Composition of Prokaryotic and Eukaryotic Microbial Communities in Waters around the Florida Reef Tract
Abstract
:1. Introduction
2. Materials and Methods
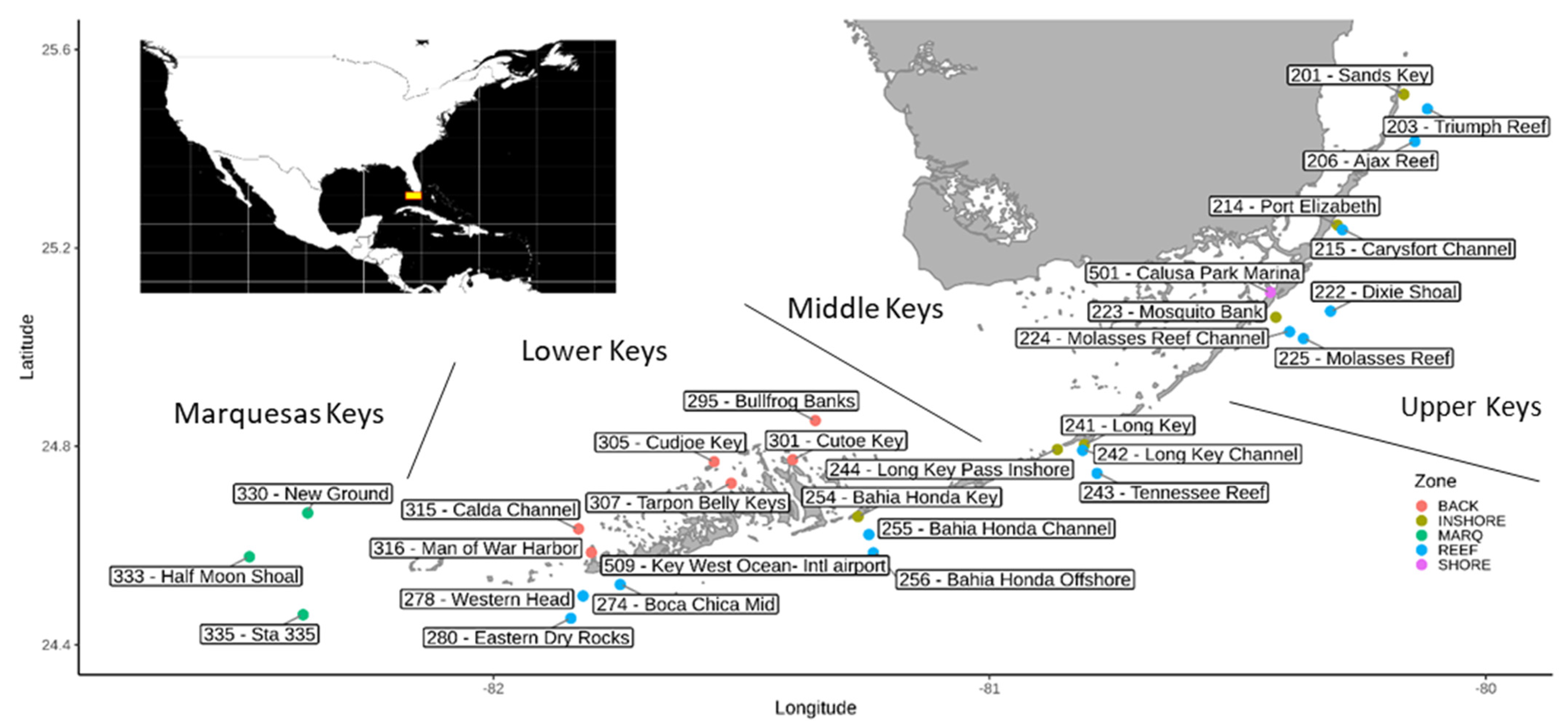
2.1. Collection of Samples and Physico-Chemical Data
2.2. Flow Cytometry
2.3. Phylogenetic Profiling
2.4. Bioinformatics
3. Results
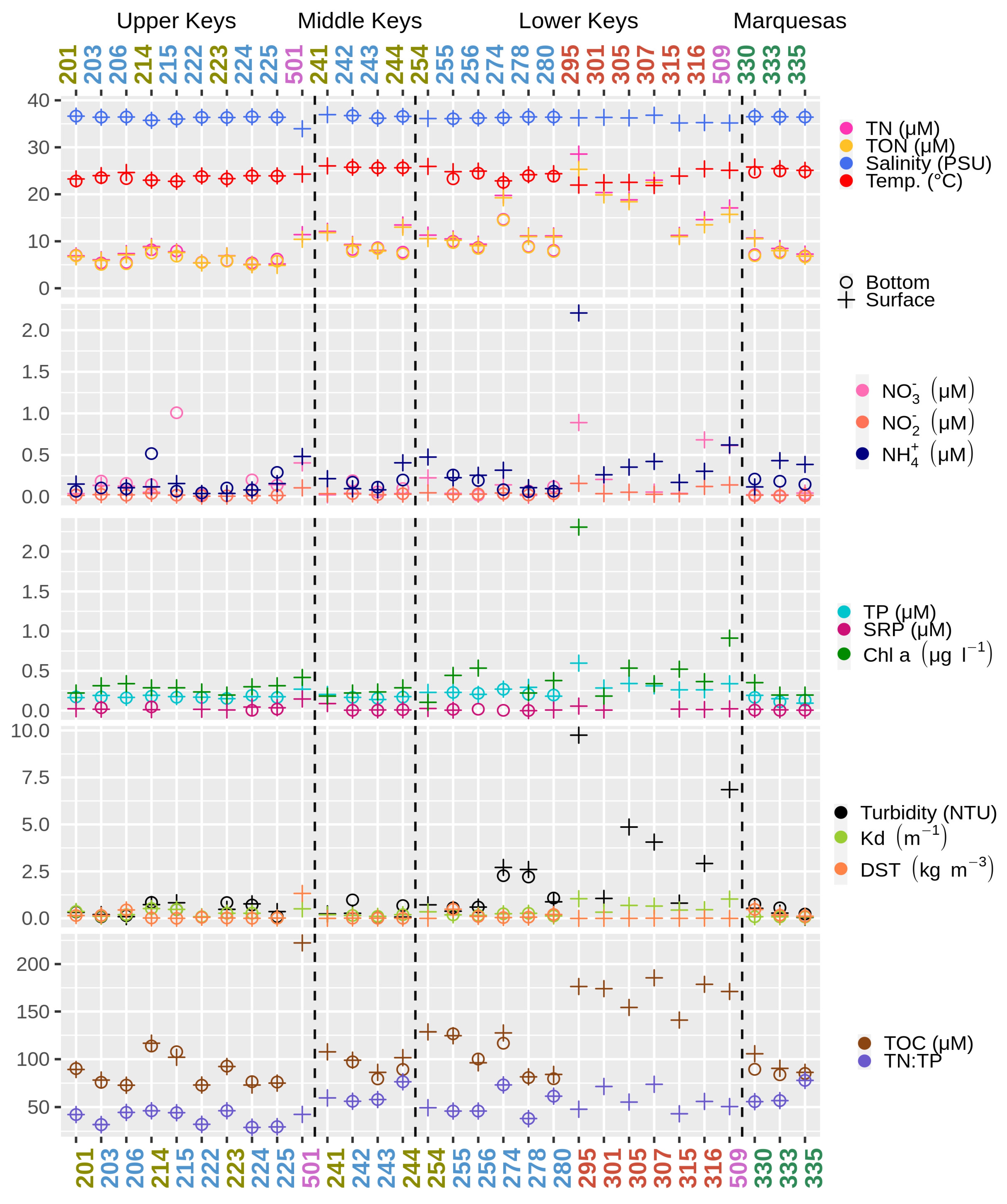
3.1. Physical and Chemical Parameters
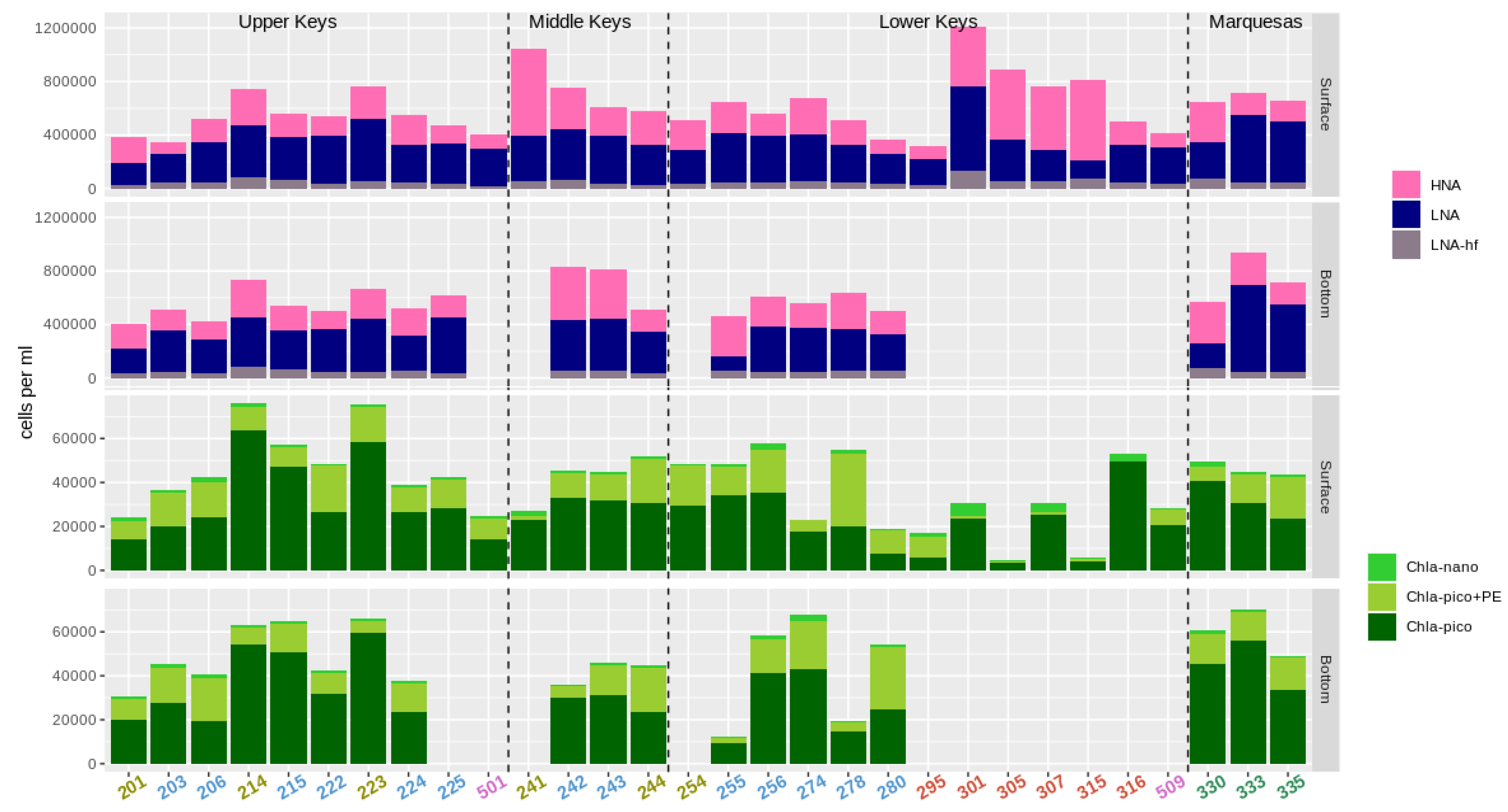
3.2. Abundance of Pico- and Nanoplankton
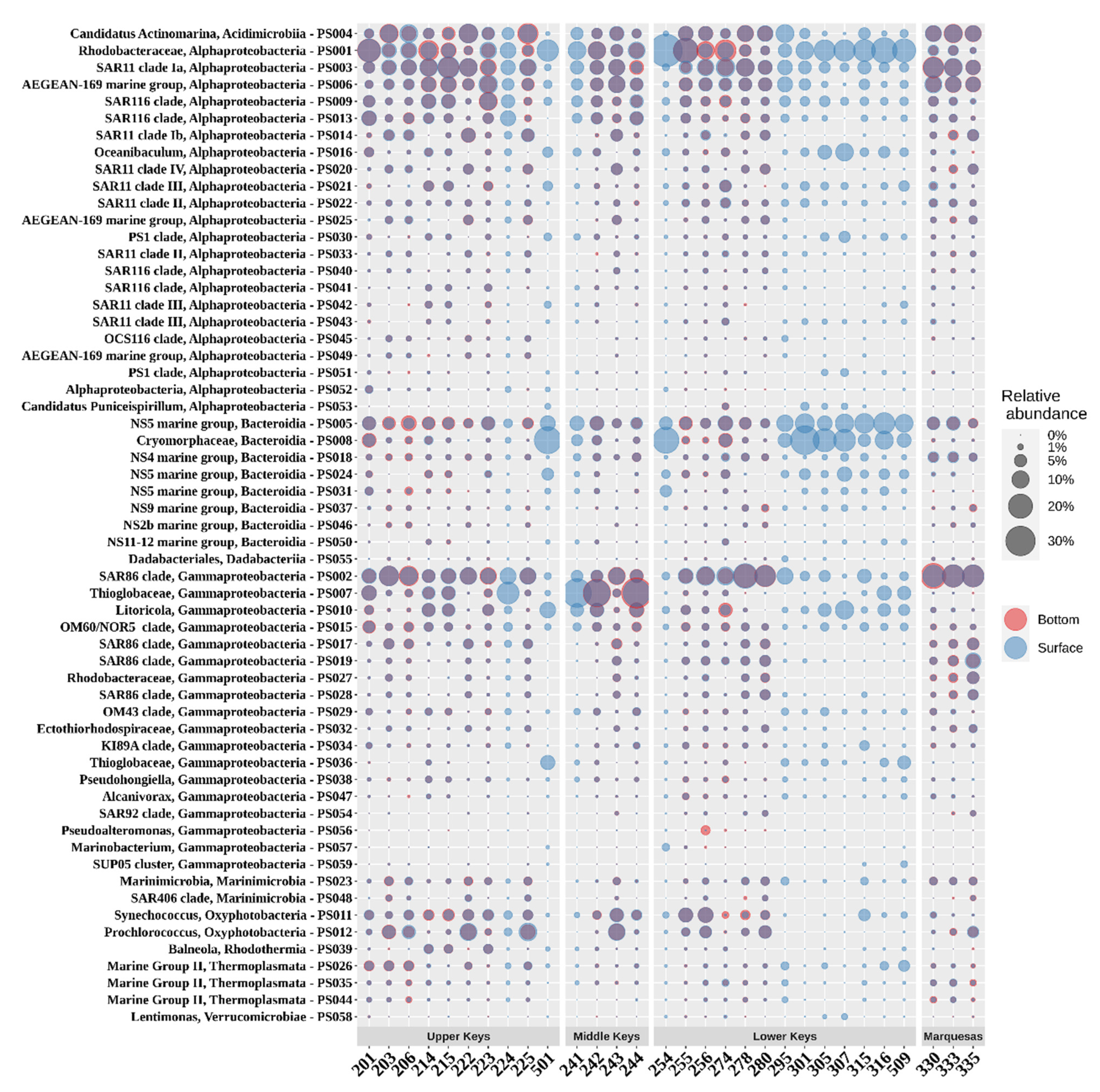
3.3. Prokaryotic Community Composition (PCC)
3.4. Eukaryotic Microbial Community (EMC)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Finkl, C.; Andrews, J. Shelf geomorphology along the southeast florida atlantic continental platform: Barrier coral reefs, nearshore bedrock, and morphosedimentary features. J. Coast. Res. 2008, 24, 823–849. [Google Scholar] [CrossRef] [Green Version]
- Moberg, F.; Folke, C. Ecological goods and services of coral reef ecosystems. Ecol. Econ. 1999, 29, 215–233. [Google Scholar] [CrossRef]
- Porter, J.; Dustan, P.; Jaap, W.; Patterson, K.; Kosmynin, V.; Meier, O.; Patterson, M.; Parsons, M. Patterns of spread of coral disease in the florida keys. Hydrobiologia 2001, 460, 1–24. [Google Scholar] [CrossRef]
- Rohmann, S.; Hayes, J.; Newhall, R.; Monaco, M.; Grigg, R. The area of potential shallow-water tropical and subtropical coral ecosystems in the united states. Coral Reefs 2005, 24, 370–383. [Google Scholar] [CrossRef]
- Rippe, J.; Kriefall, N.; Davies, S.; Castillo, K. Differential disease incidence and mortality of inner and outer reef corals of the upper florida keys in association with a white syndrome outbreak. Bull. Mar. Sci. 2019, 95, 305–316. [Google Scholar] [CrossRef]
- Meyer, J.; Castellanos-Gell, J.; Aeby, G.; Hase, C.; Ushijima, B.; Paul, V. Microbial community shifts associated with the ongoing stony coral tissue loss disease outbreak on the florida reef tract. Front. Microbiol. 2019, 10, 2244. [Google Scholar] [CrossRef]
- Aeby, G.; Ushijima, B.; Campbell, J.; Jones, S.; Williams, G.; Meyer, J.; Hase, C.; Paull, V. Pathogenesis of a tissue loss disease affecting multiple species of corals along the florida reef tract. Front. Mar. Sci. 2019, 6. [Google Scholar] [CrossRef]
- Rosales, S.; Clark, A.; Huebner, L.; Ruzicka, R.; Muller, E. Rhodobacterales and rhizobiales are associated with stony coral tissue loss disease and its suspected sources of transmission. Front. Microbiol. 2020, 11, 681. [Google Scholar] [CrossRef]
- Precht, W.; Gintert, B.; Robbart, M.; Fura, R.; van Woesik, R. Unprecedented disease-related coral mortality in southeastern florida. Sci. Rep. 2016, 6, 31374. [Google Scholar] [CrossRef] [PubMed]
- Walton, C.; Hayes, N.; Gilliam, D. Impacts of a regional, multi-year, multi-species coral disease outbreak in southeast florida. Front. Mar. Sci. 2018, 5, 323. [Google Scholar] [CrossRef]
- Lesser, M. Benthic-pelagic coupling on coral reefs: Feeding and growth of caribbean sponges. J. Exp. Mar. Biol. Ecol. 2006, 328, 277–288. [Google Scholar] [CrossRef]
- Garren, M.; Azam, F. New directions in coral reef microbial ecology. Environ. Microbiol. 2012, 14, 833–844. [Google Scholar] [CrossRef]
- Glasl, B.; Webster, N.; Bourne, D. Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Mar. Biol. 2017, 164, 91. [Google Scholar] [CrossRef]
- Zaneveld, J.R.; Burkepile, D.E.; Shantz, A.A.; Pritchard, C.E.; McMinds, R.; Payet, J.P.; Welsh, R.; Correa, A.M.; Lemoine, N.P.; Rosales, S.; et al. Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat. Commun. 2016, 7, 11833. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Agreda, A.; Gates, R.; Ainsworth, T. Defining the core microbiome in corals’ microbial soup. Trends Microbiol. 2017, 25, 125–140. [Google Scholar] [CrossRef]
- McDevitt-Irwin, J.; Baum, J.; Garren, M.; Thurber, R. Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 2017, 4, 262. [Google Scholar] [CrossRef] [Green Version]
- Maynard, J.; van Hooidonk, R.; Eakin, C.; Puotinen, M.; Garren, M.; Williams, G.; Heron, S.; Lamb, J.; Weil, E.; Willis, B.; et al. Projections of climate conditions that increase coral disease susceptibility and pathogen abundance and virulence. Nat. Clim. Chang. 2015, 5, 688–694. [Google Scholar] [CrossRef]
- Thurber, R.; Burkepile, D.; Fuchs, C.; Shantz, A.; McMinds, R.; Zaneveld, J. Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob. Chang. Biol. 2014, 20, 544–554. [Google Scholar] [CrossRef]
- Glasl, B.; Bourne, D.; Frade, P.; Thomas, T.; Schaffelke, B.; Webster, N. Microbial indicators of environmental perturbations in coral reef ecosystems. Microbiome 2019, 7, 94. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.; Clarke, M.; Williams, E.; Szmant, A.; Berger, T. Evolution of the tortugas gyre and its influence on recruitment in the Florida-keys. Bull. Mar. Sci. 1994, 54, 621–646. [Google Scholar]
- Lee, T.; Smith, N. Volume transport variability through the florida keys tidal channels. Cont. Shelf Res. 2002, 22, 1361–1377. [Google Scholar] [CrossRef]
- Briceño, H.O.; Boyer, J.N.; Castro, J.; Harlem, P. Biogeochemical classification of south Florida’s estuarine and coastal waters. Mar. Pollut. Bull. 2013, 75, 187–204. [Google Scholar] [CrossRef] [PubMed]
- Dixon, P. Vegan, a package of r functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Lebaron, P.; Servais, P.; Agogue, H.; Courties, C.; Joux, F. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl. Environ. Microbiol. 2001, 67, 1775–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.; Lauber, C.; Walters, W.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the illumina hiseq and miseq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [Green Version]
- Amaral-Zettler, L.; McCliment, E.; Ducklow, H.; Huse, S. A method for studying protistan diversity using massively parallel sequencing of v9 hypervariable regions of small-subunit ribosomal rna genes. PLoS ONE 2009, 4, e6372. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.; Dillon, M.; Bokulich, N.; Abnet, C.; Al-Ghalith, G.; Alexander, H.; Alm, E.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using qiime 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.; McMurdie, P.; Rosen, M.; Han, A.; Johnson, A.; Holmes, S. Dada2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F. The silva ribosomal rna gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Bokulich, N.; Kaehler, B.; Rideout, J.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.; Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with qiime 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Bouman, H.; Ulloa, O.; Scanlan, D.; Zwirglmaier, K.; Li, W.; Platt, T.; Stuart, V.; Barlow, R.; Leth, O.; Clementson, L.; et al. Oceanographic basis of the global surface distribution of prochlorococcus ecotypes. Science 2006, 312, 918–921. [Google Scholar] [CrossRef] [Green Version]
- Leblanc, K.; Queguiner, B.; Diaz, F.; Cornet, V.; Michel-Rodriguez, M.; de Madron, X.; Bowler, C.; Malviya, S.; Thyssen, M.; Gregori, G.; et al. Nanoplanktonic diatoms are globally overlooked but play a role in spring blooms and carbon export. Nat. Commun. 2018, 9, 953. [Google Scholar] [CrossRef] [PubMed]
- Schoener, D.; McManus, G. Plastid retention, use, and replacement in a kleptoplastidic ciliate. Aquat. Microb. Ecol. 2012, 67, 177–187. [Google Scholar] [CrossRef] [Green Version]
- Stoecker, D.; Silver, M.; Michaels, A.; Davis, L. Obligate mixotrophy in laboea-strobila, a ciliate which retains chloroplasts. Mar. Biol. 1988, 99, 415–423. [Google Scholar] [CrossRef]
- Kline, D.; Kuntz, N.; Breitbart, M.; Knowlton, N.; Rohwer, F. Role of elevated organic carbon levels and microbial activity in coral mortality. Mar. Ecol. Prog. Ser. 2006, 314, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Polz, M.; Hunt, D.; Preheim, S.; Weinreich, D. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos. Trans. R. Soc. B Biol. Sci. 2006, 361, 2009–2021. [Google Scholar] [CrossRef] [Green Version]
- Lauro, F.; McDougald, D.; Thomas, T.; Williams, T.; Egan, S.; Rice, S.; de Maere, M.; Ting, L.; Ertan, H.; Johnson, J.; et al. The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 15527–15533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frade, P.; Glasl, B.; Matthews, S.; Mellin, C.; Serrao, E.; Wolfe, K.; Mumby, P.; Webster, N.; Bourne, D. Spatial patterns of microbial communities across surface waters of the great barrier reef. Commun. Biol. 2020, 3, 442. [Google Scholar] [CrossRef]
- Campbell, A.M.; Fleisher, J.; Sinigalliano, C.; White, J.R.; Lopez, J.V. Dynamics of marine bacterial community diversity of the coastal waters of the reefs, inlets, and wastewater outfalls of southeast florida. Microbiologyopen 2015, 4, 390–408. [Google Scholar] [CrossRef]
- Muller, E.; Sartor, C.; Alcaraz, N.; van Woesik, R. Spatial epidemiology of the stony-coral-tissue-loss disease in florida. Front. Mar. Sci. 2020, 7, 163. [Google Scholar] [CrossRef] [Green Version]
- Cooney, R.; Pantos, O.; le Tissier, M.; Barer, M.; O’Donnell, A.; Bythell, J. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 2002, 4, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Frias-Lopez, J.; Zerkle, A.; Bonheyo, G.; Fouke, B. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 2002, 68, 2214–2228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landsberg, J.; Kiryu, Y.; Peters, E.; Wilson, P.; Perry, N.; Waters, Y.; Maxwell, K.; Huebner, L.; Work, T. Stony coral tissue loss disease in florida is associated with disruption of host zooxanthellae physiology. Front. Mar. Sci. 2020, 7, 1090. [Google Scholar] [CrossRef]
- Weber, M.; de Beer, D.; Lott, C.; Polerecky, L.; Kohls, K.; Abed, R.; Ferdelman, T.; Fabricius, K. Mechanisms of damage to corals exposed to sedimentation. Proc. Natl. Acad. Sci. USA 2012, 109, E1558–E1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, A.; Fairoz, M.; Kelly, L.; Nelson, C.; Dinsdale, E.; Edwards, R.; Giles, S.; Hatay, M.; Hisakawa, N.; Knowles, B.; et al. Global microbialization of coral reefs. Nat. Microbiol. 2016, 1, 16042. [Google Scholar] [CrossRef]
- Blanco, A.; Nadaoka, K.; Yamamoto, T. Planktonic and benthic microalgal community composition as indicators of terrestrial influence on a fringing reef in ishigaki island, southwest japan. Mar. Environ. Res. 2008, 66, 520–535. [Google Scholar] [CrossRef] [Green Version]
- Treguer, P.; Bowler, C.; Moriceau, B.; Dutkiewicz, S.; Gehlen, M.; Aumont, O.; Bittner, L.; Dugdale, R.; Finkel, Z.; Iudicone, D.; et al. Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 2018, 11, 27–37. [Google Scholar] [CrossRef]
- Hallegraeff, G. Harmful algal blooms in the australian region. Mar. Pollut. Bull. 1992, 25, 186–190. [Google Scholar] [CrossRef]
- Zinssmeister, C.; Soehner, S.; Facher, E.; Kirsch, M.; Meier, K.; Gottschling, M. Catch me if you can: The taxonomic identity of Scrippsiella trochoidea (f.Stein) a.R.Loebl. (Thoracosphaeraceae, Dinophyceae). Syst. Biodivers. 2011, 9, 145–157. [Google Scholar] [CrossRef]
- Muscatine, L.; Falkowski, P.; Dubinsky, Z.; Cook, P.; McCloskey, L. The effect of external nutrient resources on the population-dynamics of zooxanthellae in a reef coral. Proc. R. Soc. Ser. B Biol. Sci. 1989, 236, 311–324. [Google Scholar]
- Marubini, F.; Davies, P. Nitrate increases zooxanthellae population density and reduces skeletogenesis in corals. Mar. Biol. 1996, 127, 319–328. [Google Scholar] [CrossRef]
- Cunning, R.; Baker, A. Excess algal symbionts increase the susceptibility of reef corals to bleaching. Nat. Clim. Chang. 2013, 3, 259–262. [Google Scholar] [CrossRef]
- Shantz, A.; Lemoine, N.; Burkepile, D. Nutrient loading alters the performance of key nutrient exchange mutualisms. Ecol. Lett. 2016, 19, 20–28. [Google Scholar] [CrossRef]
- Wilson, W.; Francis, I.; Ryan, K.; Davy, S. Temperature induction of viruses in symbiotic dinoflagellates. Aquat. Microb. Ecol. 2001, 25, 99–102. [Google Scholar] [CrossRef] [Green Version]
- Davy, S.; Burchett, S.; Dale, A.; Davies, P.; Davy, J.; Muncke, C.; Hoegh-Guldberg, O.; Wilson, W. Viruses: Agents of coral disease? Dis. Aquat. Org. 2006, 69, 101–110. [Google Scholar] [CrossRef] [Green Version]
- Lohr, J.; Munn, C.; Wilson, W. Characterization of a latent virus-like infection of symbiotic zooxanthellae. Appl. Environ. Microbiol. 2007, 73, 2976–2981. [Google Scholar] [CrossRef] [Green Version]
- Roth, P.; Mikulskil, C.; Doucette, G. Influence of microbial interactions on the susceptibility of karenia spp. to algicidal bacteria. Aquat. Microb. Ecol. 2008, 50, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Mayali, X.; Azam, F. Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryot. Microbiol. 2004, 51, 139–144. [Google Scholar] [CrossRef]
- Dungca-Santos, J.; Caspe, F.; Tablizo, F.; Purganan, D.; Azanza, R.; Onda, D. Algicidal potential of cultivable bacteria from pelagic waters against the toxic dinoflagellate pyrodinium bahamense (dinophyceae). J. Appl. Phycol. 2019, 31, 3721–3735. [Google Scholar] [CrossRef]
- Montero, P.; Perez-Santos, I.; Daneri, G.; Gutierrez, M.; Igor, G.; Seguel, R.; Purdie, D.; Crawford, D. A winter dinoflagellate bloom drives high rates of primary production in a patagonian fjord ecosystem. Estuar. Coast. Shelf Sci. 2017, 199, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.; Yoo, Y.; Lee, K.; Kim, T.; Seong, K.; Kang, N.; Lee, S.; Kim, J.; Kim, S.; Yih, W. Red tides in masan bay, korea in 2004–2005: I. Daily variations in the abundance of red-tide organisms and environmental factors. Harmful Algae 2013, 30, S75–S88. [Google Scholar] [CrossRef]
- Anderson, D.; Alpermann, T.; Cembella, A.; Collos, Y.; Masseret, E.; Montresor, M. The globally distributed genus alexandrium: Multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 2012, 14, 10–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, M.; Kim, S.; Shin, E.; Yih, W.; Coats, D. Parasitism of harmful dinoflagellates in korean coastal waters. Harmful Algae 2013, 30, S62–S74. [Google Scholar] [CrossRef]
- Weber, L.; Gonzalez-Diaz, P.; Armenteros, M.; Ferrer, V.; Bretos, F.; Bartels, E.; Santoro, A.; Apprill, A. Microbial signatures of protected and impacted northern caribbean reefs: Changes from cuba to the florida keys. Environ. Microbiol. 2020, 22, 499–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambouvet, A.; Alves-de-Souza, C.; Cueff, V.; Marie, D.; Karpov, S.; Guillou, L. Interplay between the parasite Amoebophrya sp. (Alveolata) and the cyst formation of the red tide dinoflagellate Scrippsiella Trochoidea. Protist 2011, 162, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Jephcott, T.; Alves-De-Souza, C.; Gleason, F.; van Ogtrop, F.; Sime-Ngando, T.; Karpov, S.; Guillou, L. Ecological impacts of parasitic chytrids, syndiniales and perkinsids on populations of marine photosynthetic dinoflagellates. Fungal Ecol. 2016, 19, 47–58. [Google Scholar] [CrossRef]
- Skovgaard, A.; Daugbjerg, N. Identity and systematic position of paradinium poucheti and other paradinium-like parasites of marine copepods based on morphology and nuclear-encoded ssu rdna. Protist 2008, 159, 401–413. [Google Scholar] [CrossRef]
- Letcher, P.; Powell, M. A taxonomic summary and revision of rozella (cryptomycota). IMA Fungus 2018, 9, 383–399. [Google Scholar] [CrossRef] [Green Version]
- Gleason, F.; Lilje, O.; Marano, A.; Sime-Ngando, T.; Sullivan, B.; Kirchmair, M.; Neuhauser, S. Ecological functions of zoosporic hyperparasites. Front. Microbiol. 2014, 5, 244. [Google Scholar] [CrossRef] [Green Version]
- Vergeer, L.; den Hartog, C. Omnipresence of Labyrinthulaceae in seagrasses. Aquat. Bot. 1994, 48, 1–20. [Google Scholar] [CrossRef]
- Bremer, G. Lower marine fungi (Labyrinthulomycetes) and the decay of mangrove leaf-litter. Hydrobiologia 1995, 295, 89–95. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laas, P.; Ugarelli, K.; Absten, M.; Boyer, B.; Briceño, H.; Stingl, U. Composition of Prokaryotic and Eukaryotic Microbial Communities in Waters around the Florida Reef Tract. Microorganisms 2021, 9, 1120. https://doi.org/10.3390/microorganisms9061120
Laas P, Ugarelli K, Absten M, Boyer B, Briceño H, Stingl U. Composition of Prokaryotic and Eukaryotic Microbial Communities in Waters around the Florida Reef Tract. Microorganisms. 2021; 9(6):1120. https://doi.org/10.3390/microorganisms9061120
Chicago/Turabian StyleLaas, Peeter, Kelly Ugarelli, Michael Absten, Breege Boyer, Henry Briceño, and Ulrich Stingl. 2021. "Composition of Prokaryotic and Eukaryotic Microbial Communities in Waters around the Florida Reef Tract" Microorganisms 9, no. 6: 1120. https://doi.org/10.3390/microorganisms9061120
APA StyleLaas, P., Ugarelli, K., Absten, M., Boyer, B., Briceño, H., & Stingl, U. (2021). Composition of Prokaryotic and Eukaryotic Microbial Communities in Waters around the Florida Reef Tract. Microorganisms, 9(6), 1120. https://doi.org/10.3390/microorganisms9061120





