Reciprocal Inhibition of Immunogenic Performance in Mice of Two Potent DNA Immunogens Targeting HCV-Related Liver Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Synthetic Peptides
2.3. Analysis of Expression of HCV Core
2.4. In Vitro Luciferase Assay
2.5. DNA Immunization
2.6. Assessment of Cellular Immune Response
2.7. In Vivo Imaging of Luciferase Gene Expression
2.8. In Vivo Promoter Activation Assay
2.9. Ethical Statement
2.10. Statistical Analysis
3. Results
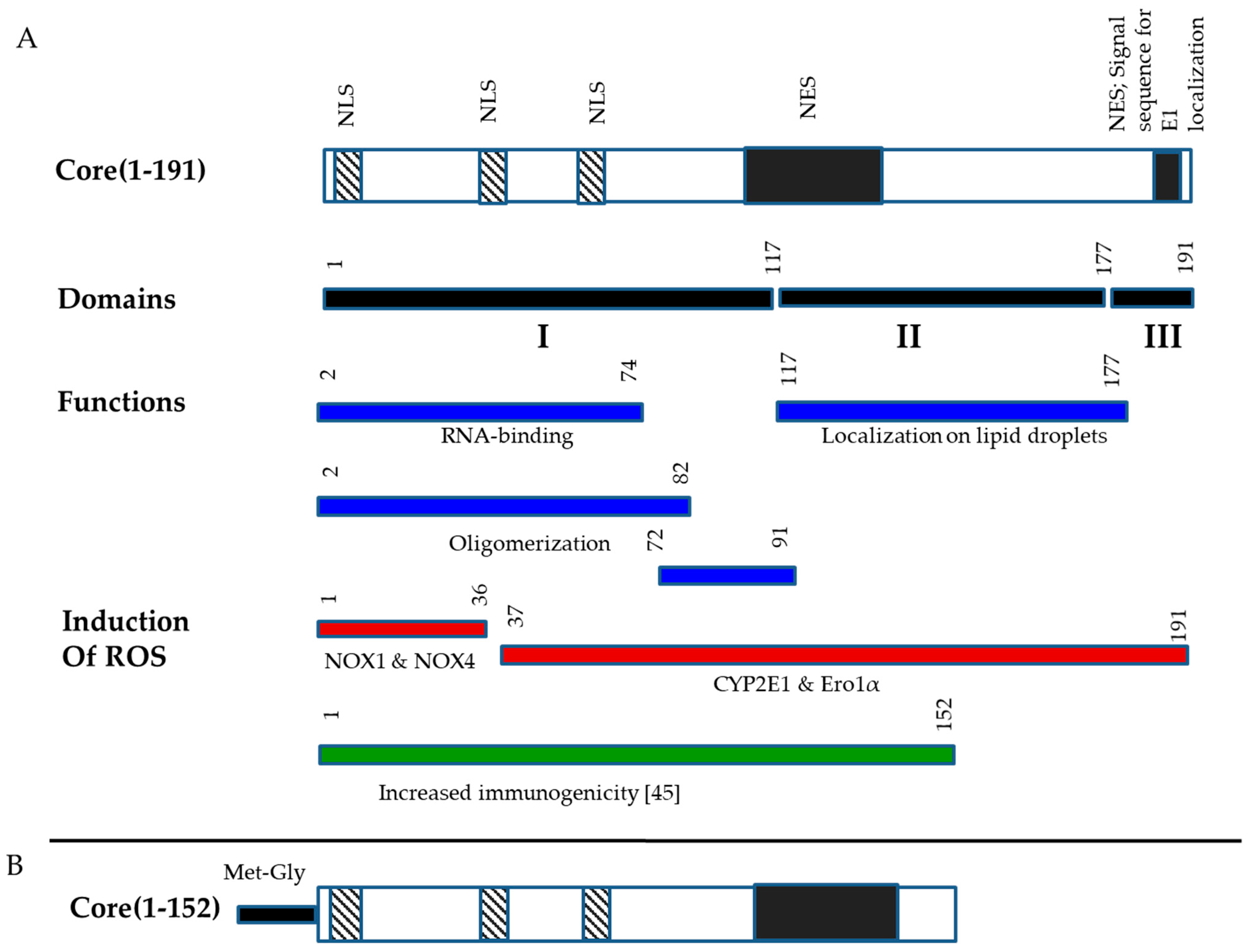
3.1. Design and Eukaryotic Expression of an Optimized HCV Core DNA Immunogen
3.2. Design of DNA Immunization Experiments
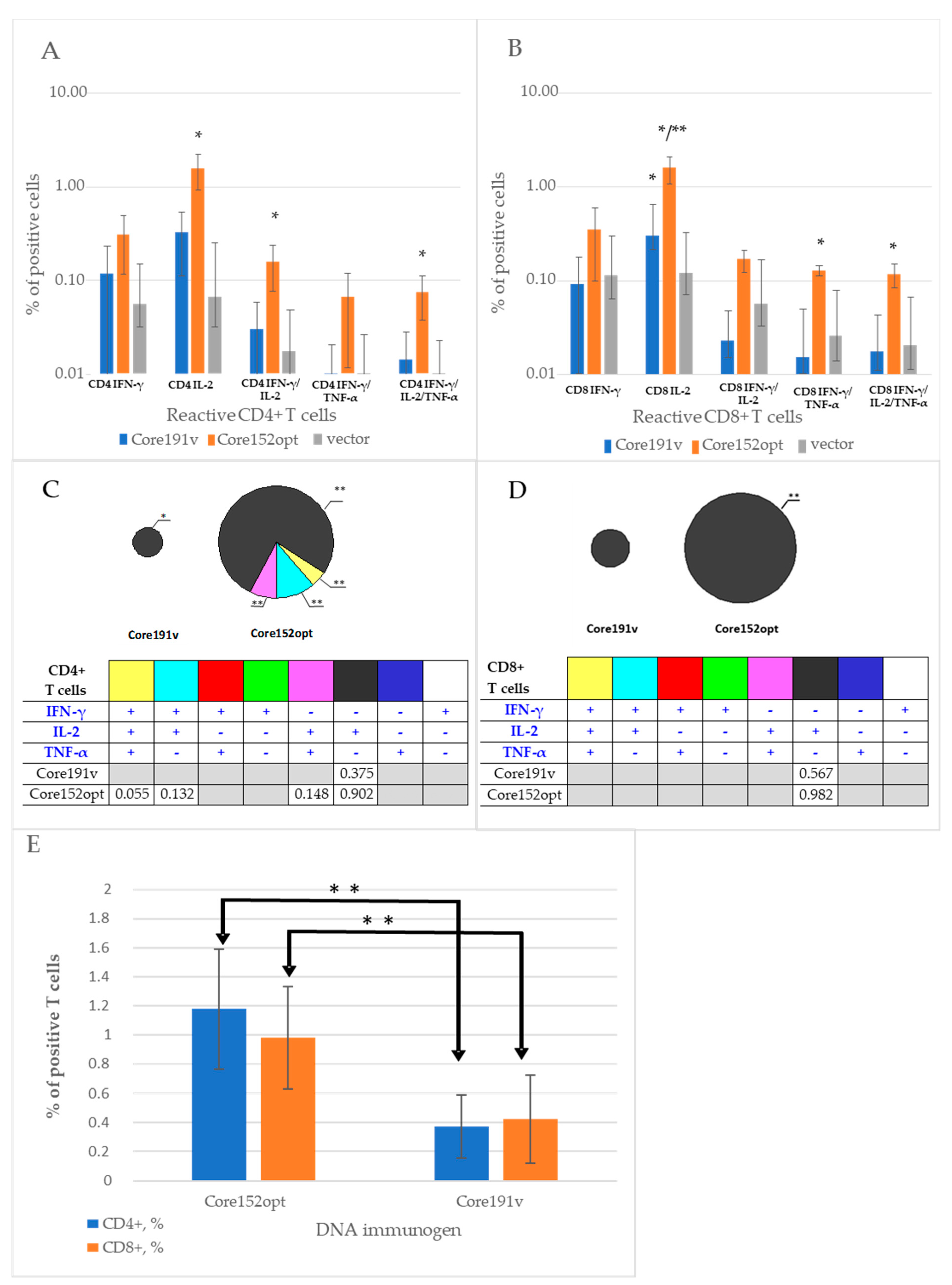
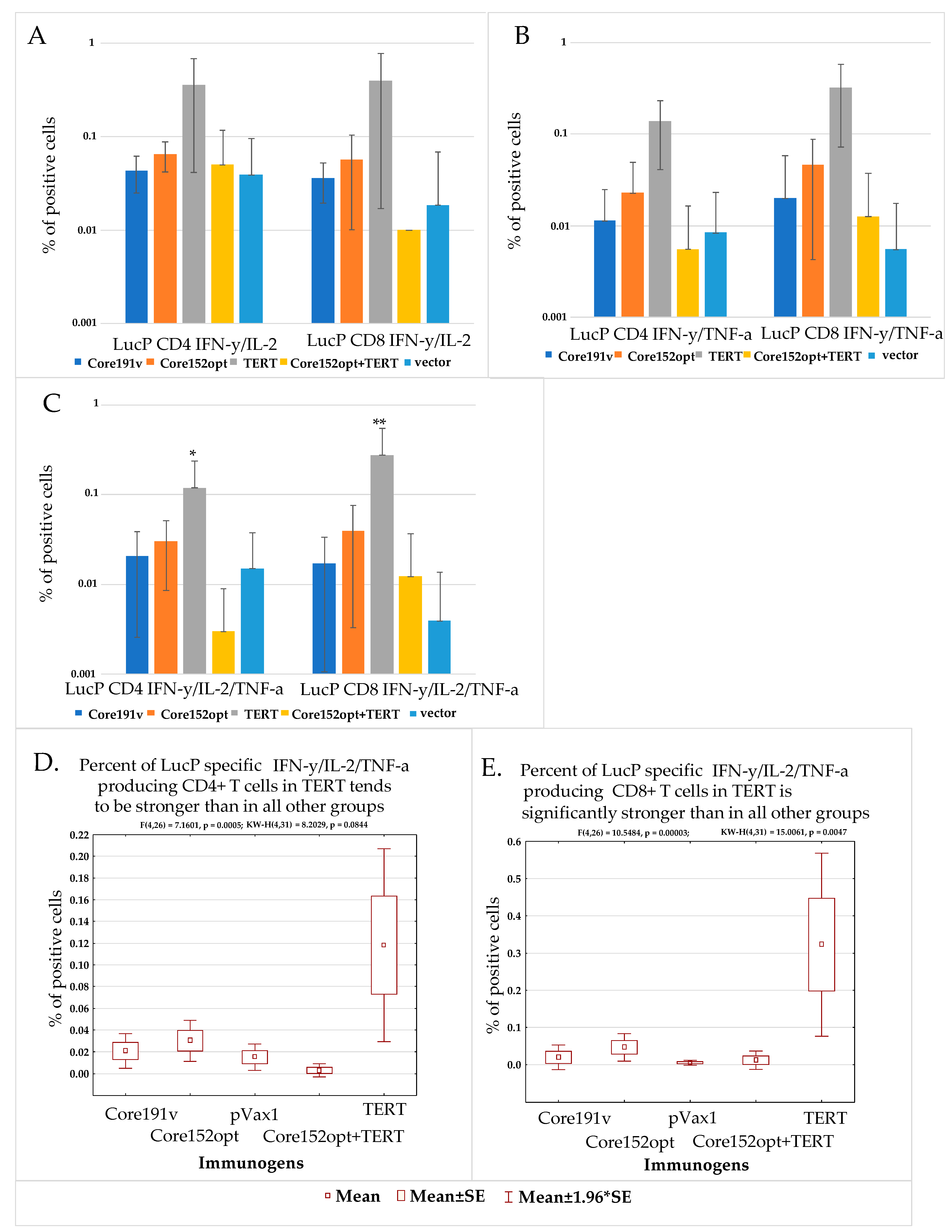
3.3. Cellular Immune Response against HCV Core
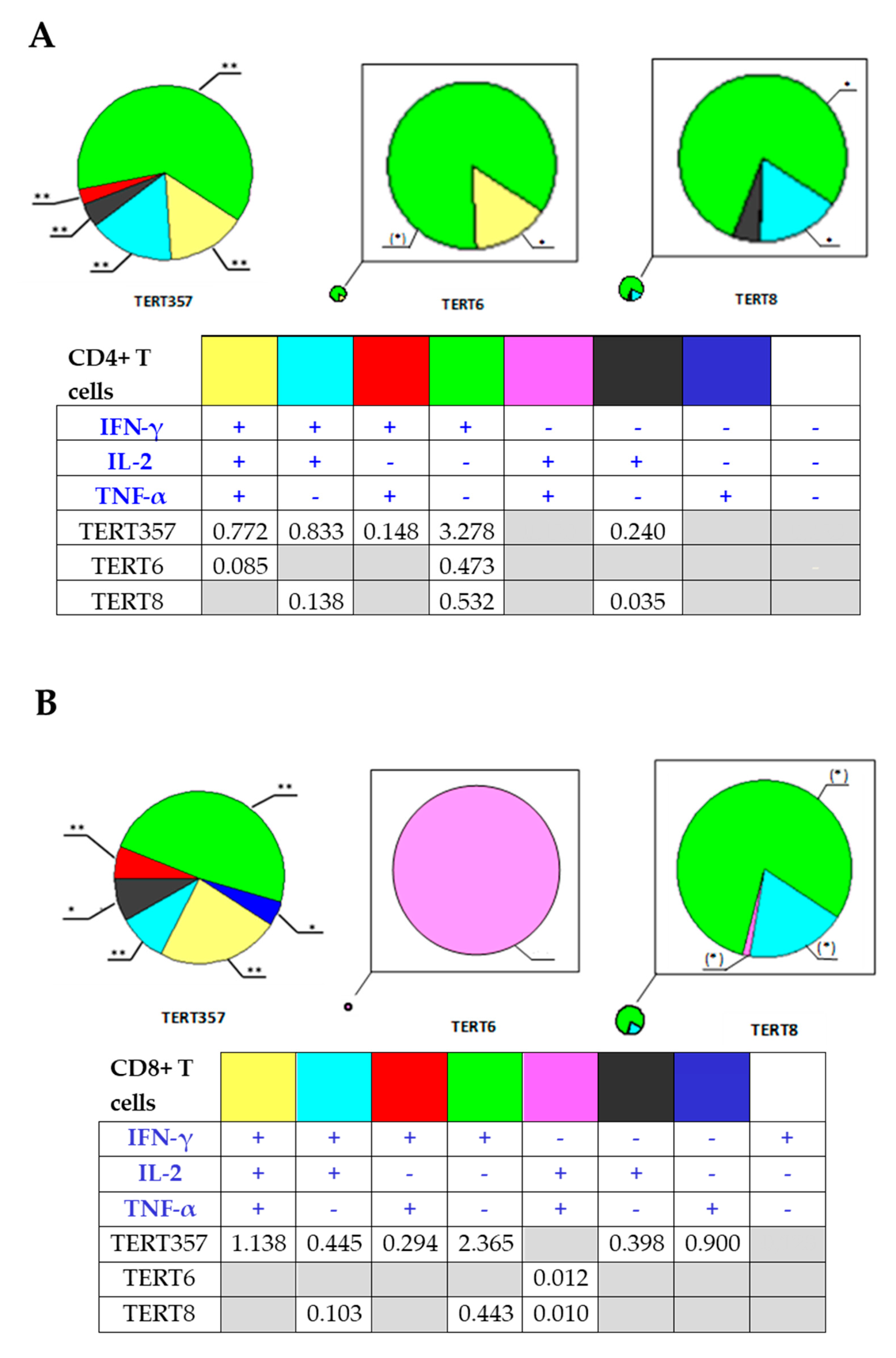
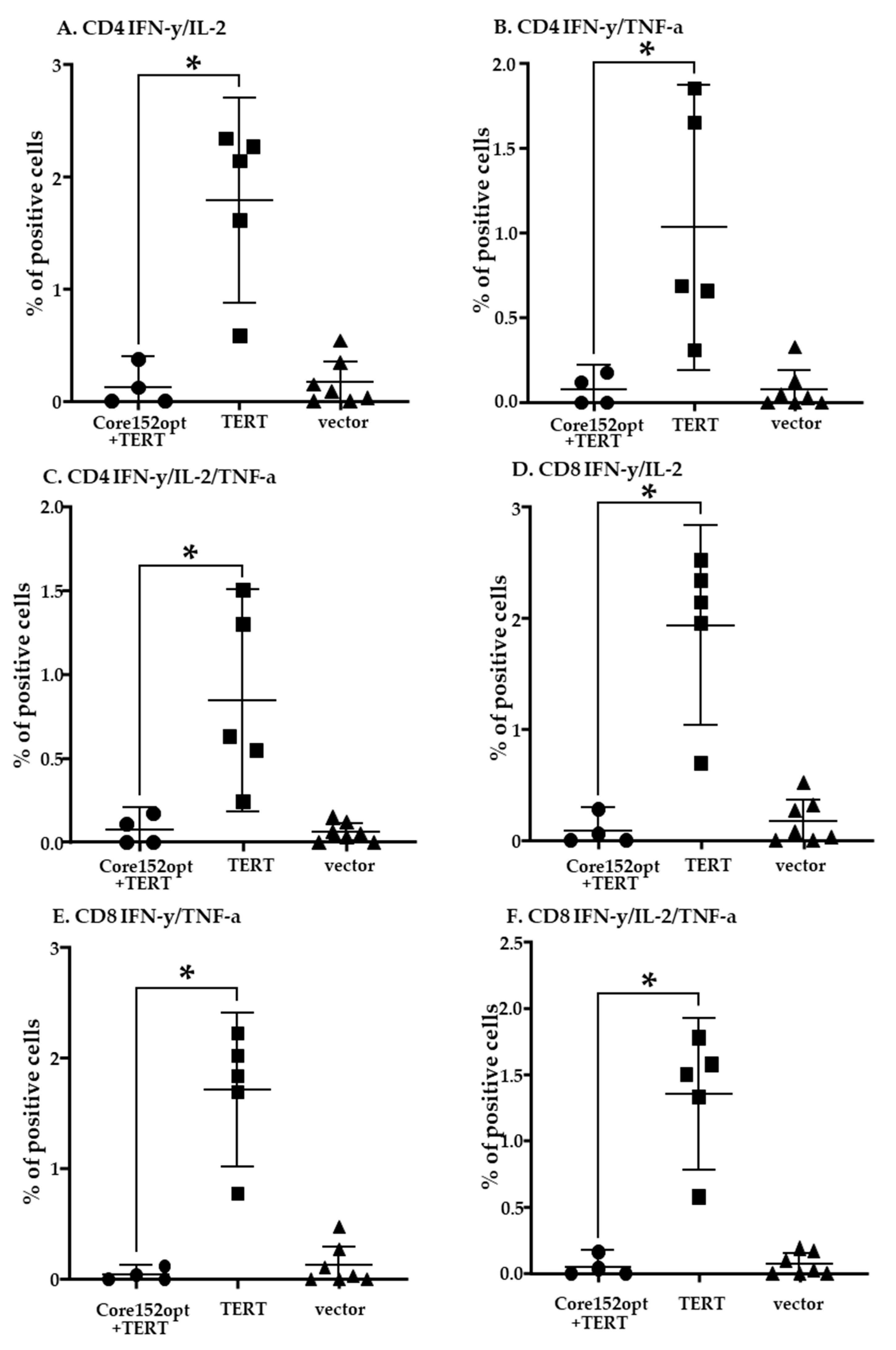
3.4. Cellular Immune Response against TERT and TERT-HCV Core Combination
3.5. Assessment of the Quality of DNA Immunization and Immune Response in Mice Receiving HCV Core and TERT
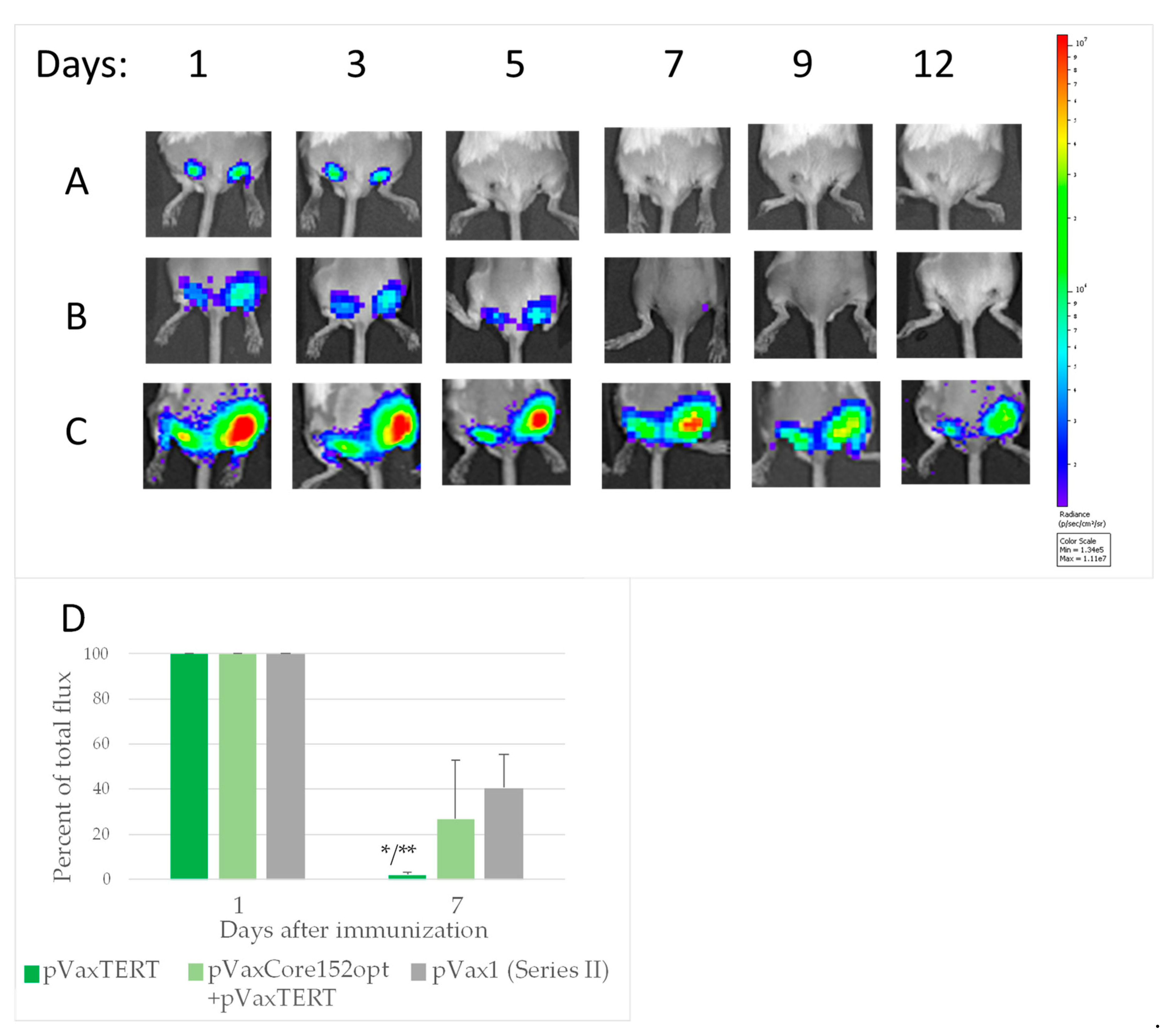
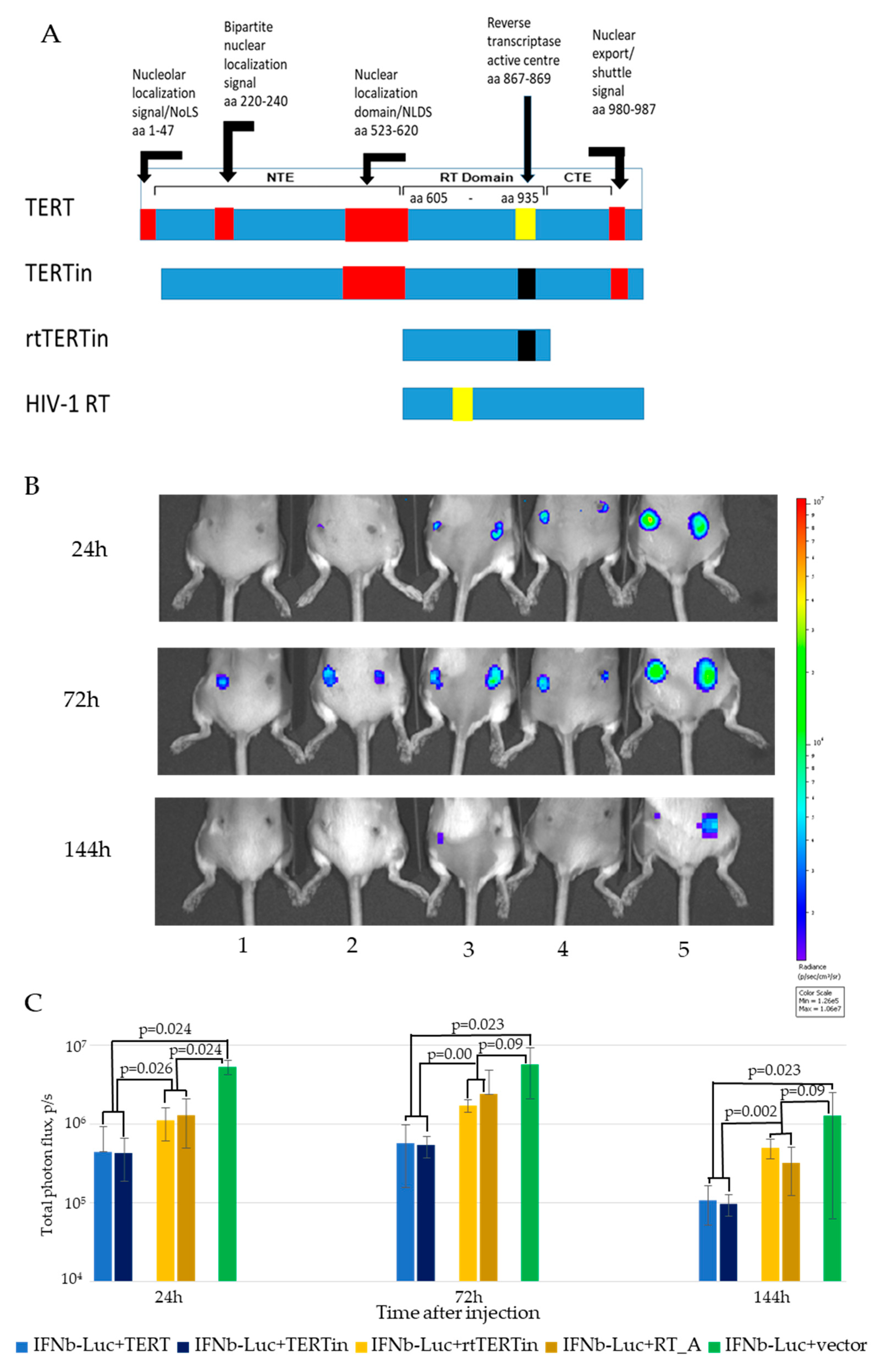
3.6. In Vivo Monitoring of Bioluminescence from the Sites of Injection of DNA Immunogens and Luciferase Reporter
3.7. In Vivo Assessment of the Effect of Co-Expression of Immunogens on the Initiation of Immune Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walker, C.M. Designing an HCV vaccine: A unique convergence of prevention and therapy? Curr. Opin. Virol. 2017, 23, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Vranjkovic, A.; Deonarine, F.; Kaka, S.; Angel, J.B.; Cooper, C.L.; Crawley, A.M. Direct-Acting Antiviral Treatment of HCV Infection Does Not Resolve the Dysfunction of Circulating CD8+ T-Cells in Advanced Liver Disease. Front. Immunol. 2019, 10, 1926. [Google Scholar] [CrossRef]
- Sacks-Davis, R.; Grebely, J.; Dore, G.J.; Osburn, W.; Cox, A.L.; Rice, T.M.; Spelman, T.; Bruneau, J.; Prins, M.; Kim, A.Y.; et al. Hepatitis C Virus Reinfection and Spontaneous Clearance of Reinfection—the InC 3 Study. J. Infect. Dis. 2015, 212, 1407–1419. [Google Scholar] [CrossRef]
- Falade-Nwulia, O.; Sulkowski, M.S.; Merkow, A.; Latkin, C.; Mehta, S.H. Understanding and addressing hepatitis C reinfection in the oral direct-acting antiviral era. J. Viral Hepat. 2018, 25, 220–227. [Google Scholar] [CrossRef]
- Swadling, L.; Capone, S.; Antrobus, R.D.; Brown, A.; Richardson, R.; Newell, E.W.; Halliday, J.; Kelly, C.; Bowen, D.; Fergusson, J.; et al. A human vaccine strategy based on chimpanzee adenoviral and MVA vectors that primes, boosts, and sustains functional HCV-specific T cell memory. Sci. Transl. Med. 2014, 6, 261ra153. [Google Scholar] [CrossRef]
- Capone, S.; Brown, A.; Hartnell, F.; Del Sorbo, M.; Traboni, C.; Vassilev, V.; Colloca, S.; Nicosia, A.; Cortese, R.; Folgori, A.; et al. Optimising T cell (re)boosting strategies for adenoviral and modified vaccinia Ankara vaccine regimens in humans. NPJ Vaccines 2020, 5, 94. [Google Scholar] [CrossRef]
- Pushparajah, D.; Jimenez, S.; Wong, S.; Alattas, H.; Nafissi, N.; Slavcev, R.A. Advances in gene-based vaccine platforms to address the COVID-19 pandemic. Adv. Drug Deliv. Rev. 2021, 170, 113–141. [Google Scholar] [CrossRef]
- Bailey, J.R.; Barnes, E.; Cox, A.L. Approaches, Progress, and Challenges to Hepatitis C Vaccine Development. Gastroenterology 2019, 156, 418–430. [Google Scholar] [CrossRef]
- Klade, C.S.; Wedemeyer, H.; Berg, T.; Hinrichsen, H.; Cholewinska, G.; Zeuzem, S.; Blum, H.; Buschle, M.; Jelovcan, S.; Buerger, V.; et al. Therapeutic Vaccination of Chronic Hepatitis C Nonresponder Patients with the Peptide Vaccine IC41. Gastroenterology 2008, 134, 1385–1395.e1. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Lajonchere, L.; Shoukry, N.H.; Grá, B.; Amador-Cañizares, Y.; Helle, F.; Bédard, N.; Guerra, I.; Drouin, C.; Dubuisson, J.; González-Horta, E.E.; et al. Immunogenicity of CIGB-230, a therapeutic DNA vaccine preparation, in HCV-chronically infected individuals in a Phase I clinical trial. J. Viral Hepat. 2009, 16, 156–167. [Google Scholar] [CrossRef]
- Pockros, P.; Jacobson Boyer, T. GI-5005 therapeutic vaccine plus PEG-IFN/ribavirin improves sustained virological response versus PEG-IFN/ ribavirin in prior non-responders with genotype-1 chronic HCV infection. Hepatology 2010, 52, 107A. [Google Scholar]
- Ray, R.B.; Ray, R. Hepatitis C virus core protein: Intriguing properties and functional relevance. FEMS Microbiol. Lett. 2001, 202, 149–156. [Google Scholar] [CrossRef]
- Ivanov, A.; Smirnova, O.; Petrushanko, I.; Ivanova, O.; Karpenko, I.; Alekseeva, E.; Sominskaya, I.; Makarov, A.; Bartosch, B.; Kochetkov, S.; et al. HCV Core Protein Uses Multiple Mechanisms to Induce Oxidative Stress in Human Hepatoma Huh7 Cells. Viruses 2015, 7, 2745–2770. [Google Scholar] [CrossRef] [PubMed]
- D’souza, S.; Lau, K.C.; Coffin, C.S.; Patel, T.R. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J. Gastroenterol. 2020, 26, 5759–5783. [Google Scholar] [CrossRef]
- Drane, D.; Maraskovsky, E.; Gibson, R.; Mitchell, S.; Barnden, M.; Moskwa, A.; Shaw, D.; Gervase, B.; Coates, S.; Houghton, M.; et al. Priming of CD4+ and CD8+ T cell responses using a HCV core ISCOMATRIXTM vaccine: A phase I study in healthy volunteers. Hum. Vaccin. 2009, 5, 151–157. [Google Scholar] [CrossRef]
- HitOmi, Y.; McDonnell, W.M.; Killeen, A.A.; Askari, F.K. Sequence analysis of the hepatitis C virus (HCV) core gene suggests the core protein as an appropriate target for HCV vaccine strategies. J. Viral Hepat. 1995, 2, 235–241. [Google Scholar] [CrossRef]
- Pavio, N.; Battaglia, S.; Boucreux, D.; Arnulf, B.; Sobesky, R.; Hermine, O.; Brechot, C. Hepatitis C virus core variants isolated from liver tumor but not from adjacent non-tumor tissue interact with Smad3 and inhibit the TGF-β pathway. Oncogene 2005, 24, 6119–6132. [Google Scholar] [CrossRef]
- Sansonno, D.; Cornacchiulo, V.; Racanelli, V.; Dammacco, F. In situ simultaneous detection of hepatitis C virus RNA and hepatitis C virus-related antigens in hepatocellular carcinoma. Cancer 1997, 80, 22–33. [Google Scholar] [CrossRef]
- Resham, S.; Saalim, M.; Manzoor, S.; Ahmad, H.; Bangash, T.A.; Latif, A.; Jaleel, S. Mechanistic study of interaction between IL-22 and HCV core protein in the development of hepatocellular carcinoma among liver transplant recipients. Microb. Pathog. 2020, 142, 104071. [Google Scholar] [CrossRef]
- Dash, S.; Aydin, Y.; Widmer, K.E.; Nayak, L. Hepatocellular Carcinoma Mechanisms Associated with Chronic HCV Infection and the Impact of Direct-Acting Antiviral Treatment. J. Hepatocell. Carcinoma 2020, 7, 45–76. [Google Scholar] [CrossRef]
- Conti, F.; Buonfiglioli, F.; Scuteri, A.; Crespi, C.; Bolondi, L.; Caraceni, P.; Foschi, F.G.; Lenzi, M.; Mazzella, G.; Verucchi, G.; et al. Early occurrence and recurrence of hepatocellular carcinoma in HCV-related cirrhosis treated with direct-acting antivirals. J. Hepatol. 2016, 65, 727–733. [Google Scholar] [CrossRef]
- Kanwal, F.; Kramer, J.; Asch, S.M.; Chayanupatkul, M.; Cao, Y.; El-Serag, H.B. Risk of Hepatocellular Cancer in HCV Patients Treated with Direct-Acting Antiviral Agents. Gastroenterology 2017, 153, 996–1005.e1. [Google Scholar] [CrossRef]
- Sandström, E.; Nilsson, C.; Hejdeman, B.; Bråve, A.; Bratt, G.; Robb, M.; Cox, J.; VanCott, T.; Marovich, M.; Stout, R.; et al. Broad Immunogenicity of a Multigene, Multiclade HIV-1 DNA Vaccine Boosted with Heterologous HIV-1 Recombinant Modified Vaccinia Virus Ankara. J. Infect. Dis. 2008, 198, 1482–1490. [Google Scholar] [CrossRef] [PubMed]
- Hobernik, D.; Bros, M. DNA Vaccines—How Far from Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef]
- Lim, M.; Badruddoza, A.Z.M.; Firdous, J.; Azad, M.; Mannan, A.; Al-Hilal, T.A.; Cho, C.-S.; Islam, M.A. Engineered Nanodelivery Systems to Improve DNA Vaccine Technologies. Pharmaceutics 2020, 12, 30. [Google Scholar] [CrossRef]
- Jiang, G.; Charoenvit, Y.; Moreno, A.; Baraceros, M.F.; Banania, G.; Richie, N.; Abot, S.; Ganeshan, H.; Fallarme, V.; Patterson, N.B.; et al. Induction of multi-antigen multi-stage immune responses against Plasmodium falciparum in rhesus monkeys, in the absence of antigen interference, with heterologous DNA prime/poxvirus boost immunization. Malar. J. 2007, 6, 135. [Google Scholar] [CrossRef]
- Bhardwaj, D.; Hora, B.; Singh, N.; Puri, S.K.; Lalitha, P.; Rupa, P.; Chauhan, V.S. Immunogenicity and protective efficacy of three DNA vaccines encoding pre-erythrocytic- and erythrocytic-stage antigens of Plasmodium cynomolgi in rhesus monkeys. FEMS Immunol. Med. Microbiol. 2002, 34, 33–43. [Google Scholar] [CrossRef]
- Churchyard, G.J.; Morgan, C.; Adams, E.; Hural, J.; Graham, B.S.; Moodie, Z.; Grove, D.; Gray, G.; Bekker, L.-G.; McElrath, M.J.; et al. A Phase IIA Randomized Clinical Trial of a Multiclade HIV-1 DNA Prime Followed by a Multiclade rAd5 HIV-1 Vaccine Boost in Healthy Adults (HVTN204). PLoS ONE 2011, 6, e21225. [Google Scholar] [CrossRef]
- Gummow, J.; Li, Y.; Yu, W.; Garrod, T.; Wijesundara, D.; Brennan, A.J.; Mullick, R.; Voskoboinik, I.; Grubor-Bauk, B.; Gowans, E.J. A Multiantigenic DNA Vaccine That Induces Broad Hepatitis C Virus-Specific T-Cell Responses in Mice. J. Virol. 2015, 89, 7991–8002. [Google Scholar] [CrossRef]
- Kjerrström, A.; Hinkula, J.; Engström, G.; Ovod, V.; Krohn, K.; Benthin, R.; Wahren, B. Interactions of Single and Combined Human Immunodeficiency Virus Type 1 (HIV-1) DNA Vaccines. Virology 2001, 284, 46–61. [Google Scholar] [CrossRef]
- Garrod, T.J.; Gargett, T.; Yu, W.; Major, L.; Burrell, C.J.; Wesselingh, S.; Suhrbier, A.; Grubor-Bauk, B.; Gowans, E.J. Loss of long term protection with the inclusion of HIV pol to a DNA vaccine encoding gag. Virus Res. 2014, 192, 25–33. [Google Scholar] [CrossRef]
- Kong, W.-P.; Huang, Y.; Yang, Z.-Y.; Chakrabarti, B.K.; Moodie, Z.; Nabel, G.J. Immunogenicity of Multiple Gene and Clade Human Immunodeficiency Virus Type 1 DNA Vaccines. J. Virol. 2003, 77, 12764–12772. [Google Scholar] [CrossRef]
- Bråve, A.; Ljungberg, K.; Boberg, A.; Rollman, E.; Isaguliants, M.; Lundgren, B.; Blomberg, P.; Hinkula, J.; Wahren, B. Multigene/Multisubtype HIV-1 Vaccine Induces Potent Cellular and Humoral Immune Responses by Needle-Free Intradermal Delivery. Mol. Ther. 2005, 12, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Mizukoshi, E.; Kaneko, S. Telomerase-Targeted Cancer Immunotherapy. Int. J. Mol. Sci. 2019, 20, 1823. [Google Scholar] [CrossRef]
- Jansons, J.; Bayurova, E.; Skrastina, D.; Kurlanda, A.; Fridrihsone, I.; Kostyushev, D.; Kostyusheva, A.; Artyuhov, A.; Dashinimaev, E.; Avdoshina, D.; et al. Expression of the Reverse Transcriptase Domain of Telomerase Reverse Transcriptase Induces Lytic Cellular Response in DNA-Immunized Mice and Limits Tumorigenic and Metastatic Potential of Murine Adenocarcinoma 4T1 Cells. Vaccines 2020, 8, 318. [Google Scholar] [CrossRef] [PubMed]
- Jansons, J.; Sominskaya, I.; Petrakova, N.; Starodubova, E.S.; Smirnova, O.A.; Alekseeva, E.; Bruvere, R.; Eliseeva, O.; Skrastina, D.; Kashuba, E.; et al. The Immunogenicity in Mice of HCV Core Delivered as DNA Is Modulated by Its Capacity to Induce Oxidative Stress and Oxidative Stress Response. Cells 2019, 8, 208. [Google Scholar] [CrossRef]
- Thalmensi, J.; Pliquet, E.; Liard, C.; Chamel, G.; Kreuz, C.; Bestetti, T.; Escande, M.; Kostrzak, A.; Pailhes-Jimenez, A.-S.; Bourges, E.; et al. A DNA telomerase vaccine for canine cancer immunotherapy. Oncotarget 2019, 10, 3361–3372. [Google Scholar] [CrossRef]
- Isaguliants, M.; Smirnova, O.; Ivanov, A.V.; Kilpelainen, A.; Kuzmenko, Y.; Petkov, S.; Latanova, A.; Krotova, O.; Engström, G.; Karpov, V.; et al. Oxidative stress induced by HIV-1 reverse transcriptase modulates the enzyme’s performance in gene immunization. Hum. Vaccin. Immunother. 2013, 9, 2111–2119. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Smirnova, O.A.; Ivanova, O.N.; Masalova, O.V.; Kochetkov, S.N.; Isaguliants, M.G. Hepatitis C Virus Proteins Activate NRF2/ARE Pathway by Distinct ROS-Dependent and Independent Mechanisms in HUH7 Cells. PLoS ONE 2011, 6, e24957. [Google Scholar] [CrossRef]
- Lin, R.; Génin, P.; Mamane, Y.; Hiscott, J. Selective DNA Binding and Association with the CREB Binding Protein Coactivator Contribute to Differential Activation of Alpha/Beta Interferon Genes by Interferon Regulatory Factors 3 and 7. Mol. Cell. Biol. 2000, 20, 6342–6353. [Google Scholar] [CrossRef]
- Isaguliants, M.G.; Iakimtchouk, K.; Petrakova, N.V.; Yermalovich, M.A.; Zuber, A.K.; Kashuba, V.I.; Belikov, S.V.; Andersson, S.; Kochetkov, S.N.; Klinman, D.M.; et al. Gene immunization may induce secondary antibodies reacting with DNA. Vaccine 2004, 22, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Latanova, A.A.; Petkov, S.; Kilpelainen, A.; Jansons, J.; Latyshev, O.E.; Kuzmenko, Y.V.; Hinkula, J.; Abakumov, M.A.; Valuev-Elliston, V.T.; Gomelsky, M.; et al. Codon optimization and improved delivery/immunization regimen enhance the immune response against wild-type and drug-resistant HIV-1 reverse transcriptase, preserving its Th2-polarity. Sci. Rep. 2018, 8, 8078. [Google Scholar] [CrossRef]
- Petkov, S.; Starodubova, E.; Latanova, A.; Kilpeläinen, A.; Latyshev, O.; Svirskis, S.; Wahren, B.; Chiodi, F.; Gordeychuk, I.; Isaguliants, M. DNA immunization site determines the level of gene expression and the magnitude, but not the type of the induced immune response. PLoS ONE 2018, 13, e0197902. [Google Scholar] [CrossRef]
- Zhu, W.; Chang, Y.; Wu, C.; Han, Q.; Pei, R.; Lu, M.; Chen, X. The Wild-Type Hepatitis C Virus Core Inhibits Initiation of Antigen-Specific T- and B-Cell Immune Responses in BALB/c Mice. Clin. Vaccine Immunol. 2010, 17, 1139–1147. [Google Scholar] [CrossRef]
- Zhu, W.; Wu, C.; Deng, W.; Pei, R.; Wang, Y.; Cao, L.; Qin, B.; Lu, M.; Chen, X. Inhibition of the HCV Core Protein on the Immune Response to HBV Surface Antigen and on HBV Gene Expression and Replication In Vivo. PLoS ONE 2012, 7, e45146. [Google Scholar] [CrossRef]
- Limberis, M.P.; Bell, C.L.; Wilson, J.M. Identification of the murine firefly luciferase-specific CD8 T-cell epitopes. Gene Ther. 2009, 16, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Baklaushev, V.P.; Kilpeläinen, A.; Petkov, S.; Abakumov, M.A.; Grinenko, N.F.; Yusubalieva, G.M.; Latanova, A.A.; Gubskiy, I.L.; Zabozlaev, F.G.; Starodubova, E.S.; et al. Luciferase Expression Allows Bioluminescence Imaging but Imposes Limitations on the Orthotopic Mouse (4T1) Model of Breast Cancer. Sci. Rep. 2017, 7, 7715. [Google Scholar] [CrossRef]
- Sominskaya, I.; Jansons, J.; Dovbenko, A.; Petrakova, N.; Lieknina, I.; Mihailova, M.; Latyshev, O.; Eliseeva, O.; Stahovska, I.; Akopjana, I.; et al. Comparative Immunogenicity in Rabbits of the Polypeptides Encoded by the 5′ Terminus of Hepatitis C Virus RNA. J. Immunol. Res. 2015, 2015, 1–12. [Google Scholar] [CrossRef]
- Isaguliants, M.G.; Petrakova, N.V.; Kashuba, E.V.; Suzdaltzeva, Y.G.; Belikov, S.V.; Mokhonov, V.V.; Prilipov, A.G.; Matskova, L.; Smirnova, I.S.; Jolivet-Reynaud, C.; et al. Immunization with hepatitis C virus core gene triggers potent T-cell response, but affects CD4+ T-cells. Vaccine 2004, 22, 1656–1665. [Google Scholar] [CrossRef]
- Dehghani, B.; Hashempour, T.; Hasanshahi, Z.; Moayedi, J. Bioinformatics Analysis of Domain 1 of HCV-Core Protein: Iran. Int. J. Pept. Res. Ther. 2020, 26, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Schwanke, H.; Stempel, M.; Brinkmann, M.M. Of Keeping and Tipping the Balance: Host Regulation and Viral Modulation of IRF3-Dependent IFNB1 Expression. Viruses 2020, 12, 733. [Google Scholar] [CrossRef]
- Çevik, R.E.; Cesarec, M.; Da Silva Filipe, A.; Licastro, D.; McLauchlan, J.; Marcello, A. Hepatitis C Virus NS5A Targets Nucleosome Assembly Protein NAP1L1 To Control the Innate Cellular Response. J. Virol. 2017, 91, e00880-17. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.J.; Sparrer, K.M.J.; van Gent, M.; Lässig, C.; Huang, T.; Osterrieder, N.; Hopfner, K.-P.; Gack, M.U. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat. Immunol. 2018, 19, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-L.; Cheng, J.-S.; Shu, C.-W.; Lai, K.-H.; Chan, H.-H.; Wu, C.-C.; Wu, J.-M.; Hsu, P.-I.; Chung, R.T.; Chang, T.-H. Asunaprevir Evokes Hepatocytes Innate Immunity to Restrict the Replication of Hepatitis C and Dengue Virus. Front. Microbiol. 2017, 8, 668. [Google Scholar] [CrossRef]
- Buckley, S.M.K.; Delhove, J.M.K.M.; Perocheau, D.P.; Karda, R.; Rahim, A.A.; Howe, S.J.; Ward, N.J.; Birrell, M.A.; Belvisi, M.G.; Arbuthnot, P.; et al. In vivo bioimaging with tissue-specific transcription factor activated luciferase reporters. Sci. Rep. 2015, 5, 11842. [Google Scholar] [CrossRef]
- Chung, J.; Khadka, P.; Chung, I.K. Nuclear import of hTERT requires a bipartite nuclear localization signal and Akt-mediated phosphorylation. J. Cell Sci. 2012, 125, 2684–2697. [Google Scholar] [CrossRef]
- Etheridge, K.T.; Banik, S.S.R.; Armbruster, B.N.; Zhu, Y.; Terns, R.M.; Terns, M.P.; Counter, C.M. The Nucleolar Localization Domain of the Catalytic Subunit of Human Telomerase. J. Biol. Chem. 2002, 277, 24764–24770. [Google Scholar] [CrossRef]
- Jacobo-Molina, A.; Ding, J.; Nanni, R.G.; Clark, A.D.; Lu, X.; Tantillo, C.; Williams, R.L.; Kamer, G.; Ferris, A.L.; Clark, P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 1993, 90, 6320–6324. [Google Scholar] [CrossRef]
- Kovalenko, O.A.; Caron, M.J.; Ulema, P.; Medrano, C.; Thomas, A.P.; Kimura, M.; Bonini, M.G.; Herbig, U.; Santos, J.H. A mutant telomerase defective in nuclear-cytoplasmic shuttling fails to immortalize cells and is associated with mitochondrial dysfunction. Aging Cell 2010, 9, 203–219. [Google Scholar] [CrossRef]
- Alekseeva, E.; Sominskaya, I.; Skrastina, D.; Egorova, I.; Starodubova, E.; Kushners, E.; Mihailova, M.; Petrakova, N.; Bruvere, R.; Kozlovskaya, T.; et al. Enhancement of the expression of HCV core gene does not enhance core-specific immune response in DNA immunization: Advantages of the heterologous DNA prime, protein boost immunization regimen. Genet. Vaccines Ther. 2009, 7, 7. [Google Scholar] [CrossRef]
- Vonderheide, R.H. Prospects and challenges of building a cancer vaccine targeting telomerase. Biochimie 2008, 90, 173–180. [Google Scholar] [CrossRef]
- in der Stroth, L.; Tharehalli, U.; Günes, C.; Lechel, A. Telomeres and Telomerase in the Development of Liver Cancer. Cancers 2020, 12, 2048. [Google Scholar] [CrossRef]
- Ningarhari, M.; Caruso, S.; Hirsch, T.Z.; Bayard, Q.; Franconi, A.; Védie, A.-L.; Noblet, B.; Blanc, J.-F.; Amaddeo, G.; Ganne, N.; et al. Telomere length is key to hepatocellular carcinoma diversity and telomerase addiction is an actionable therapeutic target. J. Hepatol. 2020, S0168–8278, 33846. [Google Scholar] [CrossRef]
- Negrini, S.; De Palma, R.; Filaci, G. Anti-Cancer Immunotherapies Targeting Telomerase. Cancers 2020, 12, 2260. [Google Scholar] [CrossRef]
- Yan, J.; Pankhong, P.; Shin, T.H.; Obeng-Adjei, N.; Morrow, M.P.; Walters, J.N.; Khan, A.S.; Sardesai, N.Y.; Weiner, D.B. Highly Optimized DNA Vaccine Targeting Human Telomerase Reverse Transcriptase Stimulates Potent Antitumor Immunity. Cancer Immunol. Res. 2013, 1, 179–189. [Google Scholar] [CrossRef]
- Thalmensi, J.; Pliquet, E.; Liard, C.; Escande, M.; Bestetti, T.; Julithe, M.; Kostrzak, A.; Pailhes-Jimenez, A.-S.; Bourges, E.; Loustau, M.; et al. Anticancer DNA vaccine based on human telomerase reverse transcriptase generates a strong and specific T cell immune response. Oncoimmunology 2016, 5, e1083670. [Google Scholar] [CrossRef]
- Teixeira, L.; Medioni, J.; Garibal, J.; Adotevi, O.; Doucet, L.; Durey, M.-A.D.; Ghrieb, Z.; Kiladjian, J.-J.; Brizard, M.; Laheurte, C.; et al. A First-in-Human Phase I Study of INVAC-1, an Optimized Human Telomerase DNA Vaccine in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2020, 26, 588–597. [Google Scholar] [CrossRef]
- Slingluff, C.L. Building on the Promise of Cancer Vaccines for Solid Tumors. Clin. Cancer Res. 2020, 26, 529–531. [Google Scholar] [CrossRef]
- Jansons, J.; Bayurova, E.; Skrastina, D.; Kurlanda, A.; Breiksa, A.; Gebrila, S.; Gordeychuk, I.I.M. DNA immunization with reverse transcriptase domain of rat TERT protects mice against challenge with murine adenocarcinoma cells expressing the immunogen. In Proceedings of the “Perspective technologies in vaccination and immunotherapy”, Moscow, Russia, 27–29 October 2020; Abstract Nr TIII. p. 32. [Google Scholar]
- Wang, Z.; Deng, Z.; Dahmane, N.; Tsai, K.; Wang, P.; Williams, D.R.; Kossenkov, A.V.; Showe, L.C.; Zhang, R.; Huang, Q.; et al. Telomeric repeat-containing RNA (TERRA) constitutes a nucleoprotein component of extracellular inflammatory exosomes. Proc. Natl. Acad. Sci. USA 2015, 112, E6293–E6300. [Google Scholar] [CrossRef]
- Wang, Z.; Lieberman, P.M. The crosstalk of telomere dysfunction and inflammation through cell-free TERRA containing exosomes. RNA Biol. 2016, 13, 690–695. [Google Scholar] [CrossRef]
- Toubiana, S.; Selig, S. DNA: RNA hybrids at telomeres—when it is better to be out of the (R) loop. FEBS J. 2018, 285, 2552–2566. [Google Scholar] [CrossRef]
- Rigby, R.E.; Webb, L.M.; Mackenzie, K.J.; Li, Y.; Leitch, A.; Reijns, M.A.M.; Lundie, R.J.; Revuelta, A.; Davidson, D.J.; Diebold, S.; et al. RNA: DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J. 2014, 33, 542–558. [Google Scholar] [CrossRef]
- Bakari, M.; Aboud, S.; Nilsson, C.; Francis, J.; Buma, D.; Moshiro, C.; Aris, E.A.; Lyamuya, E.F.; Janabi, M.; Godoy-Ramirez, K.; et al. Broad and potent immune responses to a low dose intradermal HIV-1 DNA boosted with HIV-1 recombinant MVA among healthy adults in Tanzania. Vaccine 2011, 29, 8417–8428. [Google Scholar] [CrossRef] [PubMed]
- Munseri, P.J.; Kroidl, A.; Nilsson, C.; Joachim, A.; Geldmacher, C.; Mann, P.; Moshiro, C.; Aboud, S.; Lyamuya, E.; Maboko, L.; et al. Priming with a Simplified Intradermal HIV-1 DNA Vaccine Regimen followed by Boosting with Recombinant HIV-1 MVA Vaccine Is Safe and Immunogenic: A Phase IIa Randomized Clinical Trial. PLoS ONE 2015, 10, e0119629. [Google Scholar] [CrossRef]
- Kallas, E.G.; Grunenberg, N.A.; Yu, C.; Manso, B.; Pantaleo, G.; Casapia, M.; Baden, L.R.; Valencia, J.; Sobieszczyk, M.; Van Tieu, H.; et al. Antigenic competition in CD4 + T cell responses in a randomized, multicenter, double-blind clinical HIV vaccine trial. Sci. Transl. Med. 2019, 11, eaaw1673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, C.; Feng, L.; Xiao, L.; Chen, L. Enhancement of Gag-Specific but Reduction of Env- and Pol-Specific CD8 + T Cell Responses by Simian Immunodeficiency Virus Nonstructural Proteins In Mice. AIDS Res. Hum. Retrovir. 2012, 28, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Van Montfort, T.; Sanders, R.W. Optimizing cellular immunity against HIV-1 Gag and preventing suppression by HIV-1 gp120. Expert Rev. Vaccines 2012, 11, 1175–1177. [Google Scholar] [CrossRef] [PubMed]
- Sedegah, M.; Charoenvit, Y.; Minh, L.; Belmonte, M.; Majam, V.; Abot, S.; Ganeshan, H.; Kumar, S.; Bacon, D.; Stowers, A.; et al. Reduced immunogenicity of DNA vaccine plasmids in mixtures. Gene Ther. 2004, 11, 448–456. [Google Scholar] [CrossRef]
- Bacon, D.J.; Sedegah, M. Reduced Production of RNA Transcripts from Individual DNA Plasmids Given in a Multivalent DNA Vaccine Formula. Hum. Vaccin. 2007, 3, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, H.D.M.; West, S.C.; Beattie, T.L. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010, 38, 5609–5622. [Google Scholar] [CrossRef]
- Nguyen, K.T.T.T.; Wong, J.M.Y. Telomerase Biogenesis and Activities from the Perspective of Its Direct Interacting Partners. Cancers 2020, 12, 1679. [Google Scholar] [CrossRef]
- Huang, J.J.; Lin, M.C.; Bai, Y.X.; Jing, D.D.; Wong, B.C.Y.; Han, S.W.; Lin, J.; Xu, B.; Huang, C.-F.; Kung, H.-F. Ectopic expression of a COOH-terminal fragment of the human telomerase reverse transcriptase leads to telomere dysfunction and reduction of growth and tumorigenicity in HeLa cells. Cancer Res. 2002, 62, 3226–3232. [Google Scholar]
- Salvetti, A.; Couté, Y.; Epstein, A.; Arata, L.; Kraut, A.; Navratil, V.; Bouvet, P.; Greco, A. Nuclear Functions of Nucleolin through Global Proteomics and Interactomic Approaches. J. Proteome Res. 2016, 15, 1659–1669. [Google Scholar] [CrossRef]
- Tajrishi, M.M.; Tuteja, R.; Tuteja, N. Nucleolin. Commun. Integr. Biol. 2011, 4, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Prattes; Lo; Bergler; Stanley Shaping the Nascent Ribosome: AAA-ATPases in Eukaryotic Ribosome Biogenesis. Biomolecules 2019, 9, 715. [CrossRef] [PubMed]
- Fujiwara, Y.; Fujiwara, K.; Goda, N.; Iwaya, N.; Tenno, T.; Shirakawa, M.; Hiroaki, H. Structure and Function of the N-terminal Nucleolin Binding Domain of Nuclear Valosin-containing Protein-like 2 (NVL2) Harboring a Nucleolar Localization Signal. J. Biol. Chem. 2011, 286, 21732–21741. [Google Scholar] [CrossRef]
- Nagahama, M.; Hara, Y.; Seki, A.; Yamazoe, T.; Kawate, Y.; Shinohara, T.; Hatsuzawa, K.; Tani, K.; Tagaya, M. NVL2 Is a Nucleolar AAA-ATPase that Interacts with Ribosomal Protein L5 through Its Nucleolar Localization Sequence. Mol. Biol. Cell 2004, 15, 5712–5723. [Google Scholar] [CrossRef] [PubMed]
- Dreyfuss, G.; Kim, V.N.; Kataoka, N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002, 3, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Khattar, E.; Kumar, P.; Liu, C.Y.; Akıncılar, S.C.; Raju, A.; Lakshmanan, M.; Maury, J.J.P.; Qiang, Y.; Li, S.; Tan, E.Y.; et al. Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J. Clin. Invest. 2016, 126, 4045–4060. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.A.; Iwasaki, A.; Barber, B.H.; Robinson, H.L. Differential dependence on target site tissue for gene gun and intramuscular DNA immunizations. J. Immunol. 1997, 158, 4529–4532. [Google Scholar]
- Kang, S.-M.; Kim, S.-J.; Kim, J.-H.; Lee, W.; Kim, G.-W.; Lee, K.-H.; Choi, K.-Y.; Oh, J.-W. Interaction of hepatitis C virus core protein with Hsp60 triggers the production of reactive oxygen species and enhances TNF-α-mediated apoptosis. Cancer Lett. 2009, 279, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Ware, C.F.; Lai, M.M.C. Hepatitis C Virus Core Protein Enhances FADD-Mediated Apoptosis and Suppresses TRADD Signaling of Tumor Necrosis Factor Receptor. Virology 2001, 283, 178–187. [Google Scholar] [CrossRef][Green Version]
- Mohd-Ismail, N.K.; Deng, L.; Sukumaran, S.K.; Yu, V.C.; Hotta, H.; Tan, Y.-J. The hepatitis C virus core protein contains a BH3 domain that regulates apoptosis through specific interaction with human Mcl-1. J. Virol. 2009, 83, 9993–10006. [Google Scholar] [CrossRef] [PubMed]
- Realdon, S.; Gerotto, M.; Dal Pero, F.; Marin, O.; Granato, A.; Basso, G.; Muraca, M.; Alberti, A. Proapoptotic effect of hepatitis C virus CORE protein in transiently transfected cells is enhanced by nuclear localization and is dependent on PKR activation. J. Hepatol. 2004, 40, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Ferrell, J.E. Apoptosis propagates through the cytoplasm as trigger waves. Science 2018, 361, 607–612. [Google Scholar] [CrossRef]


| Group | Nn Mice | Prime | Boost | ||
|---|---|---|---|---|---|
| Plasmids | Dose per Injection/ Number of Injections | Plasmids | Dose per Injection/ Number of Injections | ||
| Series I I-1 | 5 | pVaxCore191v | 20 µg × 2 | pVaxCore191v + pVaxLuc2 * | 15 µg × 2 |
| I-2 | 5 | pVaxCore152opt | 20 µg × 2 | pVaxCore152opt + pVaxLuc2 * | 15 µg × 2 |
| I-3 | 5 | pVax1 | 20 µg × 2 | pVaxTERT + pVaxLuc2 * | 15 µg × 2 |
| Series II II-1 | 5 | pVaxTERT | 20 µg × 2 | pVaxTERT + pVaxLuc2 * | 15 µg × 2 |
| II-2 | 5 | pVaxTERT + I have pVaxCore152opt | 10 µg each × 2 | pVaxTERT + pVaxCore152opt + pVaxLuc2 * | 7.5 µg each × 2 |
| II-3 | 5 | pVax1 | 20 µg × 2 | pVax1 + pVaxLuc2 * | 15 µg × 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jansons, J.; Skrastina, D.; Kurlanda, A.; Petkov, S.; Avdoshina, D.; Kuzmenko, Y.; Krotova, O.; Trofimova, O.; Gordeychuk, I.; Sominskaya, I.; et al. Reciprocal Inhibition of Immunogenic Performance in Mice of Two Potent DNA Immunogens Targeting HCV-Related Liver Cancer. Microorganisms 2021, 9, 1073. https://doi.org/10.3390/microorganisms9051073
Jansons J, Skrastina D, Kurlanda A, Petkov S, Avdoshina D, Kuzmenko Y, Krotova O, Trofimova O, Gordeychuk I, Sominskaya I, et al. Reciprocal Inhibition of Immunogenic Performance in Mice of Two Potent DNA Immunogens Targeting HCV-Related Liver Cancer. Microorganisms. 2021; 9(5):1073. https://doi.org/10.3390/microorganisms9051073
Chicago/Turabian StyleJansons, Juris, Dace Skrastina, Alisa Kurlanda, Stefan Petkov, Darya Avdoshina, Yulia Kuzmenko, Olga Krotova, Olga Trofimova, Ilya Gordeychuk, Irina Sominskaya, and et al. 2021. "Reciprocal Inhibition of Immunogenic Performance in Mice of Two Potent DNA Immunogens Targeting HCV-Related Liver Cancer" Microorganisms 9, no. 5: 1073. https://doi.org/10.3390/microorganisms9051073
APA StyleJansons, J., Skrastina, D., Kurlanda, A., Petkov, S., Avdoshina, D., Kuzmenko, Y., Krotova, O., Trofimova, O., Gordeychuk, I., Sominskaya, I., & Isaguliants, M. (2021). Reciprocal Inhibition of Immunogenic Performance in Mice of Two Potent DNA Immunogens Targeting HCV-Related Liver Cancer. Microorganisms, 9(5), 1073. https://doi.org/10.3390/microorganisms9051073







