The Responses to Long-Term Water Addition of Soil Bacterial, Archaeal, and Fungal Communities in a Desert Ecosystem
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Experimental Design
2.3. Measurement of Biotic and Abiotic Factors
2.4. Soil DNA Extraction and Sequencing
2.5. Statistical Analysis
3. Results
3.1. Soil and Plant Variables in Different Water Addition Treatments
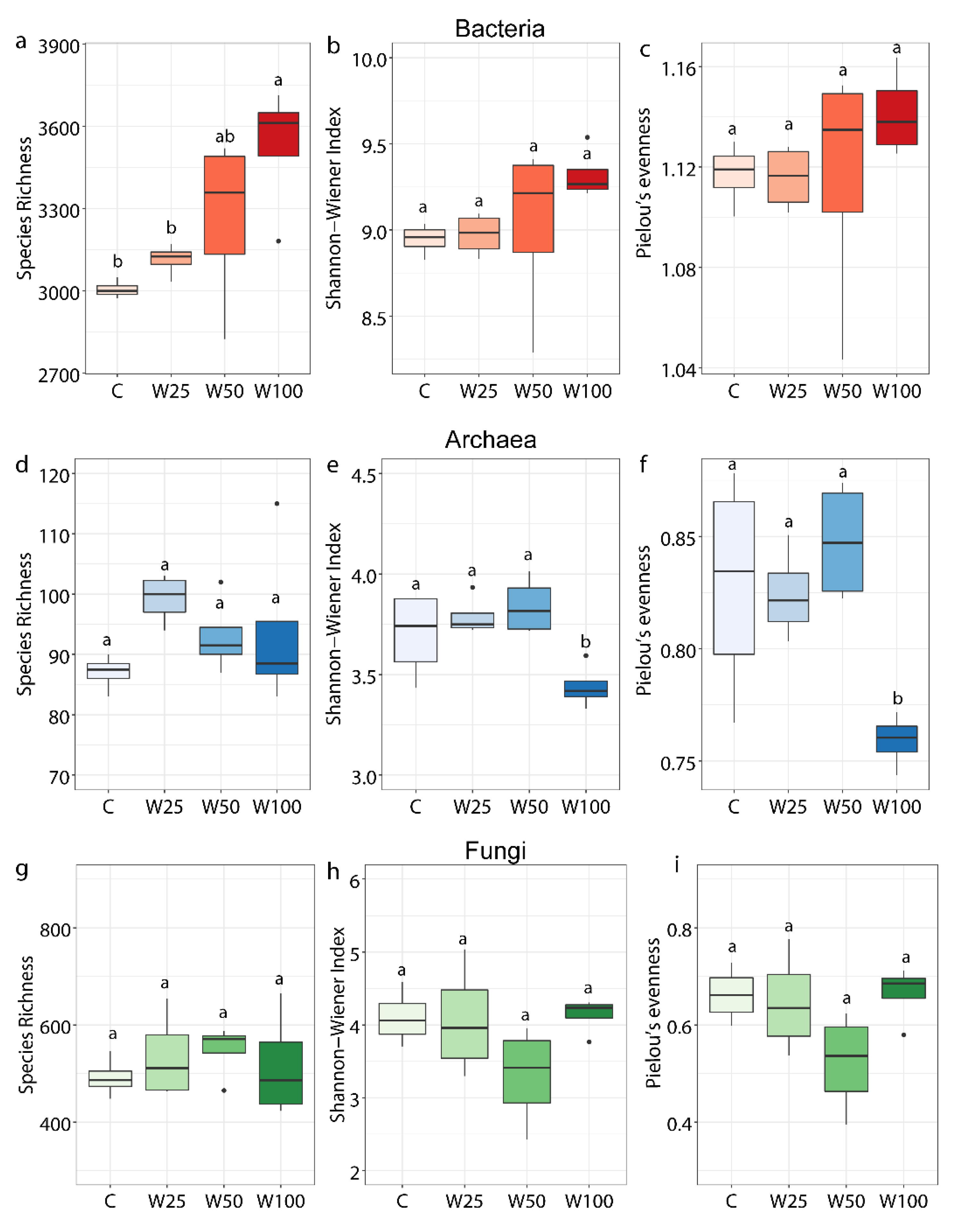
3.2. Soil Microbial Biomass, Diversity, and Community Composition
3.3. Responses of Microbial Taxa to Water Addition
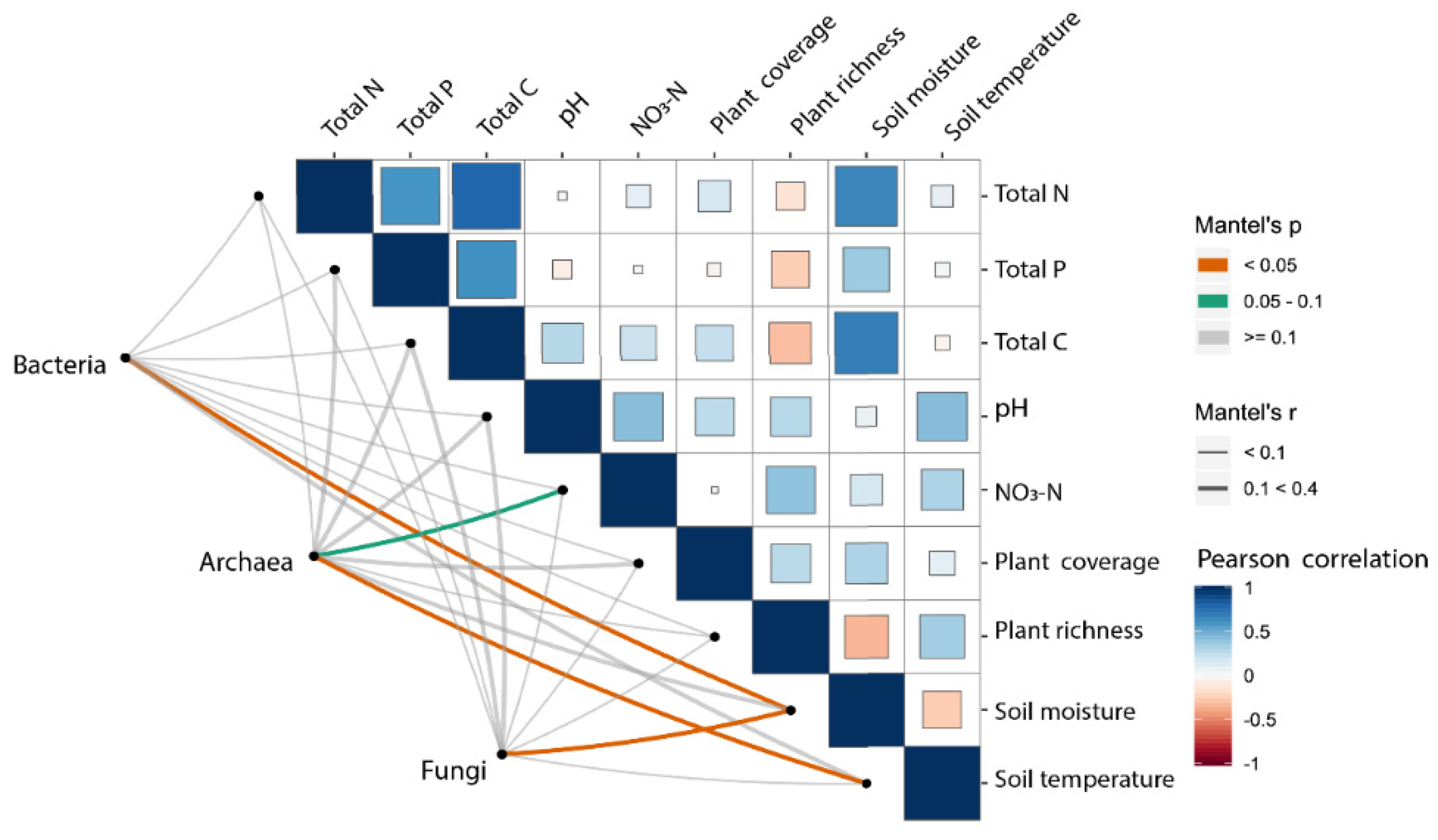
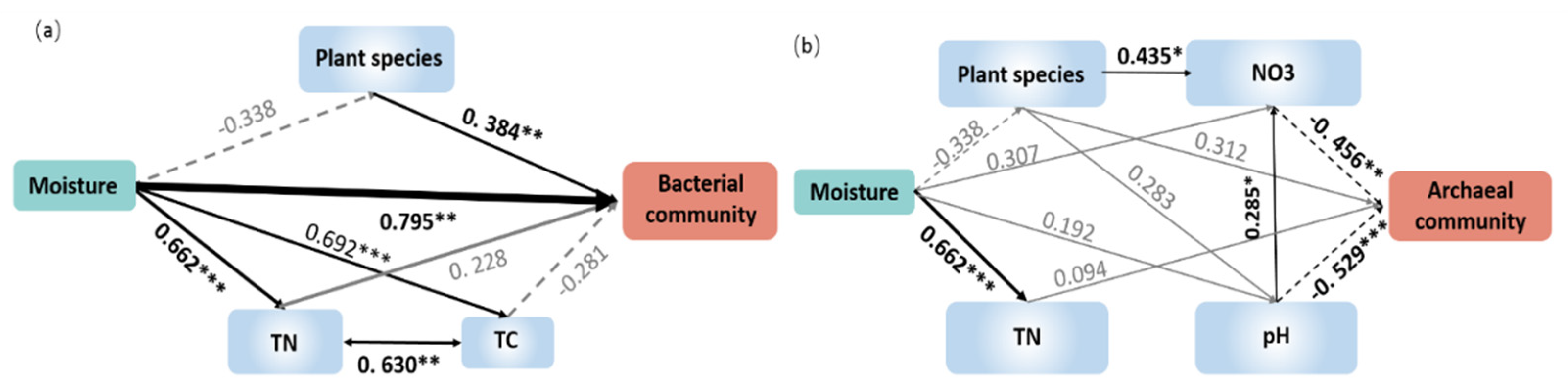
3.4. Relationships between Microbial Communities and Plant/Soil Properties
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vörösmarty, C.J.; Sahagian, D. Anthropogenic Disturbance of the Terrestrial Water Cycle. BioScience 2000, 50, 753–765. [Google Scholar] [CrossRef]
- IPCC. Climate Change 2014: The Physical Science Basis: Fifth Assessment Report of the Intergovernmental Panel on Climate Change. 2014. Available online: http://www.ipcc.ch (accessed on 10 December 2020).
- Meehl, G.A.; Stocker, T.F.; Collins, W.D.; Friedlingstein, P.; Gaye, A.T.; Gregory, J.M.; Kitoh, A.; Knutti, R.; Murphy, J.M.; Noda, A. Global Climate Projections. In Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change; Solomon, S., Qin, D., Manning, M., Chen, Z., Marquis, M., Averyt, K.B., Tignor, M., Miller, H.L., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2007. [Google Scholar]
- Nielsen, U.N.; Ball, B.A. Impacts of altered precipitation regimes on soil communities and biogeochemistry in arid and semi-arid ecosystems. Glob. Chang. Biol. 2015, 21, 1407–1421. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Hou, Y.; Zhang, L.; Liu, T.; Zhou, G. Ecosystem responses to warming and watering in typical and desert steppes. Sci. Rep. 2016, 6, 34801. [Google Scholar] [CrossRef]
- An, S.; Couteau, C.; Luo, F.; Neveu, J.; DuBow, M.S. Bacterial diversity of surface sand samples from the gobi and Taklamaken deserts. Microb. Ecol. 2013, 66, 850–860. [Google Scholar] [CrossRef]
- Lal, R. Carbon sequestration in dryland ecosystems. Environ. Manag. 2004, 33, 528–544. [Google Scholar] [CrossRef]
- Stone, R. Have desert researchers discovered a hidden loop in the carbon cycle? Science 2008, 320, 1409–1410. [Google Scholar] [CrossRef]
- Ronca, S.; Ramond, J.-B.; Jones, B.; Seely, M.; Cowan, D. Namib Desert dune/interdune transects exhibit habitat-specific edaphic bacterial communities. Front. Microbiol. 2015, 6, 845. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Xing, Z.; Zhao, X.; Pan, Y. Responses of soil microbial biomass and community composition to biological soil crusts in the revegetated areas of the Tengger Desert. Appl. Soil Ecol. 2013, 65, 52–59. [Google Scholar] [CrossRef]
- Sponseller, R.A. Precipitation pulses and soil CO2 flux in a Sonoran Desert ecosystem. Glob. Chang. Biol. 2007, 13, 426–436. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef]
- Piao, H.C.; Hong, Y.T.; Yuan, Z.Y. Seasonal changes of microbial biomass carbon related to climatic factors in soils from karst areas of southwest China. Biol. Fertil. Soils 2000, 30, 294–297. [Google Scholar] [CrossRef]
- Sorensen, P.O.; Germino, M.J.; Feris, K.P. Microbial community responses to 17 years of altered precipitation are seasonally dependent and coupled to co-varying effects of water content on vegetation and soil C. Soil Biol. Biochem. 2013, 64, 155–163. [Google Scholar] [CrossRef]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013, 7, 2229. [Google Scholar] [CrossRef]
- Placella, S.A.; Brodie, E.L.; Firestone, M.K. Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc. Natl. Acad. Sci. USA 2012, 109, 10931–10936. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.W.; Tissue, D.T.; Loik, M.E.; Wallenstein, M.D.; Acosta-Martinez, V.; Erickson, R.A.; Zak, J.C. Soil microbial and nutrient responses to 7 years of seasonally altered precipitation in a Chihuahuan Desert grassland. Glob. Chang. Biol. 2013, 20, 1657–1673. [Google Scholar] [CrossRef]
- Cregger, M.A.; Schadt, C.W.; McDowell, N.G.; Pockman, W.T.; Classen, A.T. Response of the soil microbial community to changes in precipitation in a semiarid ecosystem. Appl. Environ. Microbiol. 2012, 78, 8587–8594. [Google Scholar] [CrossRef]
- Boer, W.D.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef]
- She, W.; Bai, Y.; Zhang, Y.; Qin, S.; Feng, W.; Sun, Y.; Zheng, J.; Wu, B. Resource availability drives responses of soil microbial communities to short-term precipitation and nitrogen addition in a desert shrubland. Front. Microbiol. 2018, 9, 186. [Google Scholar] [CrossRef]
- Sher, Y.; Zaady, E.; Nejidat, A. Spatial and temporal diversity and abundance of ammonia oxidizers in semi-arid and arid soils: Indications for a differential seasonal effect on archaeal and bacterial ammonia oxidizers. FEMS Microbiol. Ecol. 2013, 86, 544–556. [Google Scholar] [CrossRef]
- Yu, T.; Wu, W.; Liang, W.; Lever, M.A.; Hinrichs, K.-U.; Wang, F. Growth of sedimentary Bathyarchaeota on lignin as an energy source. Proc. Natl. Acad. Sci. USA 2018, 115, 6022–6027. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.-K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579. [Google Scholar] [CrossRef]
- Marusenko, Y.; Bates, S.T.; Anderson, I.; Johnson, S.L.; Soule, T.; Garcia-Pichel, F. Ammonia-oxidizing archaea and bacteria are structured by geography in biological soil crusts across North American arid lands. Ecol. Process 2013, 2, 9. [Google Scholar] [CrossRef]
- Vikram, S.; Guerrero Leandro, D.; Makhalanyane Thulani, P.; Le Phuong, T.; Seely, M.; Cowan Don, A. Metagenomic analysis provides insights into functional capacity in a hyperarid desert soil niche community. Environ. Microbiol. 2015, 18, 1875–1888. [Google Scholar] [CrossRef]
- Clark, J.S.; Campbell, J.H.; Grizzle, H.; Acosta-Martìnez, V.; Zak, J.C. Soil microbial community response to drought and precipitation variability in the Chihuahuan desert. Microb. Ecol. 2009, 57, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Melillo, J.; Niu, S.; Beier, C.; Clark, J.S.; Classen, A.T.; Davidson, E.; Dukes, J.S.; Evans, R.D.; Field, C.B. Coordinated approaches to quantify long-term ecosystem dynamics in response to global change. Glob. Chang. Biol. 2011, 17, 843–854. [Google Scholar] [CrossRef]
- Li, Q.; Xu, J.; Li, H.; Wang, S.; Yan, X.; Xin, Z.; Jiang, Z.; Wang, L.; Jia, Z. Effects of aspect on clonal reproduction and biomass allocation of layering modules of Nitraria tangutorum in nebkha dunes. PLoS ONE 2013, 8, e79927. [Google Scholar] [CrossRef]
- Ying, S.; Ding, Y.H. A projection of future changes in summer precipitation and monsoon in East Asia. Sci. China Earth Sci. 2010, 53, 284–300. [Google Scholar]
- Song, W.; Chen, S.; Wu, B.; Zhu, Y.; Zhou, Y.; Li, Y.; Cao, Y.; Lu, Q.; Lin, G. Vegetation cover and rain timing co-regulate the responses of soil CO2 efflux to rain increase in an arid desert ecosystem. Soil Biol. Biochem. 2012, 49, 114–123. [Google Scholar] [CrossRef]
- Nelson, D.W.; Sommers, L.E.; Sparks, D.L.; Page, A.L.; Helmke, P.A.; Loeppert, R.H.; Soltanpour, P.N.; Tabatabai, M.A.; Johnston, C.T.; Sumner, M.E. Total carbon, organic carbon, and organic matter, Part 2 Chemical and Microbiological Properties. In Methods of Soil Analysis; American Society of Agronomy, Inc.: Madison, WI, USA, 1996; Volume 9. [Google Scholar]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Grace, J.B. Structural Equation Modeling and Natural Systems; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Computing 2014, 14, 12–21. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.3-5. 2016. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 30 March 2017).
- Martinez Arbizu, P. pairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. R package version 0.4. 2020. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 21 October 2020).
- Rosseel, Y. Lavaan: An R package for structural equation modeling. J. Stat. Softw. 2012, 48, 1–36. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Favet, J.; Lapanje, A.; Giongo, A.; Kennedy, S.; Aung, Y.-Y.; Cattaneo, A.; Davis-Richardson, A.G.; Brown, C.T.; Kort, R.; Brumsack, H.-J.; et al. Microbial hitchhikers on intercontinental dust: Catching a lift in Chad. ISME J. 2012, 7, 850. [Google Scholar] [CrossRef]
- Lin, L.; Guo, W.; Xing, Y.; Zhang, X.; Li, Z.; Hu, C.; Li, S.; Li, Y.; An, Q. The actinobacterium Microbacterium sp. 16SH accepts pBBR1-based pPROBE vectors, forms biofilms, invades roots, and fixes N2 associated with micropropagated sugarcane plants. Appl. Microbiol. Biotechnol. 2012, 93, 1185–1195. [Google Scholar] [CrossRef]
- Köberl, M.; Müller, H.; Ramadan, E.M.; Berg, G. Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS ONE 2011, 6, e24452. [Google Scholar] [CrossRef]
- Song, W.; Chen, S.; Zhou, Y.; Lin, G. Rainfall amount and timing jointly regulate the responses of soil nitrogen transformation processes to rainfall increase in an arid desert ecosystem. Geoderma 2020, 364, 114197. [Google Scholar] [CrossRef]
- Austin, A.T.; Yahdjian, L.; Stark, J.M.; Belnap, J.; Porporato, A.; Norton, U.; Ravetta, D.A.; Schaeffer, S.M. Water pulses and biogeochemical cycles in arid and semiarid ecosystems. Oecologia 2004, 141, 221–235. [Google Scholar] [CrossRef]
- Schulze-Makuch, D.; Wagner, D.; Kounaves, S.P.; Mangelsdorf, K.; Devine, K.G.; de Vera, J.-P.; Schmitt-Kopplin, P.; Grossart, H.-P.; Parro, V.; Kaupenjohann, M.; et al. Transitory microbial habitat in the hyperarid Atacama Desert. Proc. Natl. Acad. Sci. USA 2018, 115, 2670–2675. [Google Scholar] [CrossRef] [PubMed]
- Lennon, J.T.; Jones, S.E. Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 2011, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.-B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef]
- Schimel, J.; Balser Teri, C.; Wallenstein, M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 2007, 88, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Onyenwoke, R.U.; Brill, J.A.; Farahi, K.; Wiegel, J. Sporulation genes in members of the low G+C Gram-type-positive phylogenetic branch (Firmicutes). Arch. Microbiol. 2004, 182, 182–192. [Google Scholar] [CrossRef]
- Reith, F.; Brugger, J.; Zammit, C.M.; Gregg, A.L.; Goldfarb, K.C.; Andersen, G.L.; DeSantis, T.Z.; Piceno, Y.M.; Brodie, E.L.; Lu, Z.; et al. Influence of geogenic factors on microbial communities in metallogenic Australian soils. ISME J. 2012, 6, 2107. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; Peter, H.; Allison, S.D.; Baho, D.L.; Berga, M.; Bürgmann, H.; Huber, D.H.; Langenheder, S.; Lennon, J.T.; Martiny, J.B.H. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 2012, 3, 417. [Google Scholar] [CrossRef] [PubMed]
- Pointing, S.B.; Belnap, J. Microbial colonization and controls in dryland systems. Nat. Rev. Microbiol. 2012, 10, 551. [Google Scholar] [CrossRef]
- Wang, X.; Van Nostrand, J.D.; Deng, Y.; Lü, X.; Wang, C.; Zhou, J.; Han, X. Scale-dependent effects of climate and geographic distance on bacterial diversity patterns across northern China’s grasslands. FEMS Microbiol. Ecol. 2015, 91, fiv133. [Google Scholar] [CrossRef]
- Grigg, A.M.; Veneklaas, E.J.; Lambers, H. Water relations and mineral nutrition of Triodia grasses on desert dunes and interdunes. Aust. J. Bot. 2008, 56, 408–421. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, S.; Gao, Q.; Liu, S.; Ganjurjav, H.; Wang, X.; Su, X.; Wu, X. Soil bacterial and fungal diversity differently correlated with soil biochemistry in alpine grassland ecosystems in response to environmental changes. Sci. Rep. 2017, 7, 43077. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; He, D.; He, J.-S.; Myrold, D.D.; Chu, H. Ammonia-oxidizing bacteria rather than archaea respond to short-term urea amendment in an alpine grassland. Soil Biol. Biochem. 2017, 107, 218–225. [Google Scholar] [CrossRef]
- Winogradsky, S. Recherches sur les organisms de la nitrification. Ann. Inst. Pasteur 1890, 4, 213–231, 257–275, 760–771. [Google Scholar]
- Strous, M.; Fuerst, J.A.; Kramer, E.H.M.; Logemann, S.; Muyzer, G.; van de Pas-Schoonen, K.T.; Webb, R.; Kuenen, J.G.; Jetten, M.S.M. Missing lithotroph identified as new planctomycete. Nature 1999, 400, 446. [Google Scholar] [CrossRef]
- Andreas, S.; Dirk, D.B.; Armin, G.; Rudolf, A. Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ. Microbiol. 2000, 2, 680–686. [Google Scholar]
- Marusenko, Y.; Garcia-Pichel, F.; Hall, S.J. Ammonia-oxidizing archaea respond positively to inorganic nitrogen addition in desert soils. FEMS Microbiol. Ecol. 2015, 91, 1–11. [Google Scholar] [CrossRef]
- Huang, M.; Chai, L.; Jiang, D.; Zhang, M.; Zhao, Y.; Huang, Y. Increasing aridity affects soil archaeal communities by mediating soil niches in semi-arid regions. Sci. Total Environ. 2019, 647, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Zhang, Z.; Wang, X.; Zhou, Z.; Chen, D.; Zeng, H.; Zhao, S.; Chen, L.; Hu, Y.; Zhang, C.; et al. Diversity and contributions to nitrogen cycling and carbon fixation of soil salinity shaped microbial communities in tarim basin. Front. Microbiol. 2018, 9, 431. [Google Scholar] [CrossRef]
- Breidenbach, B.; Blaser Martin, B.; Klose, M.; Conrad, R. Crop rotation of flooded rice with upland maize impacts the resident and active methanogenic microbial community. Environ. Microbiol. 2015, 18, 2868–2885. [Google Scholar] [CrossRef]
- Ye, F.; Wu, S.; Jiang, Y.; Op den Camp, H.J.M.; Li, Z.; Zhu, G.; Zheng, J.; Wang, Y. Shifts of archaeal community structure in soil along an elevation gradient in a reservoir water level fluctuation zone. J. Soils Sediments 2016, 16, 2728–2739. [Google Scholar] [CrossRef]
- De Vries, F.T.; Shade, A. Controls on soil microbial community stability under climate change. Front. Microbiol. 2013, 4, 265. [Google Scholar] [CrossRef]
- Bouali, M.; Zrafi-Nouira, I.; Bakhrouf, A.; Le Paslier, D.; Chaussonnerie, S.; Ammar, E.; Sghir, A. The structure and spatio-temporal distribution of the Archaea in a horizontal subsurface flow constructed wetland. Sci. Total Environ. 2012, 435–436, 465–471. [Google Scholar] [CrossRef]
- Finstad, K.M.; Probst, A.J.; Thomas, B.C.; Andersen, G.L.; Demergasso, C.; Echeverría, A.; Amundson, R.G.; Banfield, J.F. Microbial community structure and the persistence of cyanobacterial populations in salt crusts of the hyperarid atacama desert from genome-resolved metagenomics. Front. Microbiol. 2017, 8, 1435. [Google Scholar] [CrossRef]
- Liu, P.; Klose, M.; Conrad, R. Temperature effects on structure and function of the methanogenic microbial communities in two paddy soils and one desert soil. Soil Biol. Biochem. 2018, 124, 236–244. [Google Scholar] [CrossRef]
- Treseder, K.K.; Schimel, J.P.; Garcia, M.O.; Whiteside, M.D. Slow turnover and production of fungal hyphae during a Californian dry season. Soil Biol. Biochem. 2010, 42, 1657–1660. [Google Scholar] [CrossRef]
- Zhang, X.; Wei, H.; Chen, Q.; Han, X. The counteractive effects of nitrogen addition and watering on soil bacterial communities in a steppe ecosystem. Soil Biol. Biochem. 2014, 72, 26–34. [Google Scholar] [CrossRef]
- Gordon, H.; Haygarth, P.M.; Bardgett, R.D. Drying and rewetting effects on soil microbial community composition and nutrient leaching. Soil Biol. Biochem. 2008, 40, 302–311. [Google Scholar] [CrossRef]
- Angel, R.; Soares, M.I.M.; Ungar, E.D.; Gillor, O. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. 2009, 4, 553. [Google Scholar] [CrossRef] [PubMed]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263. [Google Scholar] [CrossRef]
- Frossard, A.; Ramond, J.-B.; Seely, M.; Cowan, D.A. Water regime history drives responses of soil Namib Desert microbial communities to wetting events. Sci. Rep. 2015, 5, 12263. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhao, C.; Gao, Y.; Liu, B.; Wang, T.; Yuan, T.; Hale, L.; Van Nostrand, J.D.; Wan, S.; Zhou, J. Divergent taxonomic and functional responses of microbial communities to field simulation of aeolian soil erosion and deposition. Mol. Ecol. 2017, 26, 4186–4419. [Google Scholar] [CrossRef]
- Bates, S.T.; Berg-Lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011, 5, 908–917. [Google Scholar] [CrossRef] [PubMed]


| C | W25 | W50 | W100 | |
|---|---|---|---|---|
| September mean soil moisture (20 cm, %) | 3.17 | 3.20 | 3.85 | 6.79 |
| September mean soil temperature (20 cm, °C) | 23.58 | 24.28 | 20.09 | 22.36 |
| NO3− (×10−3 g·kg−1) | 1.50 ± 0.28 a1 | 1.19 ± 0.23 a | 0.86 ± 0.06 a | 1.49 ± 0.38 a |
| TC 2 (g·kg−1) | 0.10 ± 0.02 b | 0.10 ± 0.01 b | 0.10 ± 0.01 b | 0.16 ± 0.05 a |
| TN (g·kg−1) | 0.14 ± 0.02 b | 0.15 ± 0.01 b | 0.14 ± 0.01 b | 0.18 ± 0.03 a |
| TP (×10−4 g·kg−1) | 1.37 ± 0.05 a | 1.43 ± 0.05 a | 1.39 ± 0.09 a | 1.51 ± 0.07 a |
| pH | 7.89 ± 0.031 a | 7.89 ± 0.045 a | 7.85 ± 0.03 a | 7.89 ± 0.02 a |
| Plant coverage (%) | 25.0 ± 7.07 a | 31.25 ± 4.79 a | 26.25 ± 11.09 a | 33.75 ± 9.47 a |
| Plant species | 7.25 ± 1.50 a | 8.25 ± 0.96 a | 7.00 ± 0.82 a | 6.75 ± 0.96 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Xu, X.; Ding, J.; Bao, F.; De Costa, Y.G.; Zhuang, W.; Wu, B. The Responses to Long-Term Water Addition of Soil Bacterial, Archaeal, and Fungal Communities in a Desert Ecosystem. Microorganisms 2021, 9, 981. https://doi.org/10.3390/microorganisms9050981
Gao Y, Xu X, Ding J, Bao F, De Costa YG, Zhuang W, Wu B. The Responses to Long-Term Water Addition of Soil Bacterial, Archaeal, and Fungal Communities in a Desert Ecosystem. Microorganisms. 2021; 9(5):981. https://doi.org/10.3390/microorganisms9050981
Chicago/Turabian StyleGao, Ying, Xiaotian Xu, Junjun Ding, Fang Bao, Yashika G. De Costa, Weiqin Zhuang, and Bo Wu. 2021. "The Responses to Long-Term Water Addition of Soil Bacterial, Archaeal, and Fungal Communities in a Desert Ecosystem" Microorganisms 9, no. 5: 981. https://doi.org/10.3390/microorganisms9050981
APA StyleGao, Y., Xu, X., Ding, J., Bao, F., De Costa, Y. G., Zhuang, W., & Wu, B. (2021). The Responses to Long-Term Water Addition of Soil Bacterial, Archaeal, and Fungal Communities in a Desert Ecosystem. Microorganisms, 9(5), 981. https://doi.org/10.3390/microorganisms9050981






