Persistence of Coxsackievirus B4 in Pancreatic β Cells Disturbs Insulin Maturation, Pattern of Cellular Proteins, and DNA Methylation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Virus
2.2. Chronic Cell Infections and Viral Progeny in Supernatants
2.3. Cell Viability Assay
2.4. DNA and RNA Extraction
2.5. One-Step RT-PCR and Agarose Gel Electrophoresis
2.6. Real-Time Quantitative PCR
2.7. Quantification of Insulin, Proinsulin, and C-Peptide
2.8. Immunofluorescence Assay
2.9. Quantification of DNA Methylation
2.10. Protein Extraction and Digestion
2.11. Mass Spectrometry Data Acquisition UPLC-MS/MS
2.12. Data and Statistical Analyses
3. Results
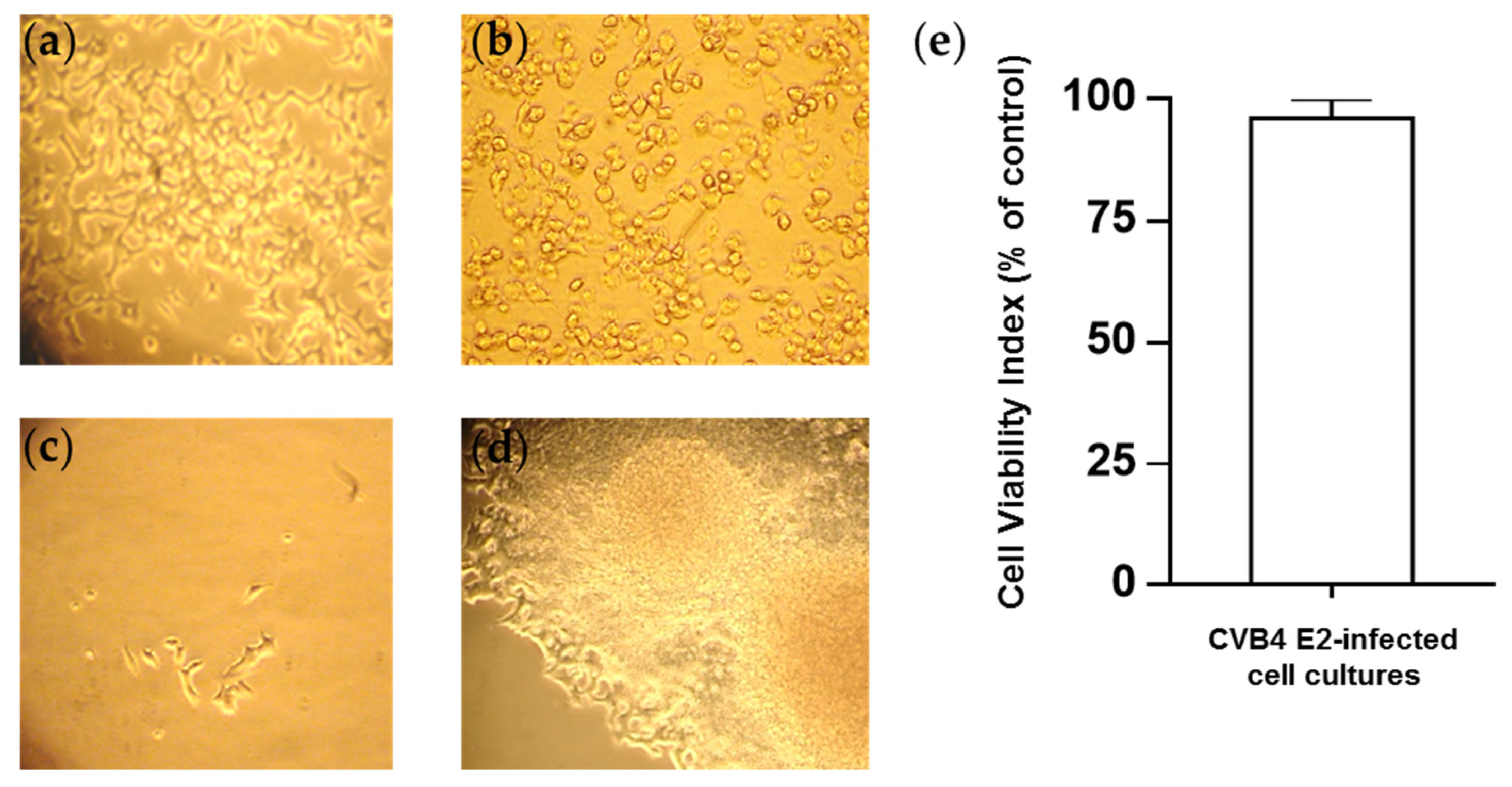
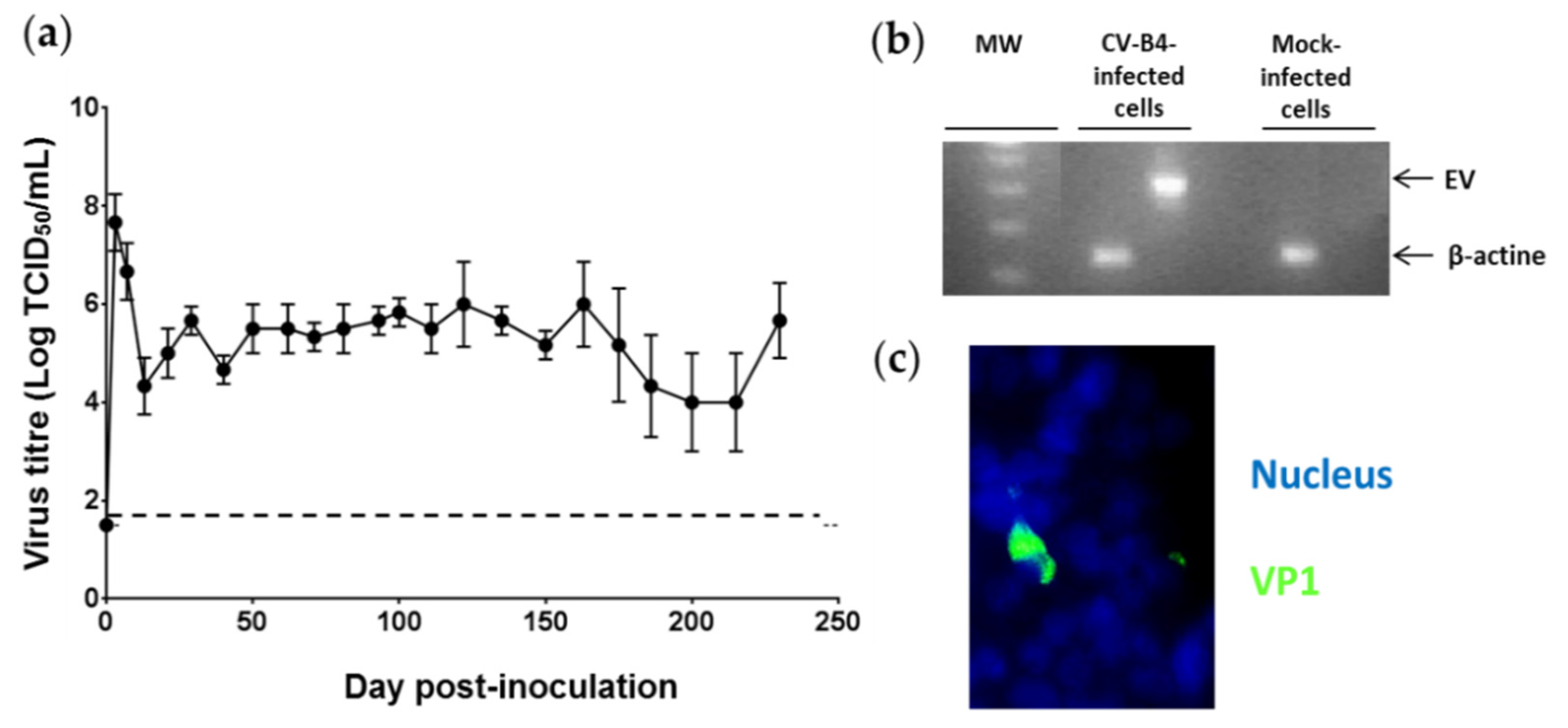
3.1. Persistent CV-B4 E2 Infection of Pancreatic β Cells
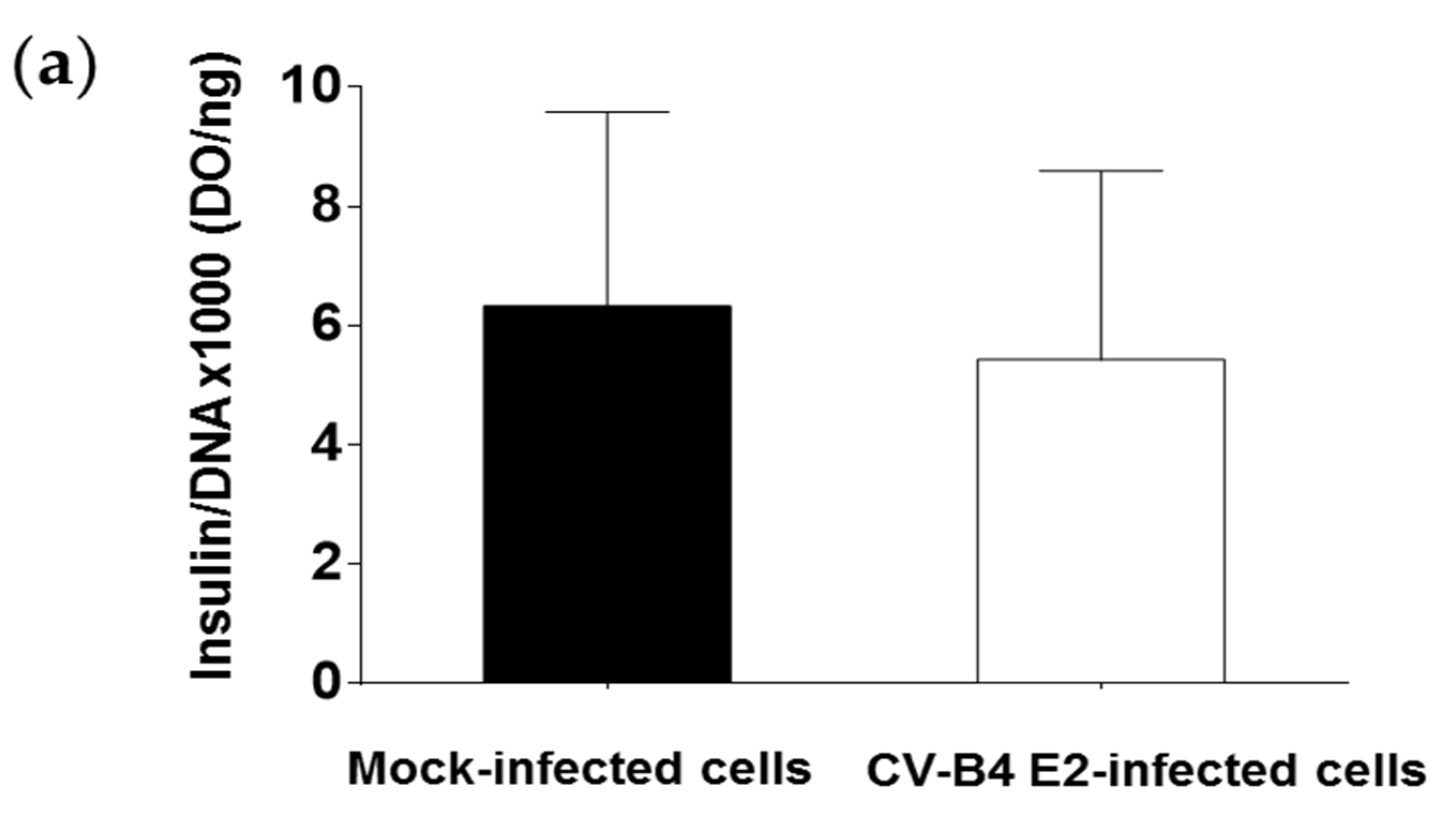
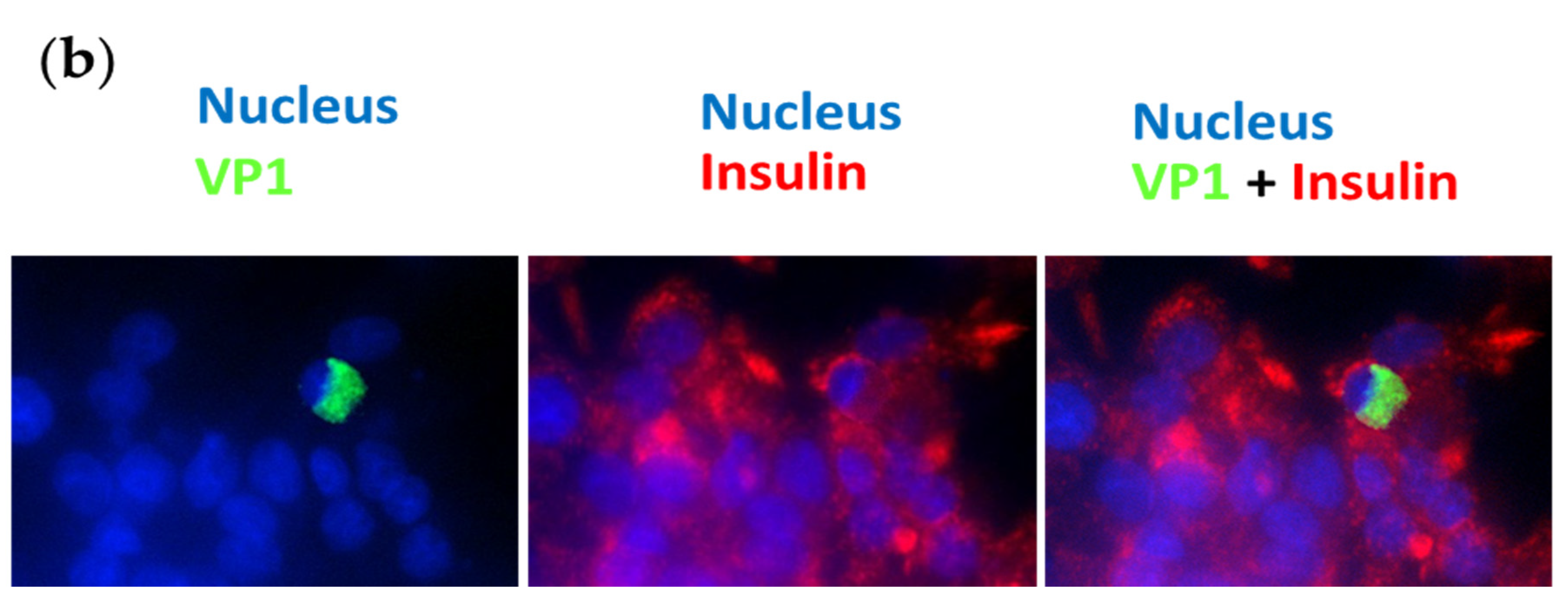
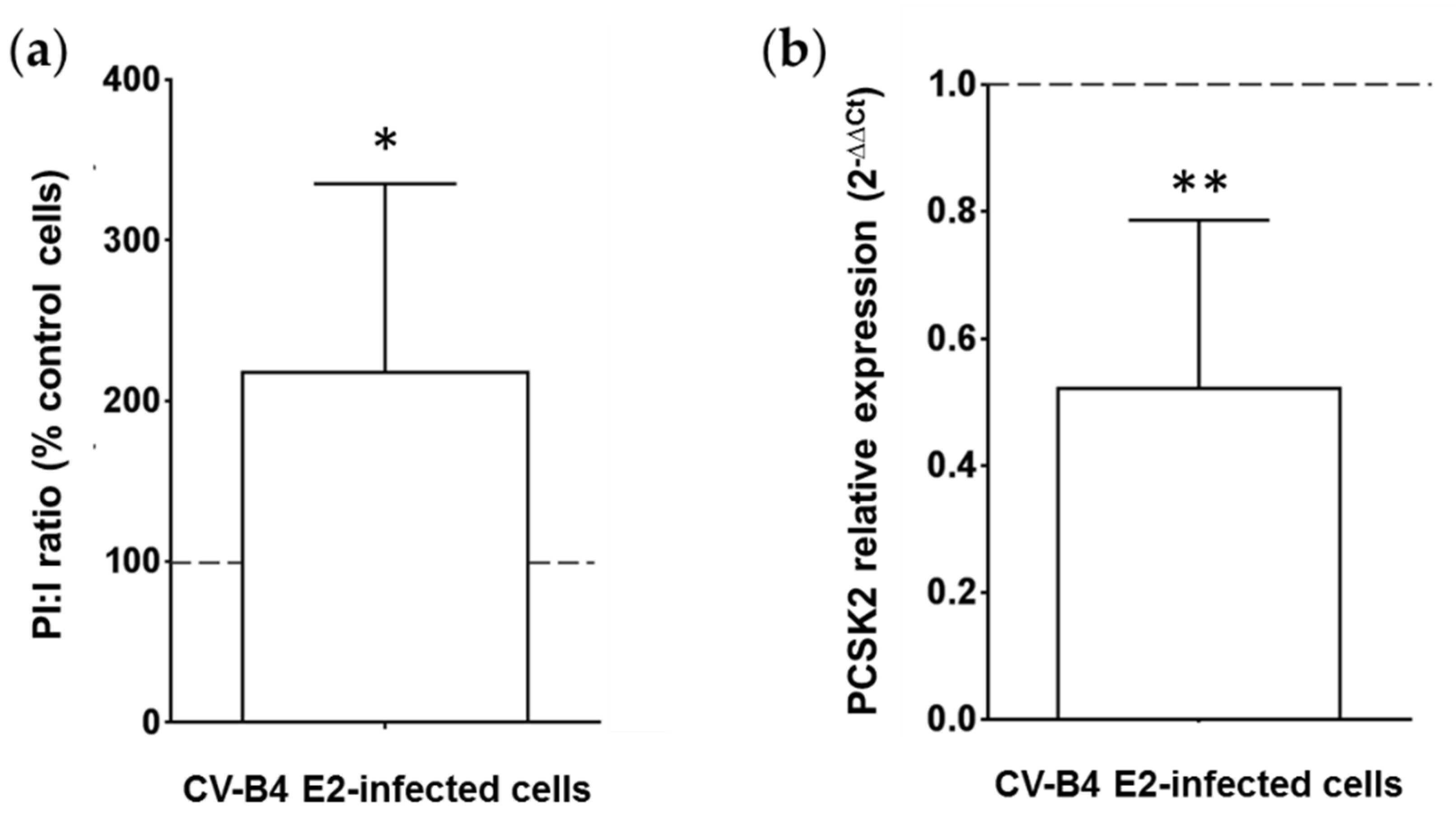
3.2. Persistent CV-B4 E2 Infection of Pancreatic β Cells Results in Change in Insulin Metabolism
3.3. Persistent CV-B4 E2 Infection of Pancreatic β Cells Changes the Expression of Cellular Proteins
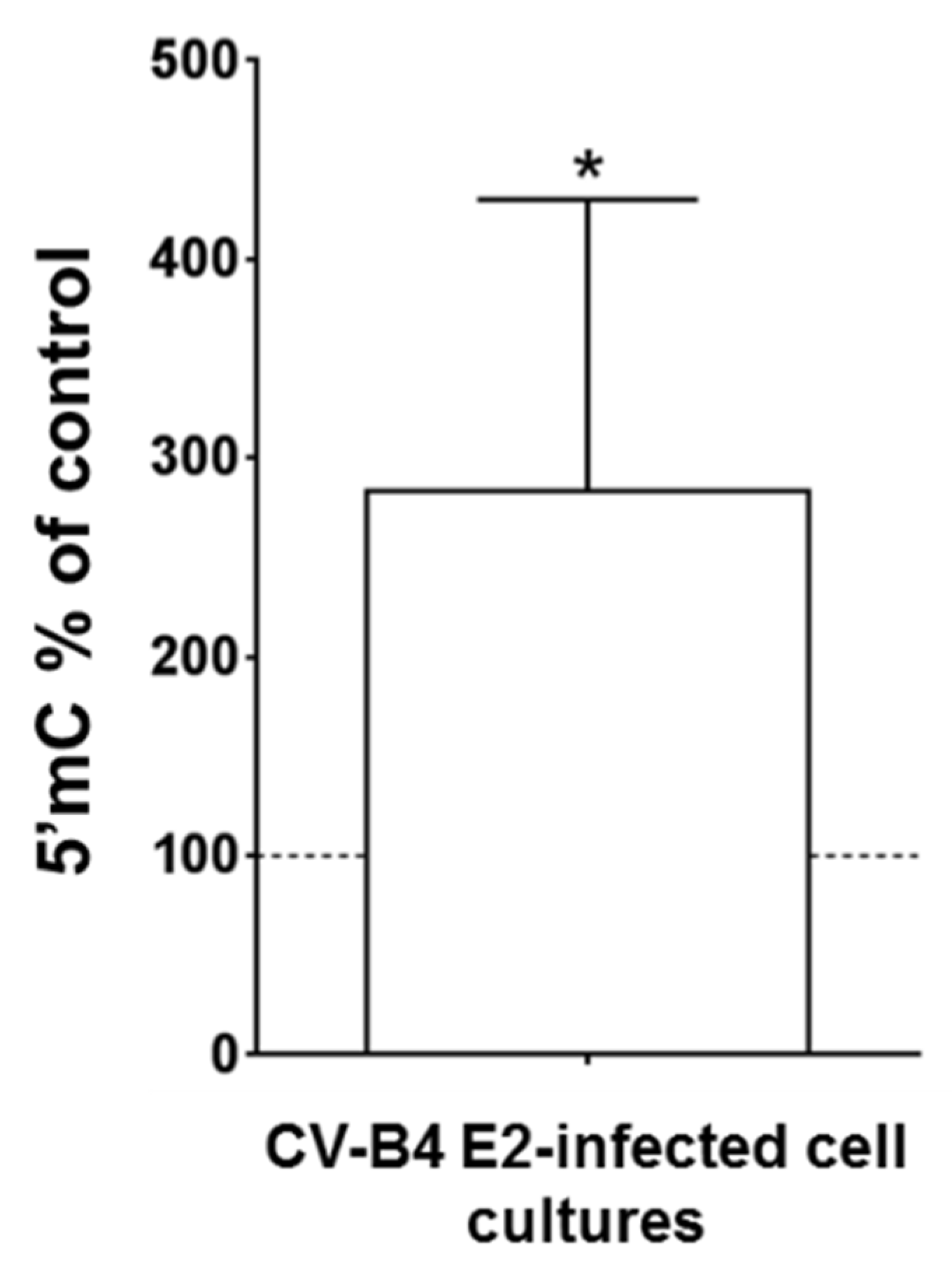
3.4. Persistent CV-B4 E2 Infection of Pancreatic β Cells Causes Changes in DNA Methylation
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tokarz, V.L.; MacDonald, P.E.; Klip, A. The cell biology of systemic insulin function. J. Cell Biol. 2018, 217, 2273–2289. [Google Scholar] [CrossRef]
- Smeekens, S.P.; Montag, A.G.; Thomas, G.; Albiges-Rizo, C.; Carroll, R.; Benig, M.; Phillips, L.A.; Martin, S.; Ohagi, S.; Gardner, P.; et al. Proinsulin processing by the subtilisin-related proprotein convertases furin, PC2, and PC3. Proc. Natl. Acad. Sci. USA 1992, 89, 8822–8826. [Google Scholar] [CrossRef]
- Davidson, H.W.; Rhodes, C.J.; Hutton, J.C. Intraorganellar calcium and pH control proinsulin cleavage in the pancreatic β cell via two distinct site-specific endopeptidases. Nature 1988, 333, 93–96. [Google Scholar] [CrossRef]
- Wasserfall, C.; Nick, H.S.; Campbell-Thompson, M.; Beachy, D.; Haataja, L.; Kusmartseva, I.; Posgai, A.; Beery, M.; Rhodes, C.; Bonifacio, E.; et al. Persistence of pancreatic insulin mRNA expression and proinsulin protein in type 1 diabetes pancreata. Cell Metab. 2017, 26, 568–575. [Google Scholar] [CrossRef]
- Liu, M.; Weiss, M.A.; Arunagiri, A.; Yong, J.; Rege, N.; Sun, J.; Haataja, L.; Kaufman, R.J.; Arvan, P. Biosynthesis, structure, and folding of the insulin precursor protein. Diabetes Obes. Metab. 2018, 20, 28–50. [Google Scholar] [CrossRef]
- Hober, D.; Sauter, P. Pathogenesis of type 1 diabetes mellitus: Interplay between enterovirus and host. Nat. Rev. Endocrinol. 2010, 6, 279–289. [Google Scholar] [CrossRef]
- Zell, R.; Delwart, E.; Gorbalenya, A.E.; Hovi, T.; King, A.M.Q.; Knowles, N.J.; Lindberg, A.M.; Pallansch, M.A.; Palmenberg, A.C.; Reuter, G.; et al. ICTV virus taxonomy profile: Picornaviridae. J. Gen. Virol. 2017, 98, 2421–2422. [Google Scholar] [CrossRef]
- Romero, J.R. Pediatric group B coxsackievirus infections. In Group B Coxsackieviruses; Springer: Berlin/Heidelberg, Germany, 2008; Volume 323, pp. 223–239. [Google Scholar]
- Tapparel, C.; Siegrist, F.; Petty, T.J.; Kaiser, L. Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. 2013, 14, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Hober, D.; Alidjinou, E.K. Enteroviral pathogenesis of type 1 diabetes: Queries and answers. Curr. Opin. Infect. Dis. 2013, 26, 263–269. [Google Scholar] [CrossRef]
- Ylipaasto, P.; Klingel, K.; Lindberg, A.M.; Otonkoski, T.; Kandolf, R.; Hovi, T.; Roivainen, M. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia 2004, 47, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Dotta, F.; Censini, S.; Van Halteren, A.G.S.; Marselli, L.; Masini, M.; Dionisi, S.; Mosca, F.; Boggi, U.; Muda, A.O.; Del Prato, S.; et al. Coxsackie B4 virus infection of β cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc. Natl. Acad. Sci. USA 2007, 104, 5115–5120. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.J.; Willcox, A.; Bone, A.J.; Foulis, A.K.; Morgan, N.G. The prevalence of enteroviral capsid protein vp1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia 2009, 52, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.J.; Leete, P.; Bone, A.J.; Foulis, A.K.; Morgan, N.G. Expression of the enteroviral capsid protein VP1 in the islet cells of patients with type 1 diabetes is associated with induction of protein kinase R and downregulation of Mcl-1. Diabetologia 2013, 56, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Willcox, A.; Richardson, S.J.; Bone, A.J.; Foulis, A.K.; Morgan, N.G. Immunohistochemical analysis of the relationship between islet cell proliferation and the production of the enteroviral capsid protein, VP1, in the islets of patients with recent-onset type 1 diabetes. Diabetologia 2011, 54, 2417–2420. [Google Scholar] [CrossRef]
- Yeung, W.-C.G.; Rawlinson, W.D.; Craig, M.E. Enterovirus infection and type 1 diabetes mellitus: Systematic review and meta-analysis of observational molecular studies. BMJ 2011, 342, d35. [Google Scholar] [CrossRef] [PubMed]
- Elshebani, A.; Olsson, A.; Westman, J.; Tuvemo, T.; Korsgren, O.; Frisk, G. Effects on isolated human pancreatic islet cells after infection with strains of enterovirus isolated at clinical presentation of type 1 diabetes. Virus Res. 2007, 124, 193–203. [Google Scholar] [CrossRef]
- Hodik, M.; Lukinius, A.; Korsgren, O.; Frisk, G. Tropism analysis of two coxsackie B5 strains reveals virus growth in human primary pancreatic islets but not in exocrine cell clusters in vitro. Open Virol. J. 2013, 7, 49–56. [Google Scholar] [CrossRef]
- Anagandula, M.; Richardson, S.J.; Oberste, M.S.; Sioofy-Khojine, A.B.; Hyöty, H.; Morgan, N.G.; Korsgren, O.; Frisk, G. Infection of human islets of langerhans with two strains of coxsackie B virus serotype 1: Assessment of virus replication, degree of cell death and induction of genes involved in the innate immunity pathway. J. Med. Virol. 2014, 86, 1402–1411. [Google Scholar] [CrossRef]
- Alidjinou, E.K.; Sané, F.; Engelmann, I.; Geenen, V.; Hober, D. Enterovirus persistence as a mechanism in the pathogenesis of type 1 diabetes. Discov. Med. 2014, 18, 273–282. [Google Scholar]
- Pinkert, S.; Klingel, K.; Lindig, V.; Dörner, A.; Zeichhardt, H.; Spiller, O.B.; Fechner, H. Virus-host coevolution in a persistently coxsackievirus B3-infected cardiomyocyte cell line. J. Virol. 2011, 85, 13409–13419. [Google Scholar] [CrossRef]
- Schmidtke, M.; Selinka, H.C.; Heim, A.; Jahn, B.; Tonew, M.; Kandolf, R.; Stelzner, A.; Zell, R. Attachment of coxsackievirus B3 variants to various cell lines: Mapping of phenotypic differences to capsid protein VP1. Virology 2000, 275, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Alidjinou, E.K.; Engelmann, I.; Bossu, J.; Villenet, C.; Figeac, M.; Romond, M.B.; Sané, F.; Hober, D. Persistence of coxsackievirus B4 in pancreatic ductal-like cells results in cellular and viral changes. Virulence 2017, 8, 1229–1244. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.E.; Messner, R.P. Molecular mechanisms of coxsackievirus persistence in chronic inflammatory myopathy: Viral RNA persists through formation of a double-stranded complex without associated genomic mutations or evolution. J. Virol. 1999, 73, 10113–10121. [Google Scholar] [CrossRef]
- Chapman, N.M.; Kim, K.-S.; Drescher, K.M.; Oka, K.; Tracy, S. 5’ terminal deletions in the genome of a coxsackievirus B2 strain occurred naturally in human heart. Virology 2008, 375, 480–491. [Google Scholar] [CrossRef]
- Tracy, S.; Smithee, S.; Alhazmi, A.; Chapman, N. Coxsackievirus can persist in murine pancreas by deletion of 5’ terminal genomic sequences. J. Med. Virol. 2015, 87, 240–247. [Google Scholar] [CrossRef]
- Kuo, H.H.; Ahmad, R.; Lee, G.Q.; Gao, C.; Chen, H.R.; Ouyang, Z.; Szucs, M.J.; Kim, D.; Tsibris, A.; Chun, T.W.; et al. Anti-apoptotic protein BIRC5 maintains survival of HIV-1-infected CD4+ T cells. Immunity 2018, 48, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Chehadeh, W.; Kerr-Conte, J.; Pattou, F.; Alm, G.; Lefebvre, J.; Wattré, P.; Hober, D. Persistent infection of human pancreatic islets by coxsackievirus B is associated with alpha interferon synthesis in β cells. J. Virol. 2000, 74, 10153–10164. [Google Scholar] [CrossRef] [PubMed]
- Nekoua, M.P.; Bertin, A.; Sane, F.; Alidjinou, E.K.; Lobert, D.; Trauet, J.; Hober, C.; Engelmann, I.; Moutairou, K.; Yessoufou, A.; et al. Pancreatic beta cells persistently infected with coxsackievirus B4 are targets of NK cell-mediated cytolytic activity. Cell. Mol. Life Sci. 2020, 77, 179–194. [Google Scholar] [CrossRef]
- De Beeck, A.O.; Eizirik, D.L. Viral infections in type 1 diabetes mellitus—Why the β cells? Nat. Rev. Endocrinol. 2016, 12, 263–273. [Google Scholar] [CrossRef]
- Lincez, P.J.; Shanina, I.; Horwitz, M.S. Reduced expression of the MDA5 Gene IFIH1 prevents autoimmune diabetes. Diabetes 2015, 64, 2184–2193. [Google Scholar] [CrossRef]
- Engelmann, I.; Alidjinou, E.K.; Bertin, A.; Bossu, J.; Villenet, C.; Figeac, M.; Sane, F.; Hober, D. Persistent coxsackievirus B4 infection induces microRNA dysregulation in human pancreatic cells. Cell. Mol. Life Sci. 2017, 74, 3851–3861. [Google Scholar] [CrossRef]
- Sane, F.; Caloone, D.; Gmyr, V.; Engelmann, I.; Belaich, S.; Kerr-Conte, J.; Pattou, F.; Desailloud, R.; Hober, D. Coxsackievirus B4 can infect human pancreas ductal cells and persist in ductal-like cell cultures which results in inhibition of Pdx1 expression and disturbed formation of islet-like cell aggregates. Cell. Mol. Life Sci. 2013, 70, 4169–4180. [Google Scholar] [CrossRef] [PubMed]
- Chick, W.L.; Warren, S.; Chute, R.N.; Like, A.A.; Lauris, V.; Kitchen, K.C. A transplantable insulinoma in the rat. Proc. Natl. Acad. Sci. USA 1977, 74, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Asfari, M.; Janjic, D.; Meda, P.; Li, G.; Halban, P.A.; Wollheim, C.B. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology 1992, 130, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Dechaumes, A.; Nekoua, M.P.; Belouzard, S.; Sane, F.; Engelmann, I.; Dubuisson, J.; Alidjinou, E.K.; Hober, D. Fluoxetine can inhibit SARS-CoV-2 in vitro. Microorganisms 2021, 9, 339. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bertin, A.; Sane, F.; Gmyr, V.; Lobert, D.; Dechaumes, A.; Kerr-Conte, J.; Pattou, F.; Hober, D. Coxsackievirus-b4 infection of human primary pancreatic ductal cell cultures results in impairment of differentiation into insulin-producing cells. Viruses 2019, 11, 597. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento, L.; Medina, A.; Aziz, K.; Anagandula, M.; Cabrera-Rode, E.; Fex, M.; Frisk, G.; Cilio, C.M. Differential effects of three echovirus strains on cell lysis and insulin secretion in beta cell derived lines. J. Med. Virol. 2016, 88, 971–978. [Google Scholar] [CrossRef]
- Yin, H.; Berg, A.-K.; Westman, J.; Hellerström, C.; Frisk, G. Complete nucleotide sequence of a Coxsackievirus B-4 strain capable of establishing persistent infection in human pancreatic islet cells: Effects on insulin release, proinsulin synthesis, and cell morphology. J. Med. Virol. 2002, 68, 544–557. [Google Scholar] [CrossRef]
- Lombardi, A.; Tomer, Y. Interferon alpha impairs insulin production in human beta cells via endoplasmic reticulum stress. J. Autoimmun. 2017, 80, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Denning, S.M.; Kurtzberg, J.; Le, P.T.; Tuck, D.T.; Singer, K.H.; Haynes, B.F. Human thymic epithelial cells directly induce activation of autologous immature thymocytes. Proc. Natl. Acad. Sci. USA 1988, 85, 3125–3129. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Leung, K.C.; Rawlinson, W.D.; Naing, Z.; Craig, M.E. Enterovirus infection induces cytokine and chemokine expression in insulin-producing cells. J. Med. Virol. 2010, 82, 1950–1957. [Google Scholar] [CrossRef] [PubMed]
- Goffard, A.; Alidjinou, E.K.; Sané, F.; Choteau, L.; Bouquillon, C.; Caloone, D.; Lobert, P.E.; Hober, D. Antibodies enhance the infection of phorbol-ester-differentiated human monocyte-like cells with coxsackievirus B4. Microbes Infect. 2013, 15, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Steenkamp, D.W.; Cacicedo, J.M.; Sahin-Efe, A.; Sullivan, C.; Sternthal, E. Preserved proinsulin secretion in long-standing type 1 diabetes. Endocr. Pract. 2017, 23, 1387–1393. [Google Scholar] [CrossRef]
- Cianciaruso, C.; Phelps, E.A.; Pasquier, M.; Hamelin, R.; Demurtas, D.; Ahmed, M.A.; Piemonti, L.; Hirosue, S.; Swartz, M.A.; De Palma, M.; et al. Primary human and rat β-Cells release the intracellular autoantigens GAD65, IA-2, and proinsulin in exosomes together with cytokine-induced enhancers of immunity. Diabetes 2017, 66, 460–473. [Google Scholar] [CrossRef]
- So, M.; Elso, C.M.; Tresoldi, E.; Pakusch, M.; Pathiraj, V.; Wentworth, J.M.; Harrison, L.C.; Krishnamurthy, B.; Thomas, H.E.; Rodda, C.; et al. Proinsulin C-peptide is an autoantigen in people with type 1 diabetes. Proc. Natl. Acad. Sci. USA 2018, 115, 10732–10737. [Google Scholar] [CrossRef]
- Michels, A.W.; Landry, L.G.; McDaniel, K.A.; Yu, L.; Campbell-Thompson, M.; Kwok, W.W.; Jones, K.L.; Gottlieb, P.A.; Kappler, J.W.; Tang, Q.; et al. Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes 2017, 66, 722–734. [Google Scholar] [CrossRef]
- Leong, W.F.; Chow, V.T.K. Transcriptomic and proteomic analyses of rhabdomyosarcoma cells reveal differential cellular gene expression in response to enterovirus 71 infection. Cell. Microbiol. 2006, 8, 565–580. [Google Scholar] [CrossRef]
- Rassmann, A.; Martin, U.; Saluz, H.P.; Peter, S.; Munder, T.; Henke, A. Identification of gene expression profiles in HeLa cells and HepG2 cells infected with Coxsackievirus B3. J. Virol. Methods 2013, 187, 190–194. [Google Scholar] [CrossRef]
- Grubman, M.J.; Moraes, M.P.; Diaz-San Segundo, F.; Pena, L.; De Los Santos, T. Evading the host immune response: How foot-and-mouth disease virus has become an effective pathogen. FEMS Immunol. Med. Microbiol. 2008, 53, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yu, B.; Lin, L.; Zhai, X.; Han, Y.; Qin, Y.; Guo, Z.; Wu, S.; Zhong, X.; Wang, Y.; et al. A functional nuclear localization sequence in the VP1 capsid protein of coxsackievirus B3. Virology 2012, 433, 513–521. [Google Scholar] [CrossRef]
- Falk, M.M.; Grigera, P.R.; Bergmann, I.E.; Zibert, A.; Multhaup, G.; Beck, E. Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J. Virol. 1990, 64, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Griger, P.R.; Tisminetzky, S.G. Histone H3 modification in BHK cells infected with foot-and-mouth disease virus. Virology 1984, 136, 10–19. [Google Scholar] [CrossRef]
- Nguyen, E.V.; Gharib, S.A.; Crothers, K.; Chow, Y.H.; Park, D.R.; Goodlett, D.R.; Schnapp, L.M. Proteomic landscape of bronchoalveolar lavage fluid in human immunodeficiency virus infection. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 306. [Google Scholar] [CrossRef]
- Hammer, E.; Goritzka, M.; Ameling, S.; Darm, K.; Steil, L.; Klingel, K.; Trimpert, C.; Herda, L.R.; Dörr, M.; Kroemer, H.K.; et al. Characterization of the human myocardial proteome in inflammatory dilated cardiomyopathy by label-free quantitative shotgun proteomics of heart biopsies. J. Proteome Res. 2011, 10, 2161–2171. [Google Scholar] [CrossRef]
- Hsu, Y.Y.; Liu, Y.N.; Lu, W.W.; Kung, S.H. Visualizing and quantifying the differential cleavages of the eukaryotic translation initiation factors eIF4GI and eIF4GII in the enterovirus-infected cell. Biotechnol. Bioeng. 2009, 104, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.S.W.; Moran-Jones, K.; Shan, J.; Munro, T.P.; Snee, M.J.; Hoek, K.S.; Smith, R. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J. Biol. Chem. 2002, 277, 18010–18020. [Google Scholar] [CrossRef]
- Jagdeo, J.M.; Dufour, A.; Fung, G.; Luo, H.; Kleifeld, O.; Overall, C.M.; Jan, E. Heterogeneous nuclear ribonucleoprotein M facilitates enterovirus infection. J. Virol. 2015, 89, 7064–7078. [Google Scholar] [CrossRef]
- Kawasaki, E. ZnT8 and type 1 diabetes. Endocr. J. 2012, 59, 531–537. [Google Scholar] [CrossRef]
| Target | Sequence | |
|---|---|---|
| β-actin | Forward | 5′-GGCACTCTTCCAGCCTTCCT-3′ |
| Reverse | 5′-GCAATGCCAGGGTACATGGT-3′ | |
| CV-B4 E2 | Forward | 5′-CAAGCACTTCTGTTTCCCCGG-3′ |
| Reverse | 5′-ATTGTCACCATAAGCAGCCA-3′ | |
| PCSK2 | Forward | 5′-CGAAACCAGCTTCACGATGAG-3′ |
| Reverse | 5′-ACGCCGGCTTAGCAAAATGGA-3′ | |
| Accession | Protein | Infected Cells Score | Controls Score | Score Difference | p-Value |
|---|---|---|---|---|---|
| P21807 | Peripherin | 0.00 ± 0.00 | 12.56 ± 4.10 | −12.56 | *** |
| Q07266-2 | Isoform E1 of Drebrin | 6.29 ± 3.00 | 17.56 ± 2.90 | −11.27 | *** |
| Q4QQS7 | Protein Umps | 0.49 ± 0.99 | 5.40 ± 1.14 | −4.91 | *** |
| P34926 | Microtubule-associated protein 1A | 2.00 ± 1.37 | 14.30 ± 5.17 | −12.3 | ** |
| Q5XIM9 | T-complex protein 1 subunit beta | 7.86 ± 3.07 | 15.90 ± 2.48 | −8.04 | ** |
| P23514 | Coatomer subunit beta | 1.05 ± 2.10 | 5.01 ± 1.13 | −3.96 | ** |
| D4A2G9 | Protein Ranbp1 | 0.00 ± 0.00 | 3.60 ± 1.79 | −3.6 | ** |
| G3V7N5 | Carnitine O-palmitoyltransferase 2, mitochondrial | 0.00 ± 0.00 | 3.38 ± 1.66 | −3.38 | ** |
| D3ZMY7 | Protein Nt5c2 | 0.43 ± 0.87 | 3.75 ± 1.12 | −3.32 | ** |
| F1LMC7 | Septin-7 | 0.44 ± 0.87 | 2.60 ± 1.37 | −2.16 | ** |
| A0A0G2K1J5 | Plectin | 4.04 ± 2.38 | 21.11 ± 13.47 | −17.07 | * |
| Q6URK4 | Heterogeneous nuclear ribonucleoprotein A3 (HNRPA3) | 0.00 ± 0.00 | 16.28 ± 13.66 | −16.28 | * |
| P12785 | Fatty acid synthase | 6.78 ± 3.89 | 22.06 ± 8.44 | −15.28 | * |
| A0A0G2JU82 | Microtubule-actin cross-linking factor 1 | 7.59 ± 5.23 | 22.14 ± 9.29 | −14.55 | * |
| Q05982 | Nucleoside diphosphate kinase A | 6.26 ± 8.23 | 20.65 ± 6.46 | −14.39 | * |
| Q6URK4-2 | Heterogeneous nuclear ribonucleoprotein A3 (HNRPA3) isoform 2 | 18.03 ± 4.53 | 4.98 ± 8.08 | +13.05 | * |
| A0A0G2K013 | Alpha-actinin-4 | 9.04 ± 1.59 | 21.08 ± 7.58 | −12.04 | * |
| P06687 | Sodium/potassium-transporting ATPase subunit alpha-3 | 0.00 ± 0.00 | 11.90 ± 7.78 | −11.9 | * |
| D4AD15 | Protein Eif4g1 | 4.66 ± 2.13 | 16.16 ± 6.89 | −11.5 | * |
| A0A0G2K0Q7 | Protein Mylk | 3.17 ± 2.13 | 11.74 ± 4.47 | −8.57 | * |
| O35314 | Secretogranin-1 | 5.04 ± 1.57 | 13.22 ± 4.74 | −8.18 | * |
| Q62667 | Major vault protein | 6.15 ± 2.75 | 14.06 ± 4.71 | −7.91 | * |
| D4AC23 | Protein Cct7 | 6.14 ± 1.77 | 13.98 ± 4.59 | −7.84 | * |
| P04692-5 | Tropomyosin alpha-1 chain isoform 5 | 1.62 ± 3.23 | 8.57 ± 5.17 | −6.95 | * |
| Q3MIE4 | Synaptic vesicle membrane protein VAT-1 homolog | 2.34 ± 2.20 | 9.00 ± 3.48 | −6.66 | * |
| D3ZRM9 | Uncharacterized protein | 0.00 ± 0.00 | 6.54 ± 3.76 | −6.54 | * |
| D3ZRM9 | Uncharacterized protein | 0.00 ± 0.00 | 6.54 ± 3.76 | −6.54 | * |
| Q62950 | Dihydropyrimidinase-related protein 1 | 2.41 ± 3.70 | 8.67 ± 4.13 | −6.26 | * |
| F1MAA1 | Ubiquitin-specific peptidase 47 | 2.26 ± 2.61 | 8.50 ± 4.06 | −6.24 | * |
| O35303-6 | Dynamin-1-like protein isoform 6 | 1.91 ± 3.83 | 7.53 ± 2.90 | −5.62 | * |
| P41562 | Isocitrate dehydrogenase [NADP] cytoplasmic | 2.57 ± 3.14 | 8.07 ± 2.98 | −5.5 | * |
| D4A0C3 | Protein Hid1 | 1.38 ± 1.78 | 6.44 ± 2.39 | −5.06 | * |
| A0A0G2JZ60 | Protein Fsd1l | 3.86 ± 3.10 | 8.89 ± 2.01 | −5.03 | * |
| A0A0G2JUN7 | Thioredoxin reductase 1, cytoplasmic | 2.77 ± 1.97 | 7.38 ± 2.33 | −4.61 | * |
| D3ZVQ0 | Protein LOC100911959 | 2.72 ± 2.66 | 7.14 ± 1.73 | −4.42 | * |
| O70593 | Small glutamine-rich tetratricopeptide repeat-containing protein alpha | 1.03 ± 1.21 | 5.43 ± 2.75 | −4.4 | * |
| P50475 | Alanine--tRNA ligase, cytoplasmic | 0.44 ± 0.88 | 4.76 ± 2.41 | −4.32 | * |
| P27008 | Poly [ADP-ribose] polymerase 1 | 1.90 ± 2.77 | 6.15 ± 2.62 | −4.25 | * |
| B2RYI2 | Signal recognition particle subunit SRP68 | 2.03 ± 1.60 | 6.04 ± 1.87 | −4.01 | * |
| Q05096-3 | Unconventional myosin-Ib isoform 3 | 1.96 ± 2.70 | 5.89 ± 1.41 | −3.93 | * |
| O88321 | Antisecretory factor | 1.47 ± 1.86 | 5.33 ± 2.30 | −3.86 | * |
| B2GV74 | Kinesin light chain 2 | 1.08 ± 2.16 | 4.89 ± 2.36 | −3.81 | * |
| Q9Z1W6-4 | Protein LYRIC isoform 4 | 0.54 ± 1.08 | 3.58 ± 2.25 | −3.04 | * |
| A0A0H2UHW4 | PEST proteolytic signal-containing nuclear protein | 0.42 ± 0.85 | 3.29 ± 2.02 | −2.87 | * |
| E9PT23 | Putative sodium-coupled neutral amino acid transporter 10 | 0.46 ± 0.93 | 3.00 ± 1.92 | −2.54 | * |
| D3Z8U5 | Metalloendopeptidase | 0.87 ± 1.73 | 3.10 ± 1.25 | −2.23 | * |
| Q5M7W6 | Protein FAM234A | 5.24 ± 0.98 | 1.88 ± 2.18 | +3.36 | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nekoua, M.P.; Bertin, A.; Sane, F.; Gimeno, J.-P.; Fournier, I.; Salzet, M.; Engelmann, I.; Alidjinou, E.K.; Hober, D. Persistence of Coxsackievirus B4 in Pancreatic β Cells Disturbs Insulin Maturation, Pattern of Cellular Proteins, and DNA Methylation. Microorganisms 2021, 9, 1125. https://doi.org/10.3390/microorganisms9061125
Nekoua MP, Bertin A, Sane F, Gimeno J-P, Fournier I, Salzet M, Engelmann I, Alidjinou EK, Hober D. Persistence of Coxsackievirus B4 in Pancreatic β Cells Disturbs Insulin Maturation, Pattern of Cellular Proteins, and DNA Methylation. Microorganisms. 2021; 9(6):1125. https://doi.org/10.3390/microorganisms9061125
Chicago/Turabian StyleNekoua, Magloire Pandoua, Antoine Bertin, Famara Sane, Jean-Pascal Gimeno, Isabelle Fournier, Michel Salzet, Ilka Engelmann, Enagnon Kazali Alidjinou, and Didier Hober. 2021. "Persistence of Coxsackievirus B4 in Pancreatic β Cells Disturbs Insulin Maturation, Pattern of Cellular Proteins, and DNA Methylation" Microorganisms 9, no. 6: 1125. https://doi.org/10.3390/microorganisms9061125
APA StyleNekoua, M. P., Bertin, A., Sane, F., Gimeno, J.-P., Fournier, I., Salzet, M., Engelmann, I., Alidjinou, E. K., & Hober, D. (2021). Persistence of Coxsackievirus B4 in Pancreatic β Cells Disturbs Insulin Maturation, Pattern of Cellular Proteins, and DNA Methylation. Microorganisms, 9(6), 1125. https://doi.org/10.3390/microorganisms9061125








