genotyping
2.8. Statistical Analysis
3. Results
3.1. Clinical Characteristics of Patients in the Analysis of HBV pgRNA Kinetics during Entecavir Therapy
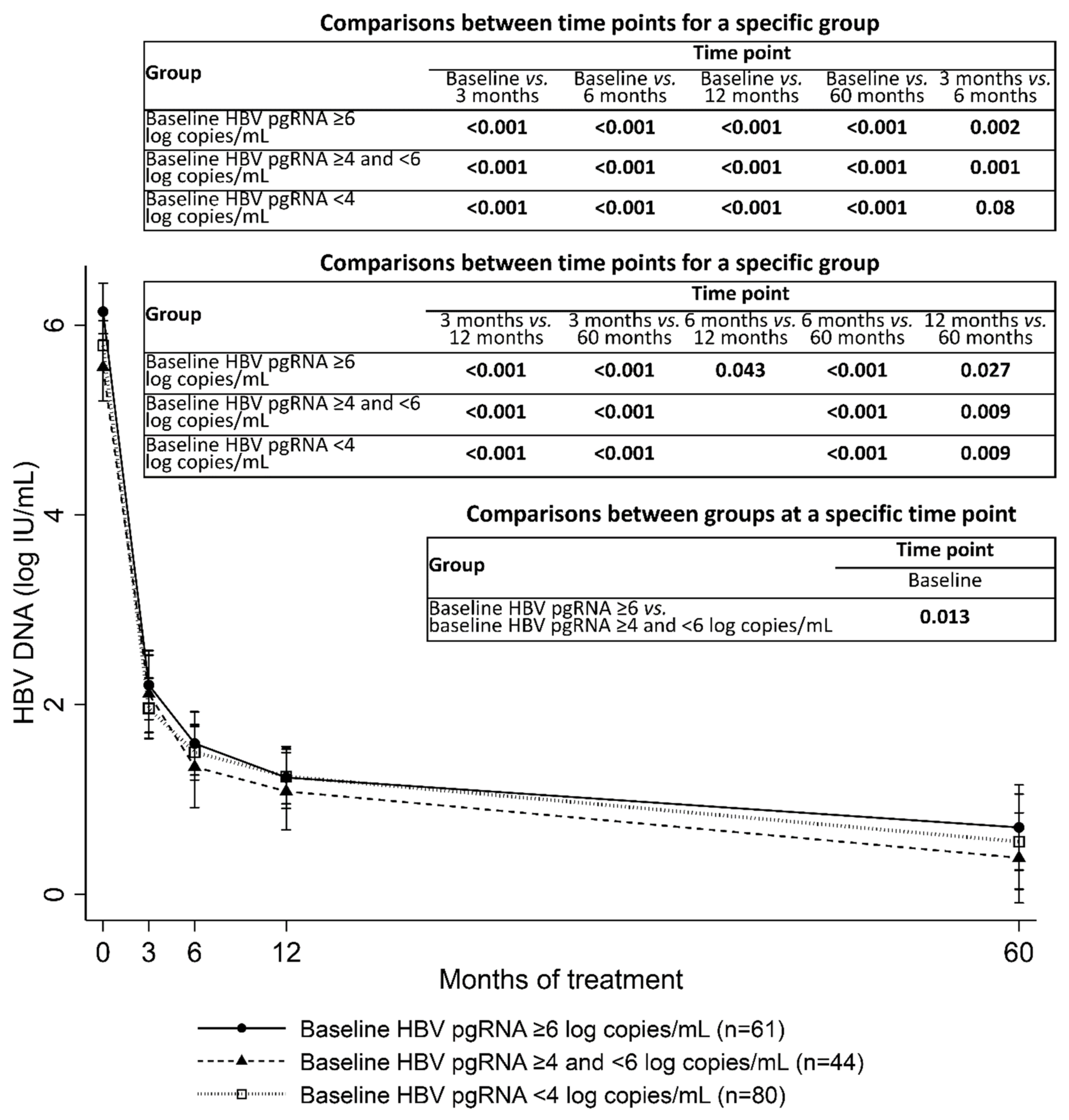
3.2. HBV pgRNA Kinetics during Entecavir Treatment
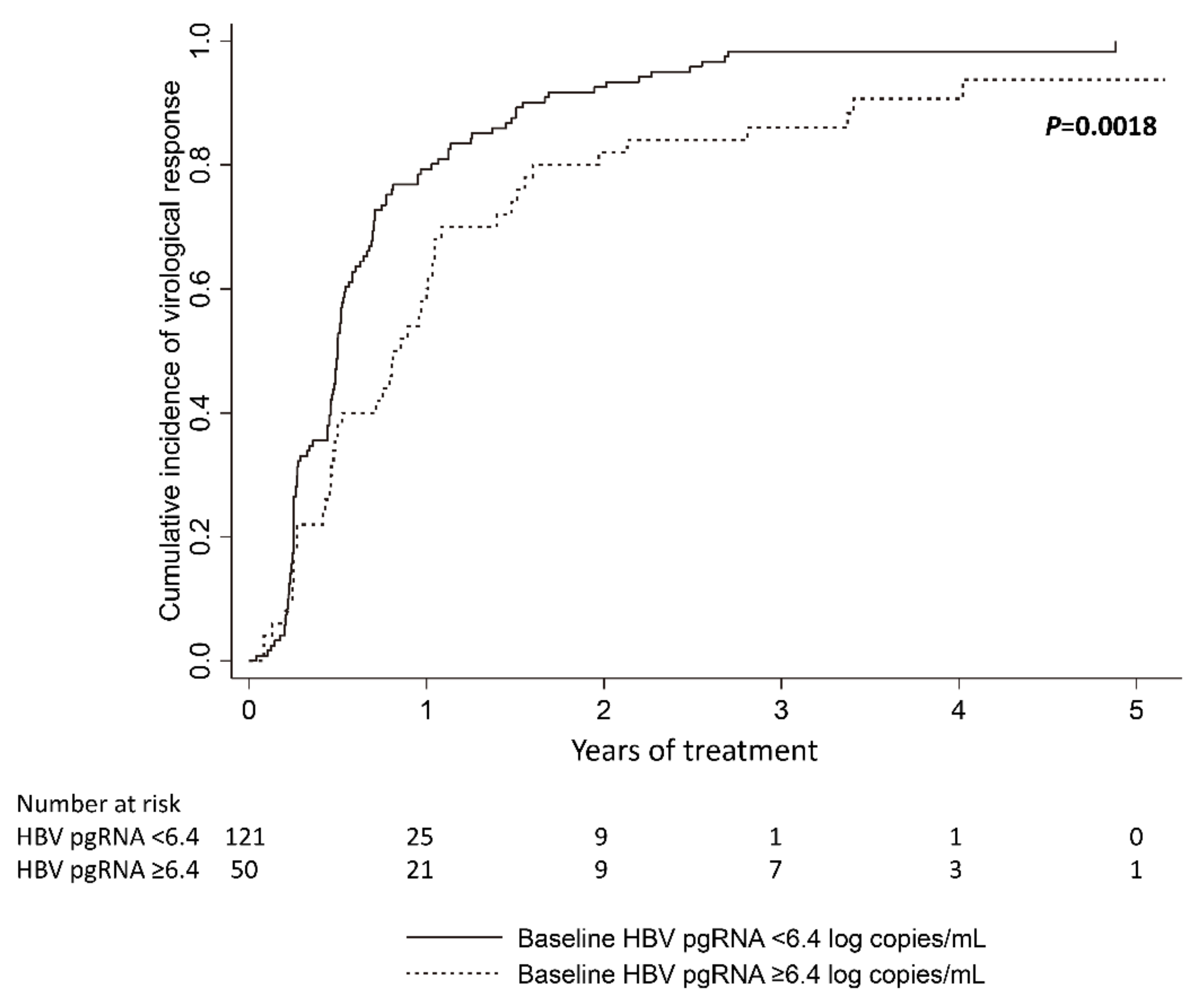
3.3. Baseline Serum HBV pgRNA and Virological Response
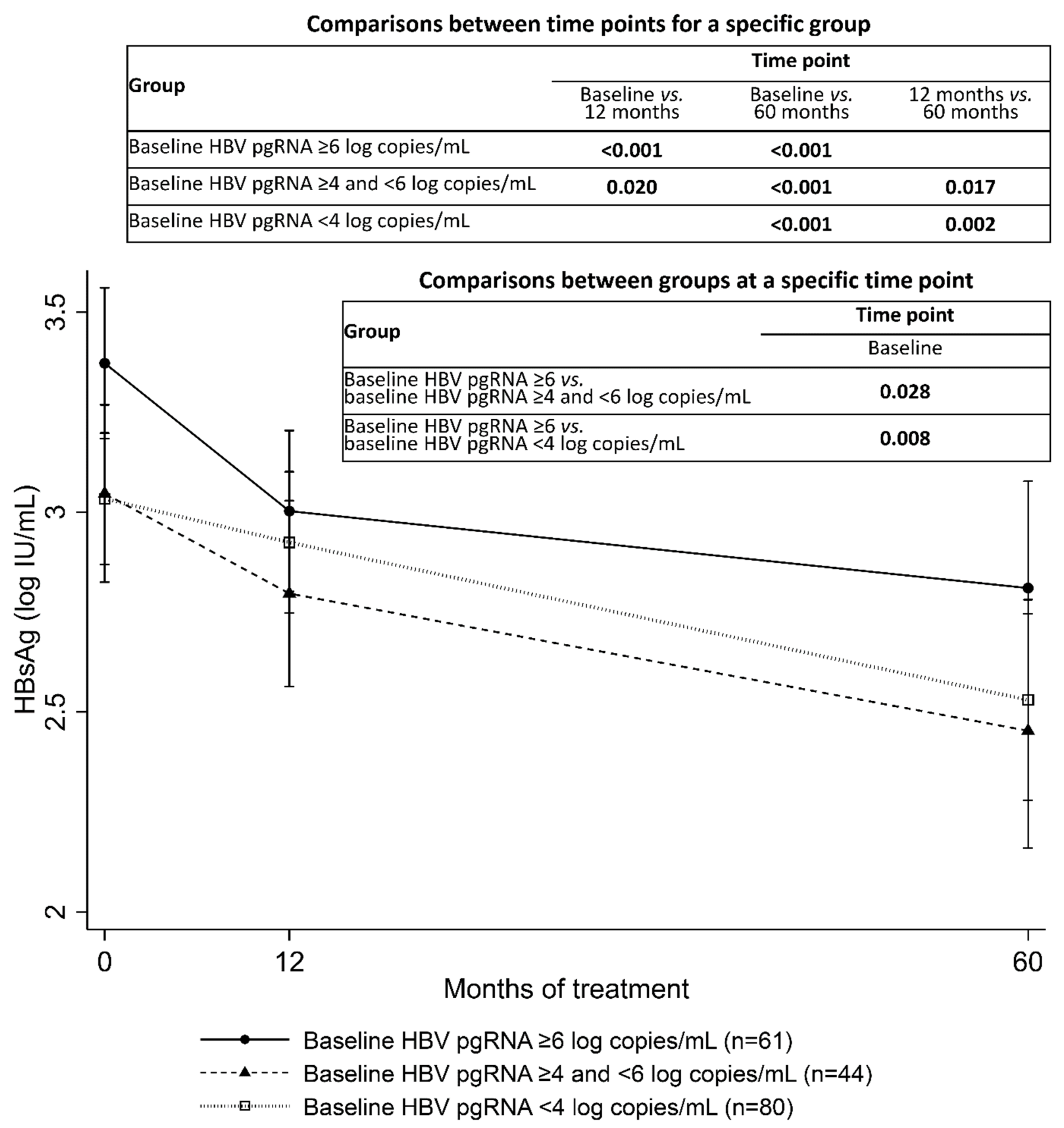
3.4. Correlation between Serum HBV pgRNA, HBV DNA, and HBsAg
3.5. Factors Associated with Virological Relapse after Cessation of Entecavir Therapy
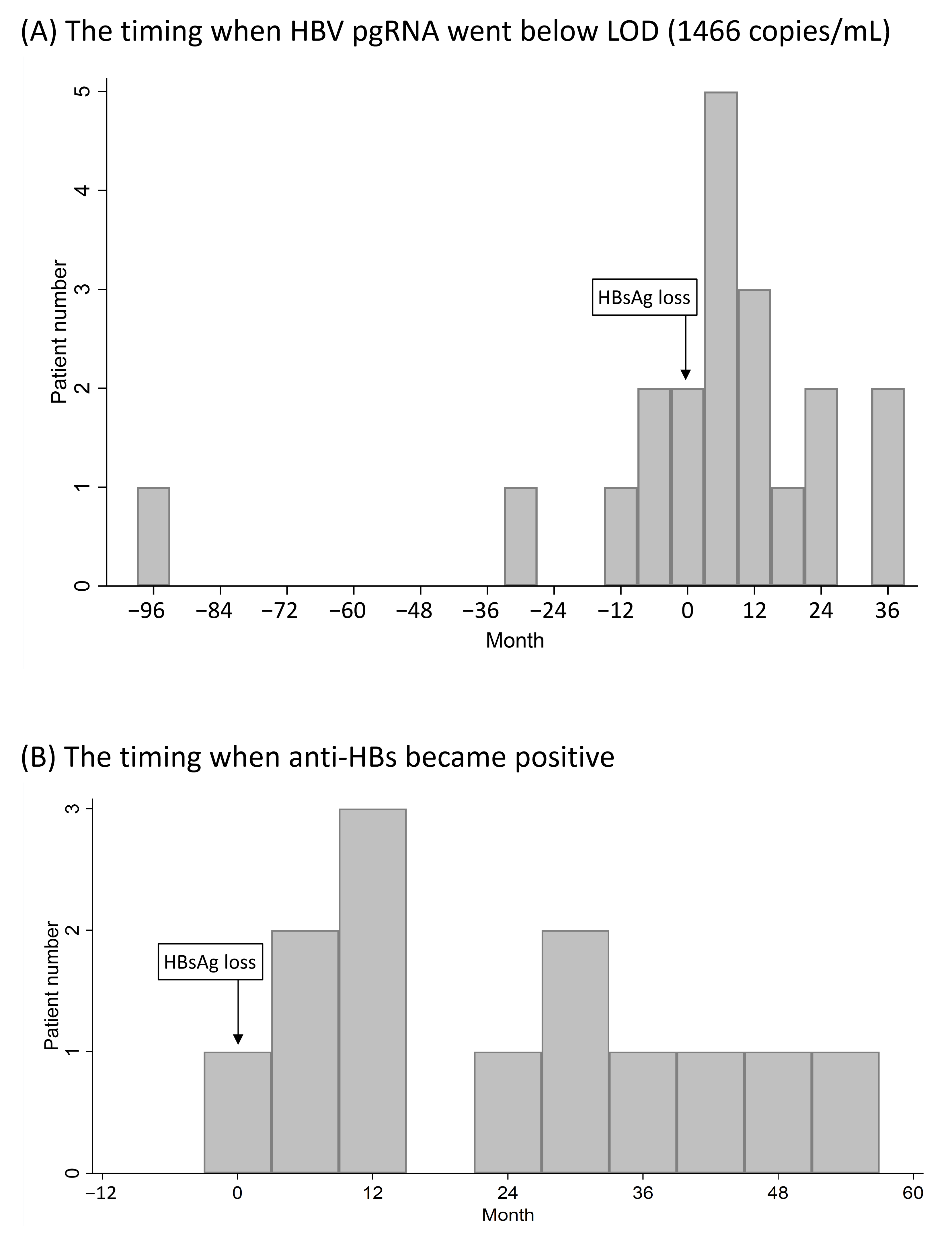
3.6. Expression of Serum HBV pgRNA after HBsAg Loss
3.7. Serum HBV pgRNA Kinetics in CHB Patients Who Achieved HBsAg Loss after Nucleos(t)ide Analogue Treatment
3.8. Factors Associated with HBsAg Loss after Nucleos(t)ide Analogue Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- WHO. Global Hepatitis Report 2017; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- European Association for the Study of the Liver. EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017, 67, 370–398. [Google Scholar] [CrossRef]
- Raffetti, E.; Fattovich, G.; Donato, F. Incidence of hepatocellular carcinoma in untreated subjects with chronic hepatitis B: A systematic review and meta-analysis. Liver Int. 2016, 36, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Terrault, N.A.; Lok, A.S.; McMahon, B.J.; Chang, K.-M.; Hwang, J.; Jonas, M.M.; Jr., R.S.B.; Bzowej, N.H.; Wong, J.B. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018, 67, 1560–1599. [Google Scholar] [CrossRef]
- Sarin, S.K.; Kumar, M.P.; Lau, G.K.; Abbas, Z.; Chan, H.L.Y.; Chen, C.J.; Chen, D.-S.; Chen, H.L.; Chien, R.N.; Dokmeci, A.; et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: A 2015 update. Hepatol. Int. 2016, 10, 1–98. [Google Scholar] [CrossRef]
- Lau, G.K.; Piratvisuth, T.; Luo, K.X.; Marcellin, P.; Thongsawat, S.; Cooksley, G.; Gane, E.; Fried, M.W.; Chow, W.C.; Paik, S.W.; et al. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N. Engl. J. Med. 2005, 352, 2682–2695. [Google Scholar] [CrossRef]
- Tao, Y.; Wu, D.; Zhou, L.; Chen, E.; Liu, C.; Tang, X.; Jiang, W.; Han, N.; Li, H.; Tang, H. Present and Future Therapies for Chronic Hepatitis B. Adv. Exp. Med. Biol. 2019, 1179, 137–186. [Google Scholar] [CrossRef]
- Buti, M.; Riveiro-Barciela, M.; Rodríguez-Frías, F.; Tabernero, D.; Esteban, R. Role of Biomarkers in Guiding Cure of Viral Hepatitis B. Semin. Liver Dis. 2019, 40, 49–60. [Google Scholar] [CrossRef]
- Coffin, C.S.; Zhou, K.; Terrault, N.A. New and Old Biomarkers for Diagnosis and Management of Chronic Hepatitis B Virus Infection. Gastroenterology 2019, 156, 355–368.e3. [Google Scholar] [CrossRef]
- Inoue, T.; Tanaka, Y. The Role of Hepatitis B Core-Related Antigen. Genes 2019, 10, 357. [Google Scholar] [CrossRef] [PubMed]
- Testoni, B.; Lebossé, F.; Scholtes, C.; Berby, F.; Miaglia, C.; Subic, M.; Loglio, A.; Facchetti, F.; Lampertico, P.; Levrero, M.; et al. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J. Hepatol. 2019, 70, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cao, X.; Wang, Z.; Gao, Y.; Deng, J.; Liu, X.; Zhuang, H. Correlation of HBcrAg with intrahepatic hepatitis B virus total DNA and covalently closed circular DNA in HBeAg-positive chronic hepatitis B patients. J. Clin. Microbiol. 2019, 57, e01303-18. [Google Scholar] [CrossRef]
- Tseng, T.-C.; Liu, C.-J.; Yang, W.-T.; Hsu, C.-Y.; Hong, C.-M.; Su, T.-H.; Tsai, C.-H.; Chen, C.-L.; Yang, H.-C.; Liu, C.-H.; et al. Serum hepatitis B core-related antigen level stratifies risk of disease progression in chronic hepatitis B patients with intermediate viral load. Aliment. Pharmacol. Ther. 2021, 53, 908–918. [Google Scholar]
- Tseng, T.-C.; Liu, C.-J.; Hsu, C.-Y.; Hong, C.-M.; Su, T.-H.; Yang, W.-T.; Chen, C.-L.; Yang, H.-C.; Huang, Y.-T.; Kuo, S.F.-T.; et al. High Level of Hepatitis B Core–Related Antigen Associated with Increased Risk of Hepatocellular Carcinoma in Patients with Chronic HBV Infection of Intermediate Viral Load. Gastroenterology 2019, 157, 1518–1529.e3. [Google Scholar] [CrossRef]
- Hosaka, T.; Suzuki, F.; Kobayashi, M.; Fujiyama, S.; Kawamura, Y.; Sezaki, H.; Akuta, N.; Suzuki, Y.; Saitoh, S.; Arase, Y.; et al. Impact of hepatitis B core-related antigen on the incidence of hepatocellular carcinoma in patients treated with nucleos(t)ide analogues. Aliment. Pharmacol. Ther. 2019, 49, 457–471. [Google Scholar] [CrossRef]
- Chuaypen, N.; Posuwan, N.; Chittmittraprap, S.; Hirankarn, N.; Treeprasertsuk, S.; Tanaka, Y.; Shinkai, N.; Poovorawan, Y.; Tangkijvanich, P. Predictive role of serum HBsAg and HBcrAg kinetics in patients with HBeAg-negative chronic hepatitis B receiving pegylated interferon-based therapy. Clin. Microbiol. Infect. 2018, 24, 306.e7–306.e13. [Google Scholar] [CrossRef]
- Wang, B.; Carey, I.; Bruce, M.; Montague, S.; Dusheiko, G.; Agarwal, K. HBsAg and HBcrAg as predictors of HBeAg seroconversion in HBeAg-positive patients treated with nucleos(t)ide analogues. J. Viral Hepat. 2018, 25, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-C.; Nguyen, M.H.; Mo, L.-R.; Wu, M.-S.; Yang, T.-H.; Chen, C.-C.; Tseng, C.-H.; Tai, C.-M.; Wu, C.-Y.; Lin, J.-T.; et al. Combining hepatitis B core-related and surface antigens at end of nucleos(t)ide analogue treatment to predict off-therapy relapse risk. Aliment. Pharmacol. Ther. 2018, 49, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.N.; Hu, T.H.; Wang, J.H.; Kuo, Y.H.; Hung, C.H.; Lu, S.N.; Jeng, W.-J.; Chen, C.-H. Incidence and factors associated with HBV relapse after cessation of entecavir or tenofovir in patients with HBsAg below 100 IU/mL. Clin. Gastroenterol. Hepatol. 2020, 18, 2803–2812.e2. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhou, B.; Valdes, J.D.; Sun, J.; Guo, H. Serum Hepatitis B Virus RNA: A New Potential Biomarker for Chronic Hepatitis B Virus Infection. Hepatology 2019, 69, 1816–1827. [Google Scholar] [CrossRef]
- Prakash, K.; Rydell, G.E.; Larsson, S.B.; Andersson, M.; Norkrans, G.; Norder, H.; Lindh, M. High serum levels of pregenomic RNA reflect frequently failing reverse transcription in hepatitis B virus particles. Virol. J. 2018, 15, 86. [Google Scholar] [CrossRef]
- Wang, J.; Shen, T.; Huang, X.; Kumar, G.R.; Chen, X.; Zeng, Z.; Zhang, R.; Chen, R.; Li, T.; Zhang, T.; et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J. Hepatol. 2016, 65, 700–710. [Google Scholar] [CrossRef]
- Wang, J.; Peiyu, Q.; Li, G.; Shen, C.; Meng, Z.; Zheng, J.-M.; Jianming, Z.; Chen, S.; Zhang, X.; Zhu, M.; et al. Relationship between serum HBV-RNA levels and intrahepatic viral as well as histologic activity markers in entecavir-treated patients. J. Hepatol. 2018, 68, 16–24. [Google Scholar] [CrossRef]
- Huang, H.; Wang, J.; Li, W.; Chen, R.; Chen, X.; Zhang, F.; Xu, D.; Lu, F. Serum HBV DNA plus RNA shows superiority in reflecting the activity of intrahepatic cccDNA in treatment-naïve HBV-infected individuals. J. Clin. Virol. 2018, 99–100, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, Y.; Meng, Q.; Zhang, Z.; Zhao, P.; Shang, Q.; Li, Y.; Su, M.; Li, T.; Liu, X.; et al. Serum Hepatitis B Virus DNA, RNA, and HBsAg: Which Correlated Better with Intrahepatic Covalently Closed Circular DNA before and after Nucleos(t)ide Analogue Treatment? J. Clin. Microbiol. 2017, 55, 2972–2982. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Peng, J.; Xie, Q.; Tan, D.; Xu, M.; Niu, J.; Wang, H.; Ren, H.; Chen, X.; Wang, M.; et al. Combining Hepatitis B Virus RNA and Hepatitis B Core–Related Antigen: Guidance for Safely Stopping Nucleos(t)ide Analogues in Hepatitis B e Antigen–Positive Patients with Chronic Hepatitis B. J. Infect. Dis. 2020, 222, 611–618. [Google Scholar] [CrossRef]
- Fan, R.; Zhou, B.; Xu, M.; Tan, D.; Niu, J.; Wang, H.; Ren, H.; Chen, X.; Wang, M.; Ning, Q.; et al. Association Between Negative Results from Tests for HBV DNA and RNA and Durability of Response After Discontinuation of Nucles(t)ide Analogue Therapy. Clin. Gastroenterol. Hepatol. 2020, 18, 719–727.e7. [Google Scholar] [CrossRef]
- Carey, I.; Gersch, J.; Wang, B.; Moigboi, C.; Kuhns, M.; Cloherty, G.; Dusheiko, G.; Agarwal, K. Pregenomic HBV RNA and Hepatitis B Core-Related Antigen Predict Outcomes in Hepatitis B e Antigen–Negative Chronic Hepatitis B Patients Suppressed on Nucleos(T)ide Analogue Therapy. Hepatology 2020, 72, 42–57. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-C.; Chiu, Y.-C.; Chiu, H.-C.; Liu, W.-C.; Cheng, P.-N.; Chen, C.-Y.; Chang, T.-T.; Wu, I.-C. Clinical utility of hepatitis B surface antigen kinetics in treatment-naïve chronic hepatitis B patients during long-term entecavir therapy. World J. Gastroenterol. 2018, 24, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-C.; Wu, I.-C.; Lee, Y.-C.; Lin, C.-P.; Cheng, J.-H.; Lin, Y.-J.; Yen, C.-J.; Cheng, P.-N.; Li, P.-F.; Cheng, Y.-T.; et al. Hepatocellular carcinoma-associated single-nucleotide variants and deletions identified by the use of genome-wide high-throughput analysis of hepatitis B virus. J. Pathol. 2017, 243, 176–192. [Google Scholar] [CrossRef]
- Liu, W.-C.; Mizokami, M.; Buti, M.; Lindh, M.; Young, K.-C.; Sun, K.-T.; Chi, Y.-C.; Li, H.-H.; Chang, T.-T. Simultaneous Quantification and Genotyping of Hepatitis B Virus for Genotypes A to G by Real-Time PCR and Two-Step Melting Curve Analysis. J. Clin. Microbiol. 2006, 44, 4491–4497. [Google Scholar] [CrossRef][Green Version]
- Gonçalves, A.; Lemenuel-Diot, A.; Cosson, V.; Jin, Y.; Feng, S.; Bo, Q.; Guedj, J. What drives the dynamics of HBV RNA during treatment? J. Viral Hepat. 2021, 28, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Dahari, H.; Shlomai, A.; Cotler, S.J. Early HBV RNA kinetics under NA treatment may reveal new insights into HBV RNA dynamics and NA mode of action-more detailed kinetic studies are needed. J. Viral. Hepat. 2021, 28, 687–688. [Google Scholar] [CrossRef] [PubMed]
- Mak, L.; Cloherty, G.; Wong, D.K.; Gersch, J.; Seto, W.; Fung, J.; Yuen, M. HBV RNA profiles in chronic hepatitis B patients under different disease phases and anti-viral therapy. Hepatology 2020. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, G.; Locarnini, S.; Pollicino, T.; Levrero, M.; Zoulim, F.; Lok, A.S.; Taormina Workshop on Occult HBV Infection Faculty Members. Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J. Hepatol. 2019, 71, 397–408. [Google Scholar] [CrossRef]
- Van Campenhout, M.J.H.; van Bommel, F.; Pfefferkorn, M.; Fischer, J.; Deichsel, D.; Boonstra, A.; van Vuuren, A.J.; Berg, T.; Hansen, B.E.; Janssen, H.L.A. Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology 2018, 68, 839–847. [Google Scholar] [CrossRef]
- Huang, Y.-W.; Takahashi, S.; Tsuge, M.; Chen, C.-L.; Wang, T.-C.; Abe, H.; Hu, J.-T.; Chen, D.-S.; Yang, S.-S.; Chayama, K.; et al. On-treatment low serum HBV RNA level predicts initial virological response in chronic hepatitis B patients receiving nucleoside analogue therapy. Antivir. Ther. 2014, 20, 369–375. [Google Scholar] [CrossRef]
- Volz, T.; Lutgehetmann, M.; Wachtler, P.; Jacob, A.; Quaas, A.; Murray, J.M.; Dandri, M.; Petersen, J. Impaired intrahepatic hepatitis B virus productivity contributes to low viremia in most HBeAg-negative patients. Gastroenterology 2007, 133, 843–852. [Google Scholar] [CrossRef]
- Wu, Y.; Wen, J.; Xiao, W.; Zhang, B. Pregenomic RNA: How to assist the management of chronic hepatitis B? Rev. Med. Virol. 2019, 29, e2051. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, G.; Shang, J.; Pan, C.; Zhang, M.; Yin, Z.; Xie, Q.; Peng, Y.; Mao, Q.; Xiao, X.; et al. Rapidly decreased HBV RNA predicts responses of pegylated interferons in HBeAg-positive patients: A longitudinal cohort study. Hepatol. Int. 2020, 14, 212–224. [Google Scholar] [CrossRef]
- Farag, M.S.; van Campenhout, M.J.H.; Pfefferkorn, M.; Fischer, J.; Deichsel, D.; Boonstra, A.; van Vuuren, A.J.; Ferenci, P.; Feld, J.J.; Berg, T.; et al. Hepatitis B virus RNA as early predictor for response to pegylated interferon alfa in HBeAg-negative chronic hepatitis B. Clin. Infect. Dis. 2020, 72, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Tan, N.; Kang, Q.; Pan, J.; Chen, H.; Xi, H.; Yu, M.; Xu, X. Hepatitis B virus pregenomic RNA status can reveal the long-term prognoses of chronic hepatitis B patients treated with nucleos(t)ide analogues. J. Viral Hepat. 2019, 27, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Van Bommel, F.; van Bommel, A.; Krauel, A.; Wat, C.; Pavlovic, V.; Yang, L.; Deichsel, D.; Berg, T.; Böhm, S. Serum HBV RNA as a predictor of peginterferon alfa-2a response in patients with HBeAg-positive chronic hepatitis B. J. Infect. Dis. 2018, 218, 1066–1074. [Google Scholar] [CrossRef]
- Seto, W.-K.; Liu, K.S.; Mak, L.-Y.; Cloherty, G.; Wong, D.K.-H.; Gersch, J.; Lam, Y.-F.; Cheung, K.-S.; Chow, N.; Ko, K.-L.; et al. Role of serum HBV RNA and hepatitis B surface antigen levels in identifying Asian patients with chronic hepatitis B suitable for entecavir cessation. Gut 2021, 70, 775–783. [Google Scholar] [CrossRef]
- Lee, I.C.; Yang, S.S.; Lee, C.J.; Su, C.W.; Wang, Y.J.; Lan, K.H.; Lin, H.C.; Hou, M.-C.; Peng, C.-Y.; Huang, Y.H. Incidence and predictors of HBsAg loss after peginterferon therapy in HBeAg-negative chronic hepatitis B: A multicenter, long-term follow-up study. J. Infect. Dis. 2018, 218, 1075–1084. [Google Scholar] [CrossRef]
- Jeng, W.-J.; Chen, Y.-C.; Chien, R.-N.; Sheen, I.-S.; Liaw, Y.-F. Incidence and predictors of hepatitis B surface antigen seroclearance after cessation of nucleos(t)ide analogue therapy in hepatitis B e antigen-negative chronic hepatitis B. Hepatology 2018, 68, 425–434. [Google Scholar] [CrossRef]
- Lim, S.G.; Phyo, W.W.; Ling, J.Z.J.; Cloherty, G.; Butler, E.K.; Kuhns, M.C.; McNamara, A.L.; Holzmayer, V.; Gersch, J.; Yang, W.L.; et al. Comparative biomarkers for HBsAg loss with antiviral therapy shows dominant influence of quantitative HBsAg (qHBsAg). Aliment. Pharmacol. Ther. 2021, 53, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhang, X.; Kozlowski, M.; Li, W.; Wu, M.; Liu, J.; Chen, L.; Zhang, J.; Huang, Y.; Yuan, Z. Extracellular Hepatitis B Virus RNAs Are Heterogeneous in Length and Circulate as Capsid-Antibody Complexes in Addition to Virions in Chronic Hepatitis B Patients. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Wang, J.; Sheng, Q.; Ding, Y.; Chen, R.; Sun, X.; Chen, X.; Dou, X.; Lu, F. HBV RNA virion-like particles produced under nucleos(t)ide analogues treatment are mainly replication-deficient. J. Hepatol. 2018, 68, 847–849. [Google Scholar] [CrossRef]
- Anderson, M.; Gersch, J.; Luk, K.-C.; Dawson, G.; Carey, I.; Agarwal, K.; Shah, P.; Dusheiko, G.; Lau, D.; Cloherty, G. Circulating Pregenomic Hepatitis B Virus RNA Is Primarily Full-length in Chronic Hepatitis B Patients Undergoing Nucleos(t)ide Analogue Therapy. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Freitas, N.; Lukash, T.; Gunewardena, S.; Chappell, B.; Slagle, B.L.; Gudima, S.O. Relative Abundance of Integrant-Derived Viral RNAs in Infected Tissues Harvested from Chronic Hepatitis B Virus Carriers. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Butler, E.K.; Gersch, J.; McNamara, A.; Luk, K.-C.; Holzmayer, V.; De Medina, M.; Schiff, E.; Kuhns, M.; Cloherty, G.A. Hepatitis B Virus Serum DNA andRNA Levels in Nucleos(t)ide Analog-Treated or Untreated Patients During Chronic and Acute Infection. Hepatology 2018, 68, 2106–2117. [Google Scholar] [CrossRef] [PubMed]
- Kostyusheva, A.; Kostyushev, D.; Brezgin, S.; Volchkova, E.; Chulanov, V. Clinical Implications of Hepatitis B Virus RNA and Covalently Closed Circular DNA in Monitoring Patients with Chronic Hepatitis B Today with a Gaze into the Future: The Field Is Unprepared for a Sterilizing Cure. Genes 2018, 9, 483. [Google Scholar] [CrossRef] [PubMed]
- Kusumoto, S.; Tanaka, Y.; Suzuki, R.; Watanabe, T.; Nakata, M.; Sakai, R.; Fukushima, N.; Fukushima, T.; Moriuchi, Y.; Itoh, K.; et al. Ultra-high sensitivity HBsAg assay can diagnose HBV reactivation following rituximab-based therapy in patients with lymphoma. J. Hepatol. 2020, 73, 285–293. [Google Scholar] [CrossRef]
- Shinkai, N.; Kusumoto, S.; Murakami, S.; Ogawa, S.; Ri, M.; Matsui, T.; Tamori, A.; Toyoda, H.; Ishida, T.; Iida, S.; et al. Novel monitoring of hepatitis B reactivation based on ultra-high sensitive hepatitis B surface antigen assay. Liver Int. 2016, 37, 1138–1147. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | Total Patients (n = 185) | HBeAg-Positive Patients (n = 54) | HBeAg-Negative Patients (n = 131) | pa |
|---|---|---|---|---|
| Age (year) | 51.0 ± 12.0 | 43.8 ± 12.5 | 54.0 ± 10.5 | <0.0001 |
| Male (%) | 130/185 (70.3%) | 36/54 (66.7%) | 94/131 (71.8%) | 0.49 |
| HBV genotype (B:C) b | 88:76 | 17:36 | 71:40 | <0.001 |
| Baseline ALT (× ULN) | 3.84 ± 5.84 | 4.12± 6.63 | 3.73 ± 5.51 | 0.68 |
| Baseline HBsAg (log IU/mL) | 3.15 ± 0.78 | 3.78 ± 0.72 | 2.89 ± 0.65 | <0.0001 |
| Baseline HBV DNA (log IU/mL) | 5.83 ± 1.72 | 7.29 ± 1.32 | 5.23 ± 1.49 | <0.0001 |
| Baseline HBV pgRNA (log copies/mL) | 5.07 ± 1.98 | 5.48 ± 2.03 | 4.89 ± 1.94 | 0.066 |
| Liver cirrhosis (%) | 62/185 (33.5%) | 12/54 (22.2%) | 50/131 (38.2%) | 0.037 |
| HCC (%) c | 29/185 (15.7%) | 5/54 (9.3%) | 24/131 (18.3%) | 0.12 |
| Duration of entecavir therapy (years) | 4.85 (1.84–11.29) | 4.43 (1.95–8.85) | 4.92 (1.84–11.29) | 0.93 |
| Virological response rate (%) d | 166/171 (97.1%) | 42/46 (91.3%) | 124/125 (99.2%) | 0.019 |
| Time to virological response (years) | 0.51 (0.04–4.88) | 1.01 (0.22–4.02) | 0.46 (0.04–4.88) | 0.0001 |
| HBeAg seroconversion rate (%) | 19/54 (35.2%) | |||
| Time to HBeAg seroconversion (years) | 1.24 (0.21–7.49) | |||
| Virological response and HBeAg seroconversion rate (%) | 15/45 (33.3%) | |||
| Time to virological response and HBeAg seroconversion (years) | 1.07 (0.49–4.06) |
| Factors | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p Value | HR | 95% CI | p | |
| Age (<50 vs. ≥50 years old) | 0.91 | 0.67–1.23 | 0.54 | 1.54 | 1.03–2.30 | 0.034 |
| Sex (male vs. female) | 0.90 | 0.65–1.25 | 0.53 | 0.73 | 0.50–1.07 | 0.11 |
| Baseline HBeAg (negative vs. positive) | 2.61 | 1.81–3.77 | <0.001 | 2.03 | 1.27–3.25 | 0.003 |
| HBV genotype (C vs. B) | 0.86 | 0.62–1.19 | 0.36 | 0.96 | 0.64–1.42 | 0.83 |
| ALT (×ULN) | 0.99 | 0.96–1.02 | 0.55 | 0.99 | 0.96–1.03 | 0.64 |
| Baseline HBsAg (<4 vs. ≥4 log IU/mL) | 2.56 | 1.57–4.17 | <0.001 | 1.74 | 0.93–3.25 | 0.08 |
| Baseline HBV DNA (<5 vs. ≥5 log IU/mL) | 1.98 | 1.44–2.74 | <0.001 | 1.28 | 0.86–1.92 | 0.23 |
| Baseline HBV pgRNA (<6.4 vs. ≥6.4 log copies/mL) | 1.73 | 1.22–2.45 | 0.002 | 1.57 | 1.06–2.31 | 0.023 |
| Liver cirrhosis (yes vs. no) | 1.33 | 0.96–1.84 | 0.09 | 1.29 | 0.85–1.93 | 0.23 |
| Characteristics | CHB Patients Achieved HBsAg Loss after Nucleos(t)ide Analogue Treatment (n = 20) | CHB Patients Received Nucleos(t)ide Analogue Treatment without HBsAg Loss (n = 141) | pa |
|---|---|---|---|
| Age at baseline (year) b | 48.73 ± 9.51 | 51.4 1± 11.97 | 0.34 |
| Male (%) | 17/20 (85%) | 94/141 (66.7%) | 0.12 |
| HBeAg-positive at baseline b | 6/20 (30%) | 45/141 (31.9%) | 1.00 |
| HBV genotype (B:C) c | 8:2 | 60:63 | 0.10 |
| Baseline ALT (×ULN) b | 6.75 ± 11.03 | 3.43 ± 5.27 | 0.35 |
| Baseline HBsAg (log IU/mL) b | 2.94 ± 1.28 | 3.17 ± 0.73 | 0.45 |
| Baseline HBV DNA (log IU/mL) b | 5.99 ± 2.38 | 5.87 ± 1.68 | 0.88 |
| Baseline HBV pgRNA (log copies/mL) b | 5.14 ± 2.05 | 5.08 ± 1.94 | 0.92 |
| Liver cirrhosis (%) | 2/20 (10%) | 51/141 (36.2%) | 0.021 |
| Nucleos(t)ide analogue | |||
| ETV (%) | 18/20 (90%) | 141/141 (100%) | |
| TDF (%) | 2/20 (10%) | 0 (0%) | |
| Treatment time (year) | 5.12 (2.39–14.10) | 4.91 (1.84–11.29) | 0.24 |
| Age at HBsAg loss (year) | 54.45 ± 10.07 | ||
| HBV DNA at HBsAg loss (log IU/mL) | |||
| not detected (%) | 20/20 (100%) | ||
| HBV pgRNA at HBsAg loss | |||
| not detected (%) | 0/20 (0%) | ||
| <LOD (1466 copies/mL) (%) | 7/20 (35%) | ||
| ≥LOD (1466 copies/mL) (%) | 13/20 (65%) 5.90 (4.81–7.34) log copies/mL | ||
| HBV pgRNA at the last follow-up (%) | |||
| not detected (%) | 0/20 (0%) | ||
| <LOD (1466 copies/mL) (%) | 20/20 (100%) | ||
| Anti-HBs-positive at the last follow-up (%) | 13/20 (65%) |
| Factors | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p Value | HR | 95% CI | p | |
| Age (<50 vs. ≥50 years old) | 1.23 | 0.51–2.97 | 0.64 | 9.81 | 1.23–77.94 | 0.031 |
| Sex (male vs. female) | 2.72 | 0.80–9.28 | 0.11 | 2.10 | 0.34–13.10 | 0.43 |
| Baseline HBeAg (negative vs. positive) | 1.19 | 0.46–3.10 | 0.72 | 1.53 | 0.11–20.81 | 0.75 |
| HBV genotype (C vs. B) | 0.25 | 0.05–1.17 | 0.08 | 0.41 | 0.06–3.02 | 0.38 |
| Baseline ALT (× ULN) | 1.04 | 0.99–1.10 | 0.13 | 1.00 | 0.92–1.10 | 0.97 |
| Baseline HBsAg (<4 vs. ≥4 log IU/mL) | 0.60 | 0.19–1.86 | 0.38 | 0.87 | 0.07–10.46 | 0.91 |
| Baseline HBV DNA (<5 vs. ≥5 log IU/mL) | 1.42 | 0.43–4.64 | 0.57 | 6.84 | 0.78–60.40 | 0.08 |
| Baseline HBV pgRNA (<6.4 vs. ≥6.4 log copies/mL) | 1.06 | 0.33–3.39 | 0.92 | 3.35 | 0.43–26.42 | 0.25 |
| Liver cirrhosis (yes vs. no) | 0.21 | 0.05–0.89 | 0.035 | 0.62 | 0.06–6.85 | 0.70 |
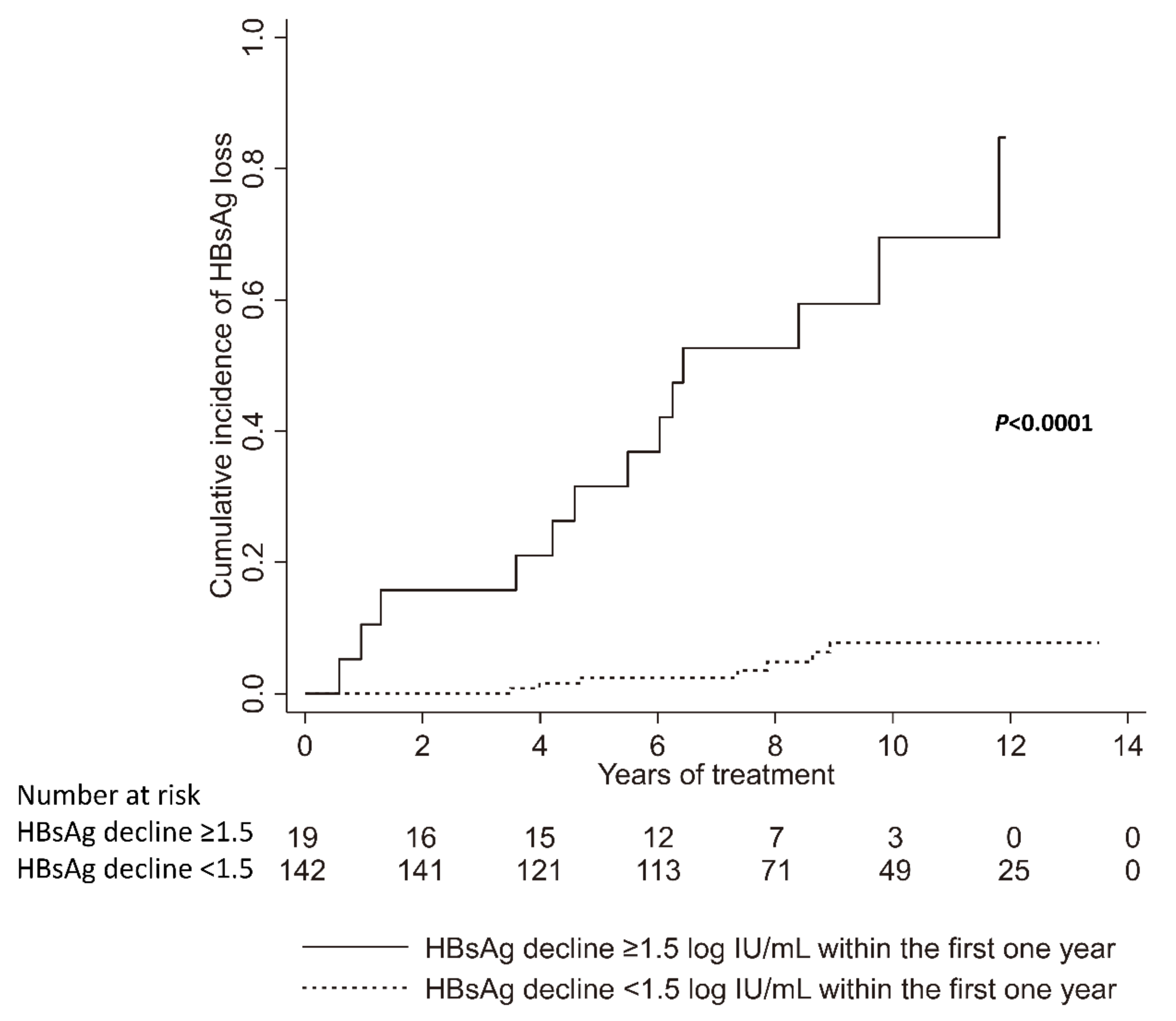
| HBsAg decline within the first one year (≥1.5 vs. <1.5 log IU/mL) | 17.57 | 6.99–44.17 | <0.001 | 53.59 | 3.83–749.5 | 0.003 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, I.-C.; Liu, W.-C.; Chiu, Y.-C.; Chiu, H.-C.; Cheng, P.-N.; Chang, T.-T. Clinical Implications of Serum Hepatitis B Virus Pregenomic RNA Kinetics in Chronic Hepatitis B Patients Receiving Antiviral Treatment and Those Achieving HBsAg Loss. Microorganisms 2021, 9, 1146. https://doi.org/10.3390/microorganisms9061146
Wu I-C, Liu W-C, Chiu Y-C, Chiu H-C, Cheng P-N, Chang T-T. Clinical Implications of Serum Hepatitis B Virus Pregenomic RNA Kinetics in Chronic Hepatitis B Patients Receiving Antiviral Treatment and Those Achieving HBsAg Loss. Microorganisms. 2021; 9(6):1146. https://doi.org/10.3390/microorganisms9061146
Chicago/Turabian StyleWu, I-Chin, Wen-Chun Liu, Yen-Cheng Chiu, Hung-Chih Chiu, Pin-Nan Cheng, and Ting-Tsung Chang. 2021. "Clinical Implications of Serum Hepatitis B Virus Pregenomic RNA Kinetics in Chronic Hepatitis B Patients Receiving Antiviral Treatment and Those Achieving HBsAg Loss" Microorganisms 9, no. 6: 1146. https://doi.org/10.3390/microorganisms9061146
APA StyleWu, I.-C., Liu, W.-C., Chiu, Y.-C., Chiu, H.-C., Cheng, P.-N., & Chang, T.-T. (2021). Clinical Implications of Serum Hepatitis B Virus Pregenomic RNA Kinetics in Chronic Hepatitis B Patients Receiving Antiviral Treatment and Those Achieving HBsAg Loss. Microorganisms, 9(6), 1146. https://doi.org/10.3390/microorganisms9061146






