Different csrA Expression Levels in C versus K-12 E. coli Strains Affect Biofilm Formation and Impact the Regulatory Mechanism Presided by the CsrB and CsrC Small RNAs
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria, Plasmids, and Growth Media
2.2. Gene Expression Determination
2.3. Luciferase Activity Assay
2.4. PNAG Detection
2.5. Western Blotting
2.6. Statistical Analysis
3. Results
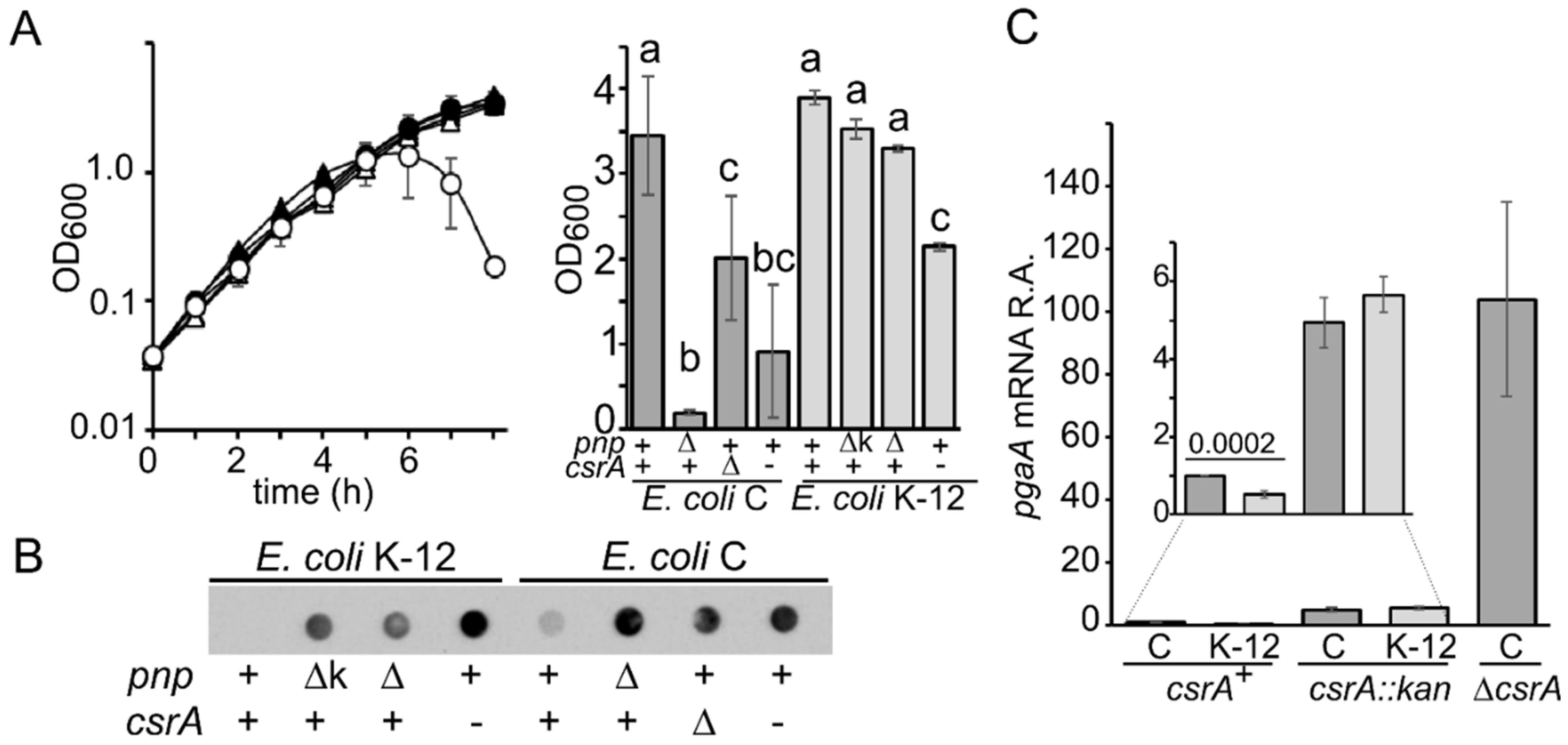
3.1. E. coli C Produces More PNAG than E. coli K-12
3.2. CsrA-Dependent Regulation of pgaABCD Operon Is More Stringent in E. coli K-12 than in E. coli C
3.3. PNPase-Dependent Regulation of pga Operon Is More Stringent in E. coli C than in E. coli K-12
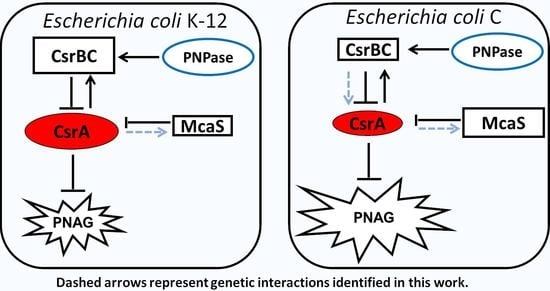
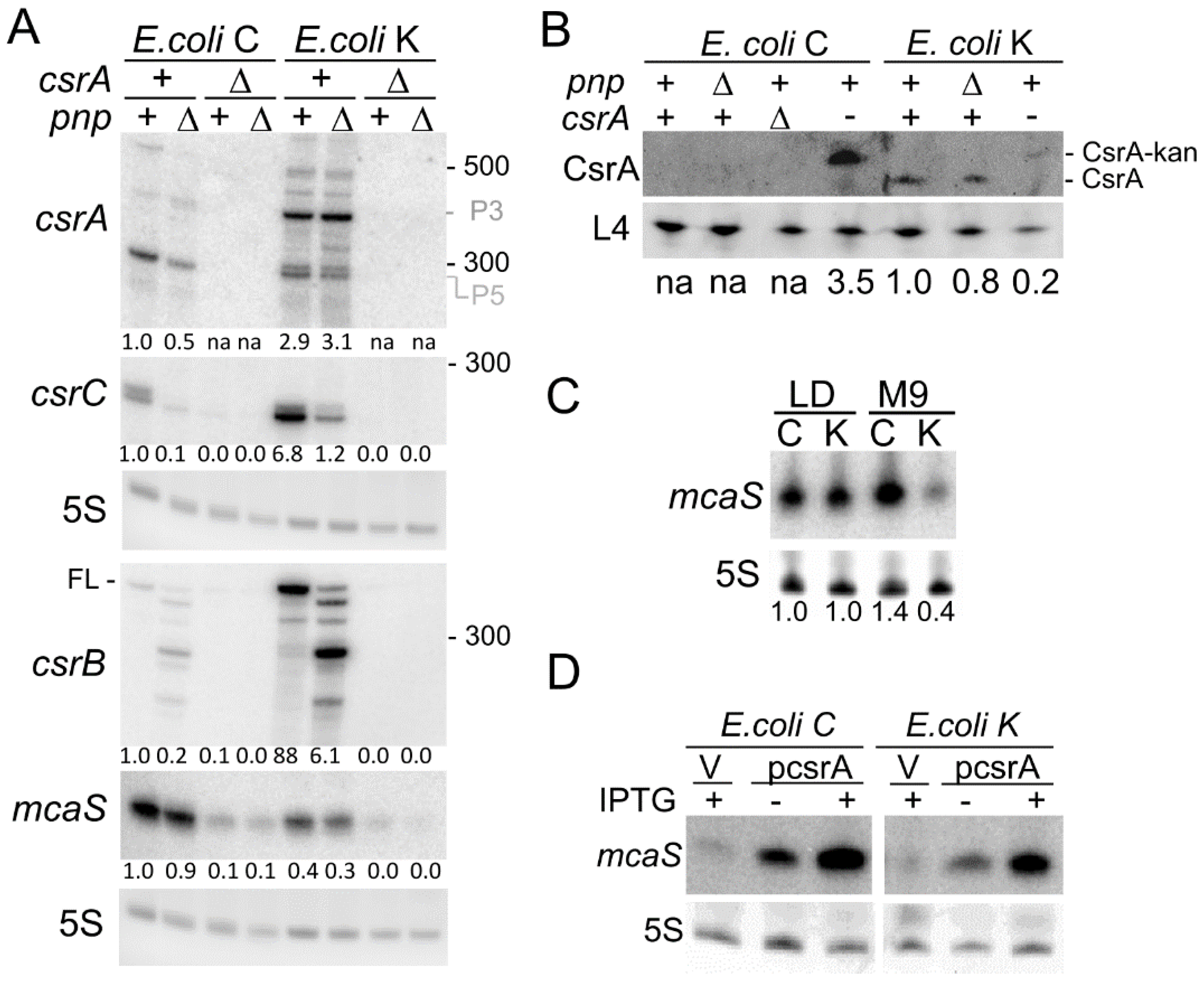
3.4. Expression Profile of the csrA Gene and of sRNAs Regulating CsrA Activity in E. coli C and K-12
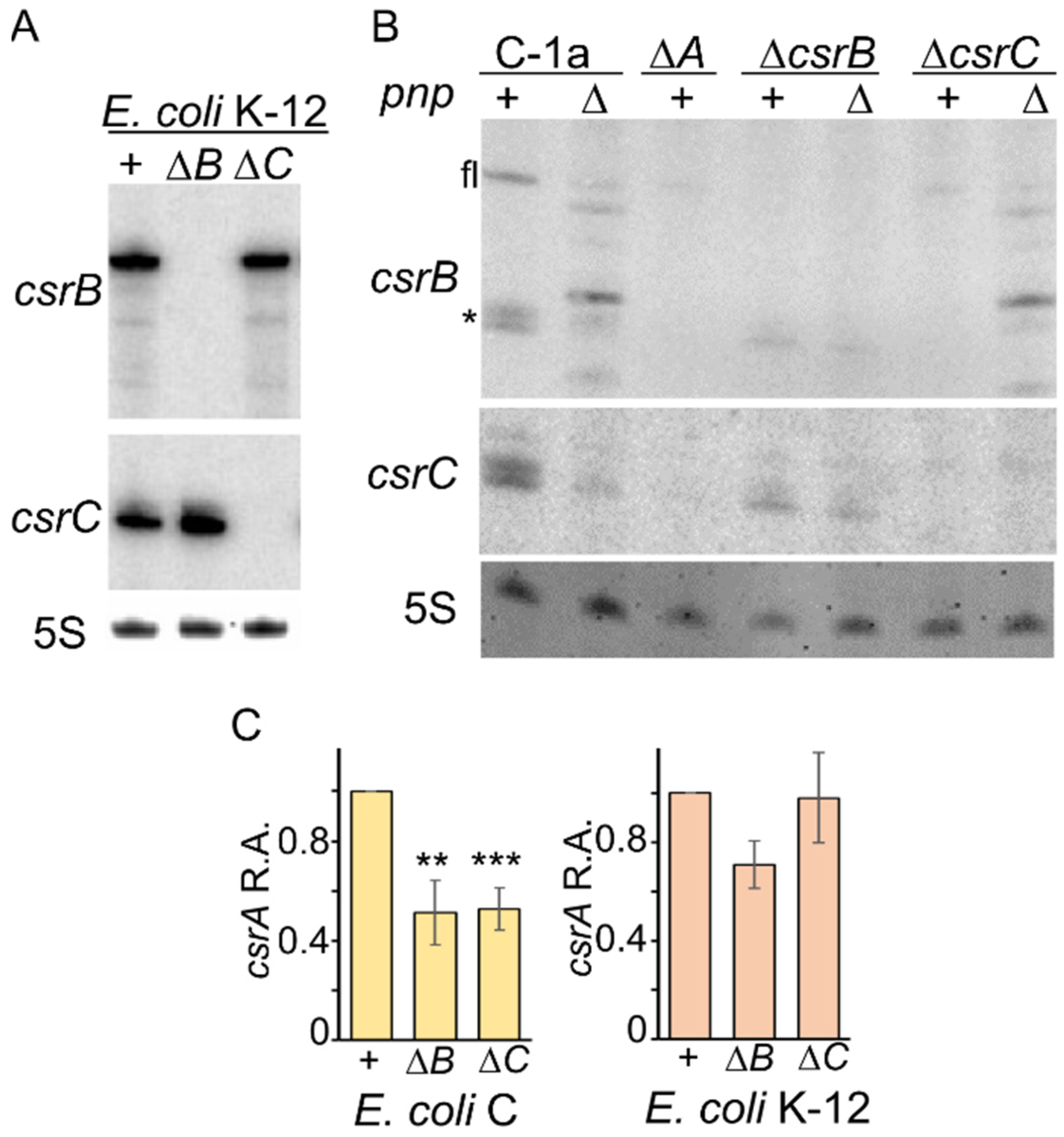
3.5. Compensatory Regulation of CsrB and CsrC Does Not Occur in E. coli C
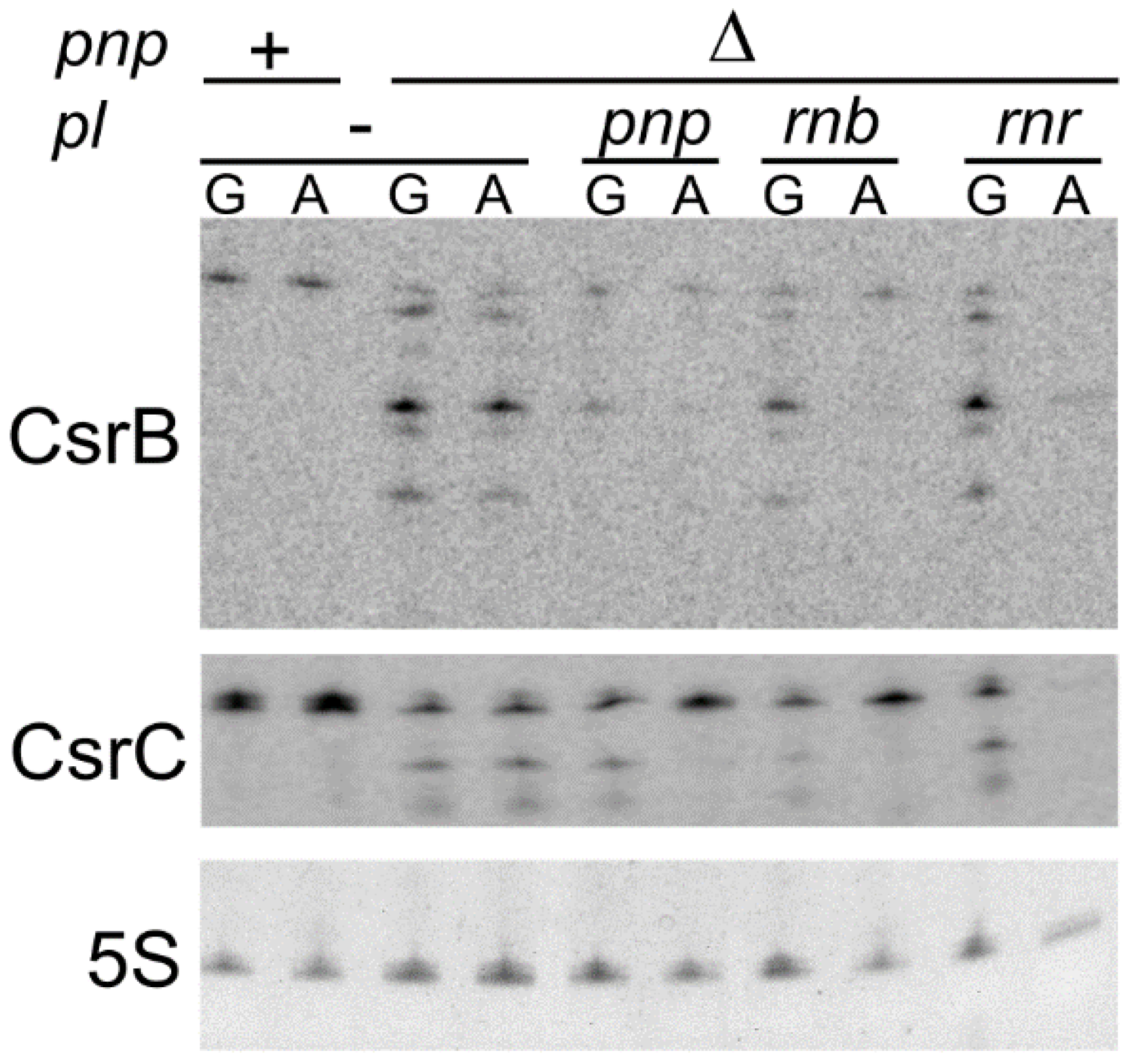
3.6. Ectopically Expressed RNase II Restores CsrB and CsrC Production in C-1a Δpnp
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Romeo, T.; Vakulskas, C.A.; Babitzke, P. Post-transcriptional regulation on a global scale: Form and function of Csr/Rsm systems. Environ. Microbiol. 2013, 15, 313–324. [Google Scholar] [CrossRef]
- Romeo, T. Global regulation by the small RNA-binding protein CsrA and the non- coding RNA molecule CsrB. Mol. Microbiol. 1998, 29, 1321–1330. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.K.; Baker, C.S.; Romeo, T.; Babitzke, P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 2005, 11, 1579–1587. [Google Scholar] [CrossRef]
- Baker, C.S.; Eöry, L.A.; Yakhnin, H.; Mercante, J.; Romeo, T.; Babitzke, P. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J. Bacteriol. 2007, 189, 5472–5481. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Gui, G.; Wei, B.; Preston, J.F.; Oakford, L.; Yüksel, Ü.; Giedroc, D.P.; Romeo, T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997, 272, 17502–17510. [Google Scholar] [CrossRef] [PubMed]
- Weilbacher, T.; Suzuki, K.; Dubey, A.K.; Wang, X.; Gudapaty, S.; Morozov, I.; Baker, C.S.; Georgellis, D.; Babitzke, P.; Romeo, T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 2003, 48, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, M.G.; Thomason, M.K.; Havelund, J.; Valentin-Hansen, P.; Storz, G. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev. 2013, 27, 1132–1145. [Google Scholar] [CrossRef]
- Suzuki, K.; Wang, X.; Weilbacher, T.; Pernestig, A.K.; Melefors, Ö.; Georgellis, D.; Babitzke, P.; Romeo, T. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 2002, 184, 5130–5140. [Google Scholar] [CrossRef]
- Adamson, D.N.; Lim, H.N. Rapid and robust signaling in the CsrA cascade via RNA-protein interactions and feedback regulation. Proc. Natl. Acad. Sci. USA 2013, 110, 13120–13125. [Google Scholar] [CrossRef]
- Gudapaty, S.; Suzuki, K.; Wang, X.; Babitzke, P.; Romeo, T. Regulatory interactions of Csr components: The RNA binding protein CsrA activates csrB transcription in Escherichia coli. J. Bacteriol. 2001, 183, 6017–6027. [Google Scholar] [CrossRef]
- Camacho, M.I.; Alvarez, A.F.; Chavez, R.G.; Romeo, T.; Merino, E.; Georgellis, D. Effects of the global regulator CsrA on the BarA/UvrY two-Component signaling system. J. Bacteriol. 2015, 197, 983–991. [Google Scholar] [CrossRef]
- Suzuki, K.; Babitzke, P.; Kushner, S.R.; Romeo, T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006, 20, 2605–2617. [Google Scholar] [CrossRef]
- Yakhnin, H.; Yakhnin, A.V.; Baker, C.S.; Sineva, E.; Berezin, I.; Romeo, T.; Babitzke, P. Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by Eσ70 and EσS, and indirect transcriptional activation by CsrA. Mol. Microbiol. 2011, 81, 689–704. [Google Scholar] [CrossRef]
- Król, J.E.; Hall, D.C.; Balashov, S.; Pastor, S.; Sibert, J.; McCaffrey, J.; Lang, S.; Ehrlich, R.L.; Earl, J.; Mell, J.C.; et al. Genome rearrangements induce biofilm formation in Escherichia coli C - An old model organism with a new application in biofilm research. BMC Genomics 2019, 20. [Google Scholar] [CrossRef]
- Romeo, T.; Gong, M.; Liu, M.Y.; Brun-Zinkernagel, A.M. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 1993, 175, 4744–4755. [Google Scholar] [CrossRef]
- Conway, T.; Creecy, J.P.; Maddox, S.M.; Grissom, J.E.; Conkle, T.L.; Shadid, T.M.; Teramoto, J.; Miguel, P.S.; Shimada, T.; Ishihama, A.; et al. Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. mBio 2014, 5. [Google Scholar] [CrossRef]
- Pourciau, C.; Lai, Y.J.; Gorelik, M.; Babitzke, P.; Romeo, T. Diverse Mechanisms and Circuitry for Global Regulation by the RNA-Binding Protein CsrA. Front. Microbiol. 2020, 11, 601352. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dubey, A.K.; Suzuki, K.; Baker, C.S.; Babitzke, P.; Romeo, T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 2005, 56, 1648–1663. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Campoccia, D.; Ravaioli, S.; Montanaro, L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell. Infect. Microbiol. 2015, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Roux, D.; Cywes-Bentley, C.; Zhang, Y.F.; Pons, S.; Konkol, M.; Kearns, D.B.; Little, D.J.; Howell, P.L.; Skurnik, D.; Pier, G.B. Identification of Poly-N-acetylglucosamine as a major polysaccharide component of the Bacillus subtilis biofilm matrix. J. Biol. Chem. 2015, 290, 19261–19272. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, G.B.; Howell, P.L. The Matrix Revisited: Opening night for the Pel polysaccharide across eubacterial kingdoms. Microbiol. Insights 2021, 14, 117863612098858. [Google Scholar] [CrossRef]
- Wang, X.; Preston, J.F.; Romeo, T. The pgaABCD Locus of Escherichia coli Promotes the Synthesis of a Polysaccharide Adhesin Required for Biofilm Formation. J. Bacteriol. 2004, 186, 2724–2734. [Google Scholar] [CrossRef]
- Goller, C.; Wang, X.; Itoh, Y.; Romeo, T. The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesin poly-β-1,6-N-acetyl-D-glucosamine. J. Bacteriol. 2006, 188, 8022–8032. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Bossi, N.; Schwartz, A.; Guillemardet, B.; D’Heygère, F.; Bossi, L.; Boudvillain, M. RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev. 2014, 28, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Carzaniga, T.; Antoniani, D.; Dehò, G.; Briani, F.; Landini, P. The RNA processing enzyme polynucleotide phosphorylase negatively controls biofilm formation by repressing poly-N-acetylglucosamine (PNAG) production in Escherichia coli C. BMC Microbiol. 2012, 12, 270. [Google Scholar] [CrossRef] [PubMed]
- Briani, F.; Carzaniga, T.; Dehò, G. Regulation and functions of bacterial PNPase. Wiley Interdiscip. Rev. RNA 2016, 7, 241–258. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Carzaniga, T.; Sbarufatti, G.; Briani, F.; Dehò, G. Polynucleotide phosphorylase is implicated in homologous recombination and DNA repair in Escherichia coli. BMC Microbiol. 2017, 17, 81. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, I.; Bertani, G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J. Gen. Microbiol. 1965, 40, 365–376. [Google Scholar] [CrossRef][Green Version]
- Regonesi, M.E.; Del Favero, M.; Basilico, F.; Briani, F.; Benazzi, L.; Tortora, P.; Mauri, P.; Dehò, G. Analysis of the Escherichia coli RNA degradosome composition by a proteomic approach. Biochimie 2006, 88, 151–161. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef]
- Blattner, F.R. The Complete Genome Sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef]
- Lessl, M.; Balzer, D.; Lurz, R.; Waters, V.L.; Guiney, D.G.; Lanka, E. Dissection of IncP conjugative plasmid transfer: Definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 1992, 174, 2493–2500. [Google Scholar] [CrossRef]
- Jaspers, M.C.M.; Suske, W.A.; Schmid, A.; Goslings, D.A.M.; Kohler, H.P.E.; Van Der Meer, J.R. HbpR, a new member of the Xylr/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2- hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J. Bacteriol. 2000, 182, 405–417. [Google Scholar] [CrossRef]
- Wall, J.D.; Harriman, P.D. Phage P1 mutants with altered transducing abilities for Escherichia coli. Virology 1974, 59, 532–544. [Google Scholar] [CrossRef]
- Briani, F.; Del Favero, M.; Capizzuto, R.; Consonni, C.; Zangrossi, S.; Greco, C.; De Gioia, L.; Tortora, P.; Dehò, G. Genetic analysis of polynucleotide phosphorylase structure and functions. Biochimie 2007, 89, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Briani, F.; Curti, S.; Rossi, F.; Carzaniga, T.; Mauri, P.; Dehò, G. Polynucleotide phosphorylase hinders mRNA degradation upon ribosomal protein S1 overexpression in Escherichia coli. RNA 2008, 14, 2417–2429. [Google Scholar] [CrossRef][Green Version]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Briani, F.; Zangrossi, S.; Ghisotti, D.; Dehò, G. A Rho-dependent transcription termination site regulated by bacteriophage P4 RNA immunity factor. Virology 1996, 223, 57–67. [Google Scholar] [CrossRef][Green Version]
- Gualdi, L.; Tagliabue, L.; Landini, P. Biofilm formation-gene expression relay system in Escherichia coli: Modulation of σs-dependent gene expression by the CsgD regulatory protein via σs protein stabilization. J. Bacteriol. 2007, 189, 8034–8043. [Google Scholar] [CrossRef] [PubMed]
- Maira-Litrán, T.; Kropec, A.; Abeygunawardana, C.; Joyce, J.; Mark, G.; Goldmann, D.A.; Pier, G.B. Immunochemical properties of the Staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 2002, 70, 4433–4440. [Google Scholar] [CrossRef] [PubMed]
- Carzaniga, T.; Briani, F.; Zangrossi, S.; Merlino, G.; Marchi, P.; Dehò, G. Autogenous regulation of Escherichia coli polynucleotide phosphorylase expression revisited. J. Bacteriol. 2009, 191, 1738–1748. [Google Scholar] [CrossRef] [PubMed]
- Fontanella, L.; Pozzuolo, S.; Costanzo, A.; Favaro, R.; Dehò, G.; Tortora, P. Photometric assay for polynucleotide phosphorylase. Anal. Biochem. 1999, 269, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, J.; Van Melderen, L. Conditional essentiality of the csrA gene in Escherichia coli. J. Bacteriol. 2009, 191, 1722–1724. [Google Scholar] [CrossRef]
- Jin, Y.; Watt, R.M.; Danchin, A.; Huang, J.D. Use of a riboswitch-controlled conditional hypomorphic mutation to uncover a role for the essential csrA gene in bacterial autoaggregation. J. Biol. Chem. 2009, 284, 28738–28745. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Yakhnin, H.; Connolly, M.; Romeo, T.; Babitzke, P. CsrA participates in a PNPase autoregulatory mechanism by selectively repressing translation of pnp transcripts that have been previously processed by RNase III and PNPase. J. Bacteriol. 2015, 197, 3751–3759. [Google Scholar] [CrossRef]
- Thomason, M.K.; Fontaine, F.; De Lay, N.; Storz, G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol. 2012, 84, 17–35. [Google Scholar] [CrossRef]
- Jørgensen, M.G.; Nielsen, J.S.; Boysen, A.; Franch, T.; Møller-Jensen, J.; Valentin-Hansen, P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol. Microbiol. 2012, 84, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Raneri, M.; Pinatel, E.; Peano, C.; Rampioni, G.; Leoni, L.; Bianconi, I.; Jousson, O.; Dalmasio, C.; Ferrante, P.; Briani, F. Pseudomonas aeruginosa mutants defective in glucose uptake have pleiotropic phenotype and altered virulence in non-mammal infection models. Sci. Rep. 2018, 8, 16912. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Leng, Y.; Abe, H.; Amaki, T.; Okayama, A.; Babitzke, P.; Suzuki, K.; Romeo, T. Antagonistic control of the turnover pathway for the global regulatory sRNA CsrB by the CsrA and CsrD proteins. Nucleic Acids Res. 2016, 44, 7896–7910. [Google Scholar] [CrossRef]
- Viegas, S.C.; Pfeiffer, V.; Sittka, A.; Silva, I.J.; Vogel, J.; Arraiano, C.M. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007, 35, 7651–7664. [Google Scholar] [CrossRef]
- Kazazian, H.H. Mobile Elements: Drivers of Genome Evolution. Science 2004, 303, 1626–1632. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.; Lenski, R.E. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res. Microbiol. 2004, 155, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Brükner, R.; Titgemeyer, F. Carbon catabolite repression in bacteria: Choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 2002, 209, 141–148. [Google Scholar] [CrossRef] [PubMed]


| Name | Relevant Characters a | Source/Reference |
|---|---|---|
| Bacterial Strains | ||
| C-1a | E. coli C, prototrophic | [29] |
| C-5691 | C-1a ∆pnp751 | [30] |
| C-5736 | ∆pgaC | This work |
| C-5737 | C-5691 ∆pgaC | This work |
| C-5738 | C-5736 ∆csrA::kan | This work |
| C-5739 | C-5737 ∆csrA::kan | This work |
| C-5741 | C-1a csrA::kan | This work |
| C-5936 | ∆pgaC::kan | [25] |
| C-5937 | C-5691 ∆pgaC::kan | [25] |
| C-5938 | C-1a ∆csrA::kan | This work |
| C-5939 | C-1a ∆csrA | This work |
| C-5940 | ∆csrB::kan | [25] |
| C-5942 | C-5691 ∆csrB::kan | [25] |
| C-5944 | ∆csrC::cat | [25] |
| C-5946 | C-5691 ∆csrC::cat | [25] |
| JW1007 | BW25113 ∆pgaC::kan | [31] |
| KG-206 | MG1655 ∆pnp751::kan | This work |
| KG-211 | MG1655 ∆pnp751 | This work |
| KG-292 | MG1655 ∆pgaC::kan | This work |
| KG-293 | KG-211 ∆pgaC::kan | This work |
| KG-294 | MG1655∆pgaC ∆csrA::kan | This work |
| KG-295 | KG-211 ∆pgaC ∆csrA::kan | This work |
| KG-299 | MG1655∆csrC::cat | This work |
| KG-302 | MG1655∆csrB::kan | This work |
| KG-305 | KG-211 ∆csrB | This work |
| KG-307 | MG1655 ∆pgaC | This work |
| KG-308 | KG-211 ∆pgaC | This work |
| KG-309 | KG-211 ∆csrC::cat | This work |
| MG1655 | rph- | [32] |
| MG1655 csrA::kan | csrA::kan | [15] |
| Plasmids | ||
| pBAD21Δ1 | Cloning vector | [25] |
| pBADpnp | pBAD24 derivative; carries the pnp gene | [25] |
| pBADrnb | pBAD24 derivative; carries the rnb gene | [25] |
| pBADrnr | pBAD24 derivative; carries the rnr gene | [25] |
| pCP20 | FLP encoding plasmid | [27] |
| pCSRA | pGZ119HE derivative; carries 28191692818929 region of MG1655 genome under pTAC promoter | This work |
| pΔLpga | pJAMA8 derivative, harbors the −116 to +32 region relative to the pgaA transcription start site cloned into the SphI/XbaI sites | [25] |
| pGZ119HE | Cloning vector | [33] |
| pJAMA8 | Cloning vector | [34] |
| pKD13 | RED mutagenesis system plasmid | [27] |
| pKD3 | RED mutagenesis system plasmid | [27] |
| pKD46 | RED mutagenesis system plasmid | [27] |
| pLpga2 | pJAMA8 derivative; carries -116 to +249 relative to transcription start of pgaA promoter translationally fused with luxA | [25] |
| Phage P1 hft | High transduction frequency phage P1 derivative | [35] |
| E. coli C | E. coli K-12 | |||||||
|---|---|---|---|---|---|---|---|---|
| - | wt | Δpnp | ΔcsrA | Δpnp ΔcsrA | wt | Δpnp | ΔcsrA | Δpnp ΔcsrA |
| LUX R.A. a | 1.0 | 14.0 ± 0.3 | 26.3 ± 3.0 | 37.9 ± 1.2 | 0.1 ± 0.0 | 0.3 ± 0.0 | 42.0 ± 13.5 | 65.1 ± 27.2 |
| Pwt b | na | 9 × 10−8 | 6 × 10−5 | 4 × 10−7 | na | 1 × 10−5 | 3 × 10−3 | 7 × 10−3 |
| PC-K c | 3 × 10−8 | 7 × 10−8 | ns | ns | 3 × 10−8 | 7 × 10−8 | ns | ns |
| Gene | RNA Relative Amount a | |
|---|---|---|
| C-5691 | MG1655 | |
| csrAb | 0.3 ± 0.1 | 3.5 ± 1.3 |
| csrB | 0.2 ± 0.2 | 57.6 ± 9.9 |
| csrC | 0.1 ± 0.1 | 10.5 ± 5.2 |
| mcaS | 0.7 ± 0.1 | 0.2 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carzaniga, T.; Falchi, F.A.; Forti, F.; Antoniani, D.; Landini, P.; Briani, F. Different csrA Expression Levels in C versus K-12 E. coli Strains Affect Biofilm Formation and Impact the Regulatory Mechanism Presided by the CsrB and CsrC Small RNAs. Microorganisms 2021, 9, 1010. https://doi.org/10.3390/microorganisms9051010
Carzaniga T, Falchi FA, Forti F, Antoniani D, Landini P, Briani F. Different csrA Expression Levels in C versus K-12 E. coli Strains Affect Biofilm Formation and Impact the Regulatory Mechanism Presided by the CsrB and CsrC Small RNAs. Microorganisms. 2021; 9(5):1010. https://doi.org/10.3390/microorganisms9051010
Chicago/Turabian StyleCarzaniga, Thomas, Federica A. Falchi, Francesca Forti, Davide Antoniani, Paolo Landini, and Federica Briani. 2021. "Different csrA Expression Levels in C versus K-12 E. coli Strains Affect Biofilm Formation and Impact the Regulatory Mechanism Presided by the CsrB and CsrC Small RNAs" Microorganisms 9, no. 5: 1010. https://doi.org/10.3390/microorganisms9051010
APA StyleCarzaniga, T., Falchi, F. A., Forti, F., Antoniani, D., Landini, P., & Briani, F. (2021). Different csrA Expression Levels in C versus K-12 E. coli Strains Affect Biofilm Formation and Impact the Regulatory Mechanism Presided by the CsrB and CsrC Small RNAs. Microorganisms, 9(5), 1010. https://doi.org/10.3390/microorganisms9051010