Genetic Mutations That Confer Fluoride Resistance Modify Gene Expression and Virulence Traits of Streptococcus mutans
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Streptococcus mutans Fluoride-Resistant (FR) Strains
2.2. Growth Measurements
2.3. Visualization of Cell Morphology and Biofilm
2.4. Genomic DNA Extraction and Whole-Genome Sequencing
2.5. RNA Extraction and Transcriptome Analysis
2.6. Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
2.7. PEP-Dependent PTS Assay
2.8. pH Drop Assay
2.9. Statistical Analysis
3. Results
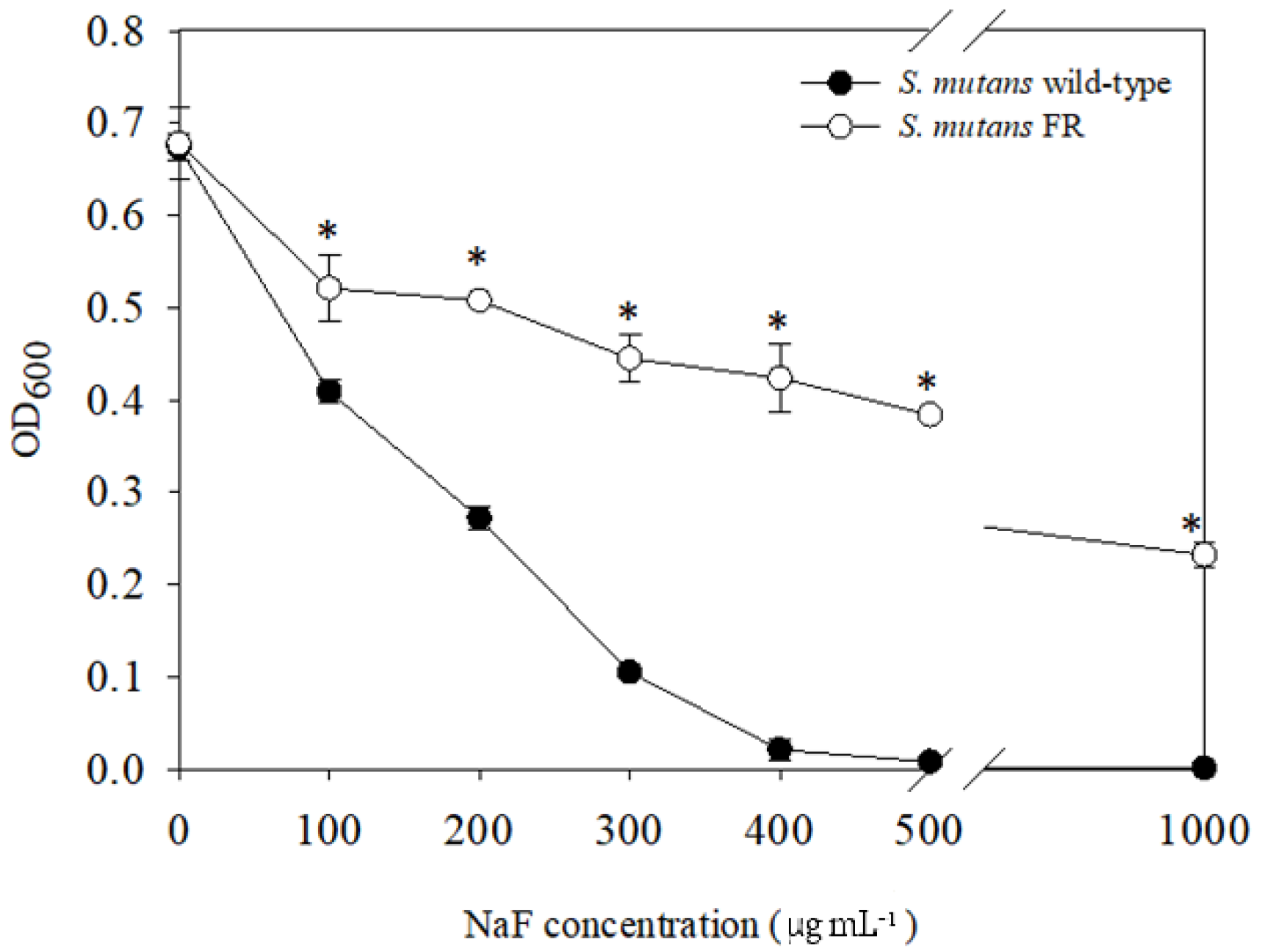
3.1. Isolation of a Streptococcus mutans Fluoride-Resistant (FR) Strain
3.2. Genomic Analysis Revealed Six Genetic Mutations in the FR Strain
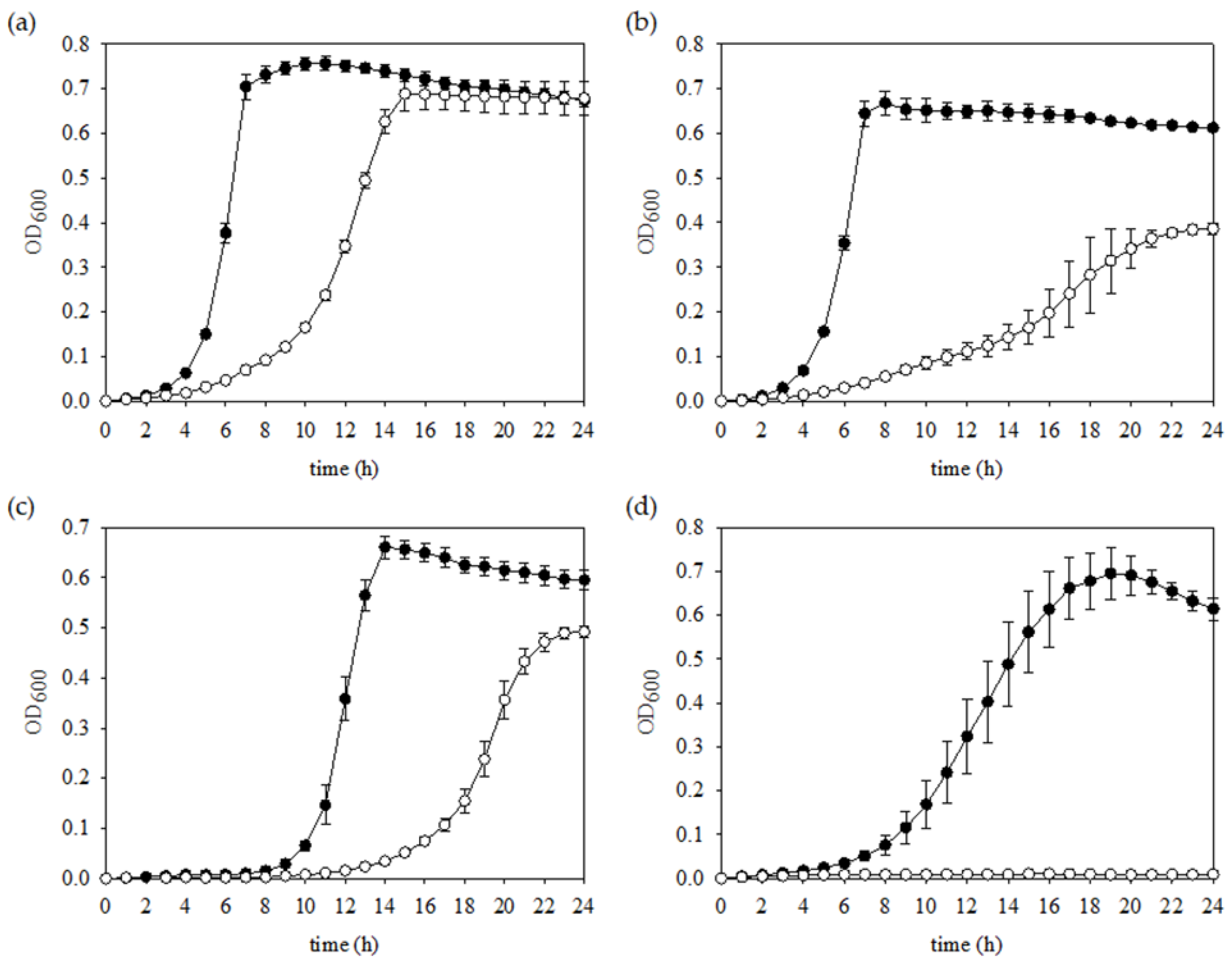
3.3. FR Strain Shows Impaired Growth and Low Stress Tolerance
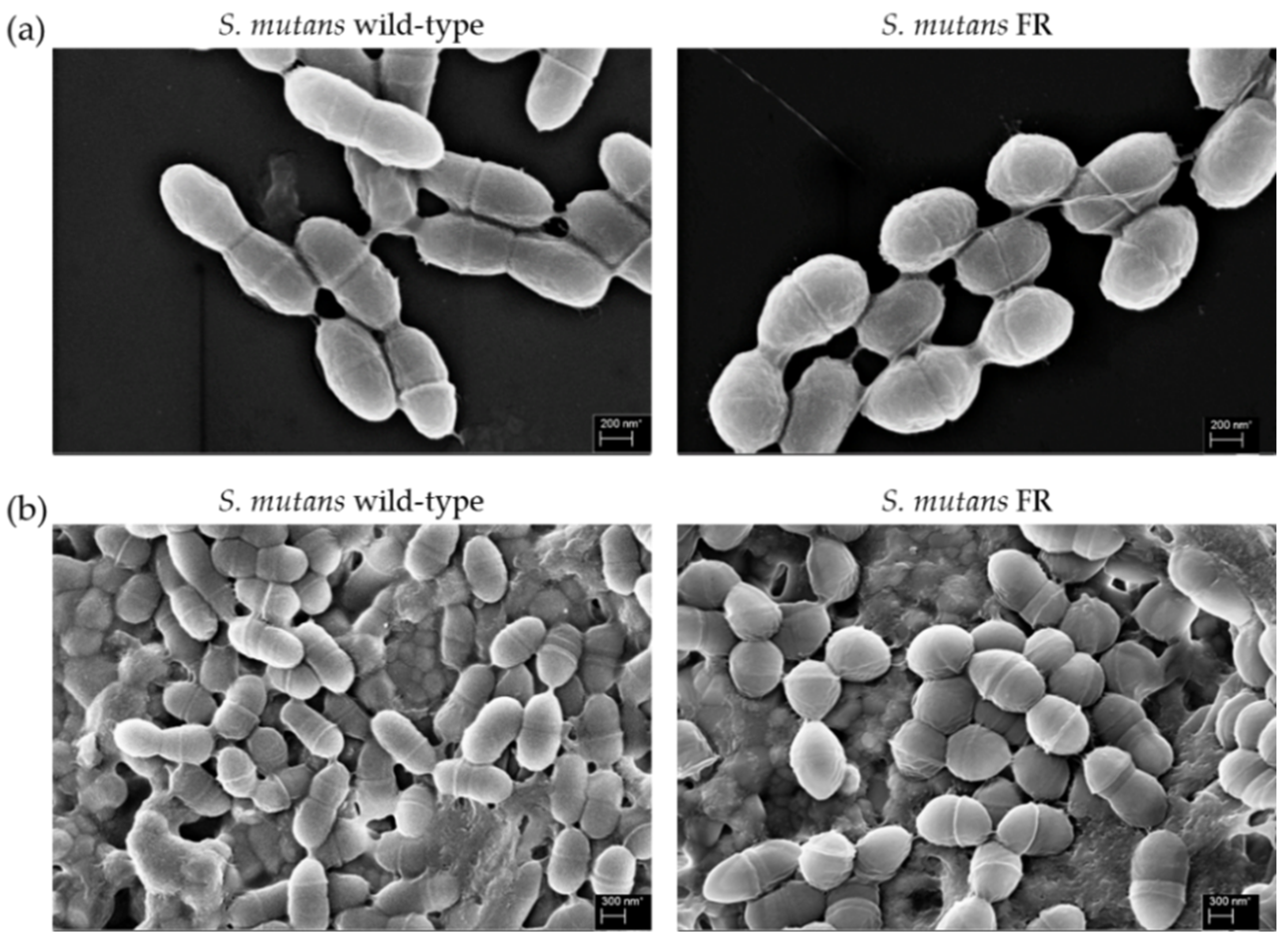
3.4. Morphological Change of the FR Strain
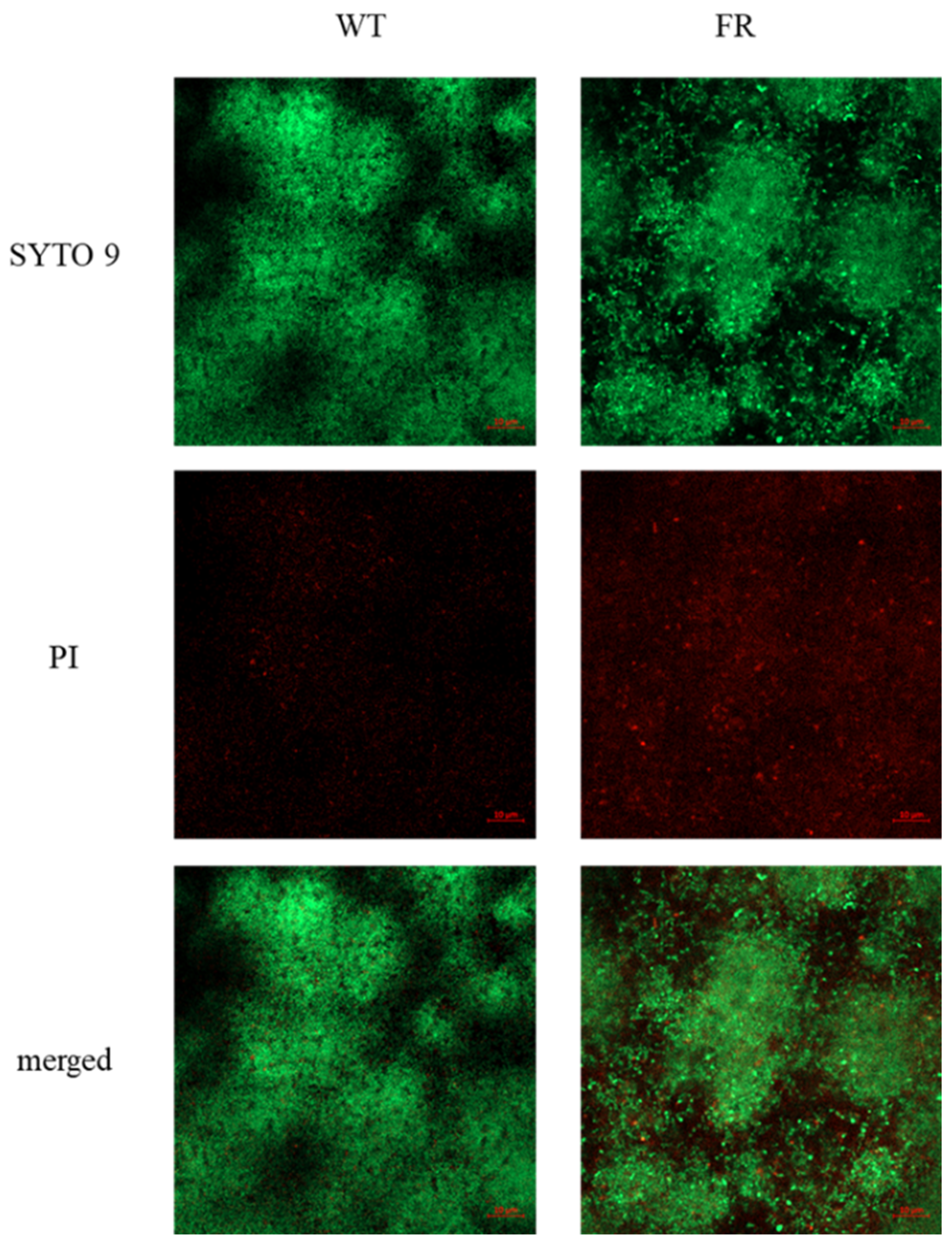
3.5. FR Strain Lowers the Ability to Form Biofilms
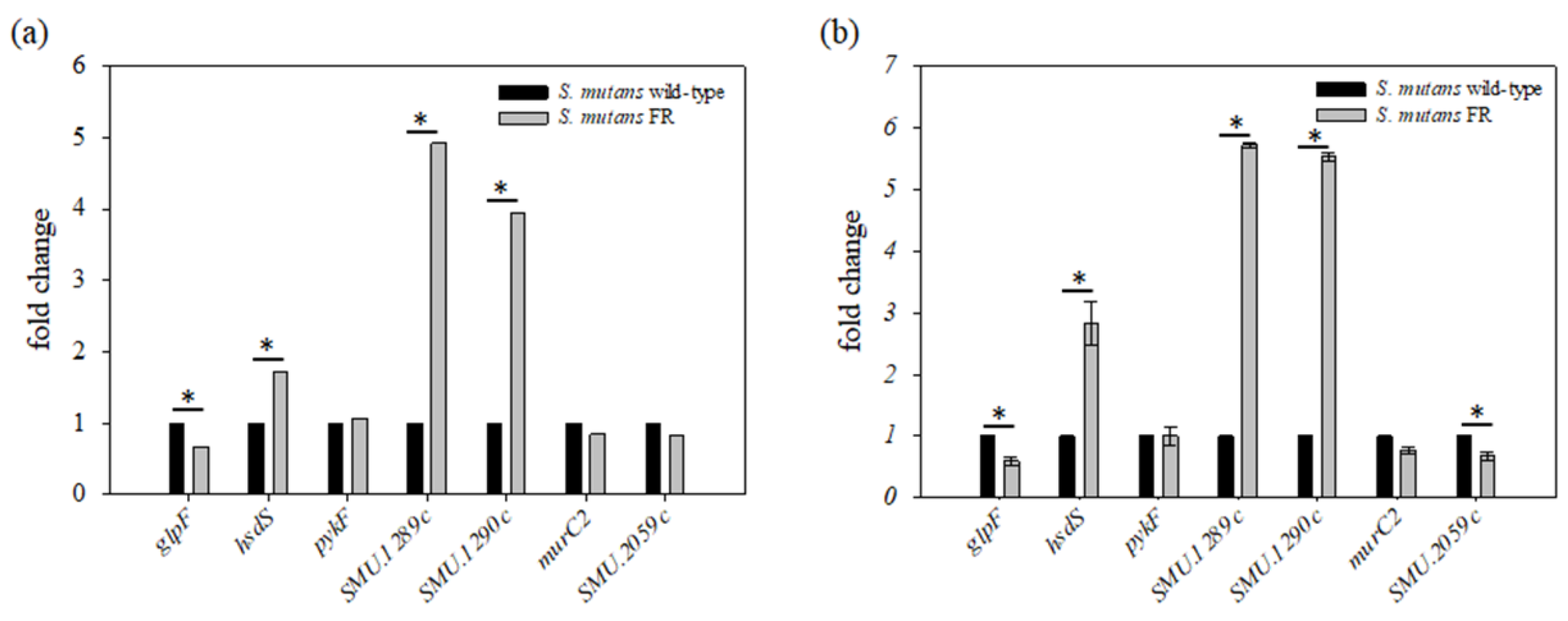
3.6. Transcription Profiling of an FR Strain
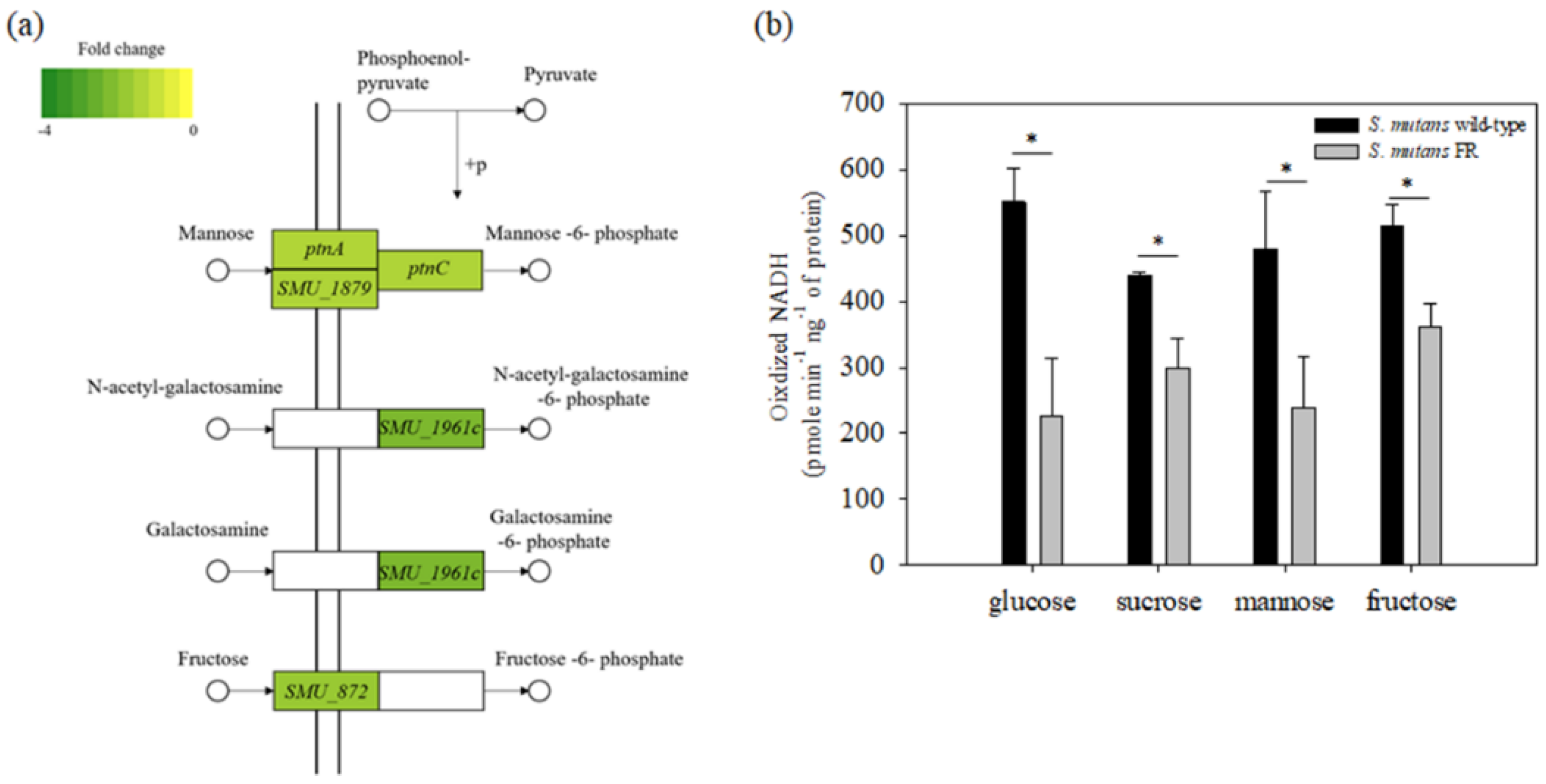
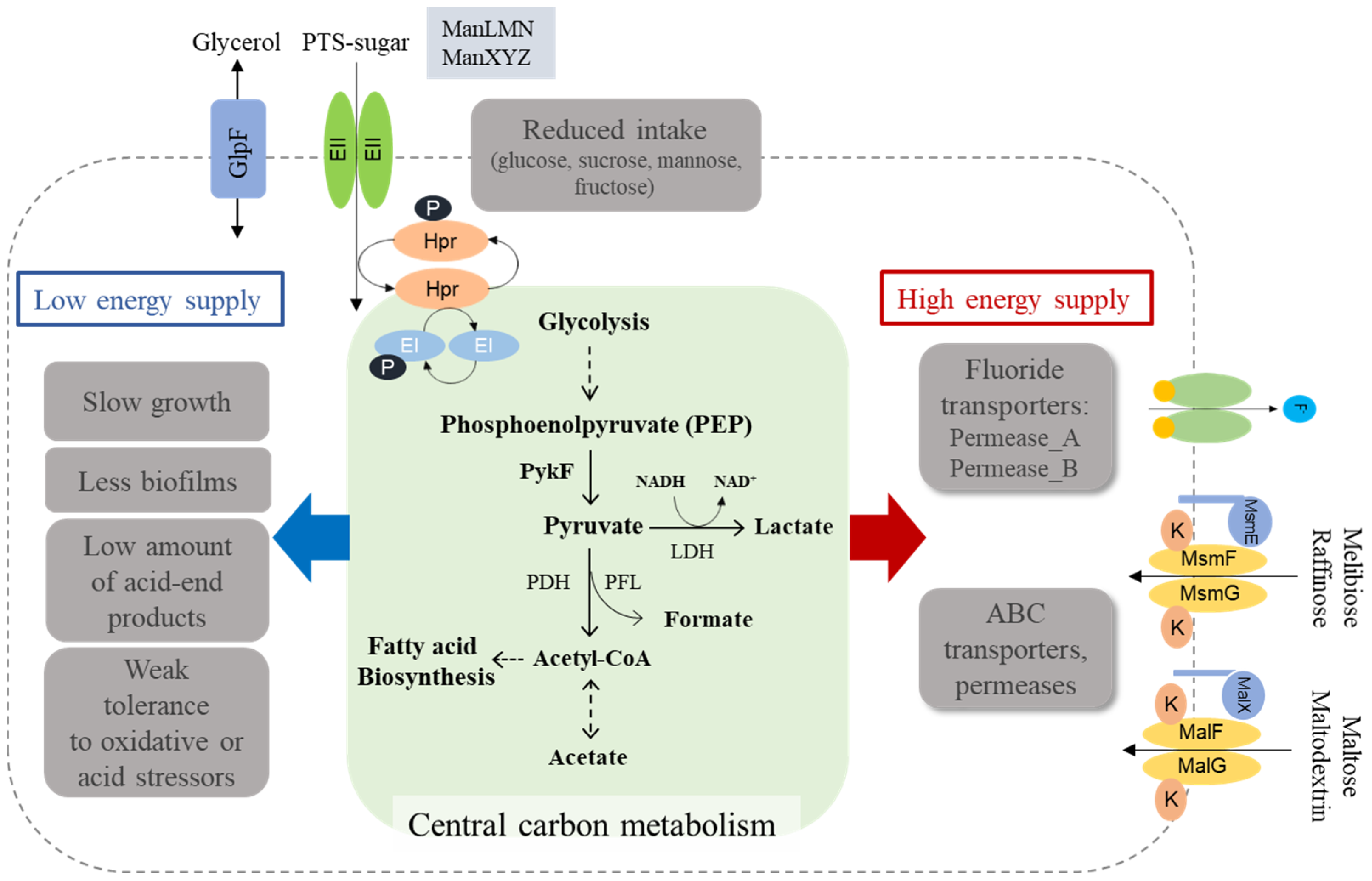
3.7. Fluoride Resistance Affects Carbohydrate Uptake
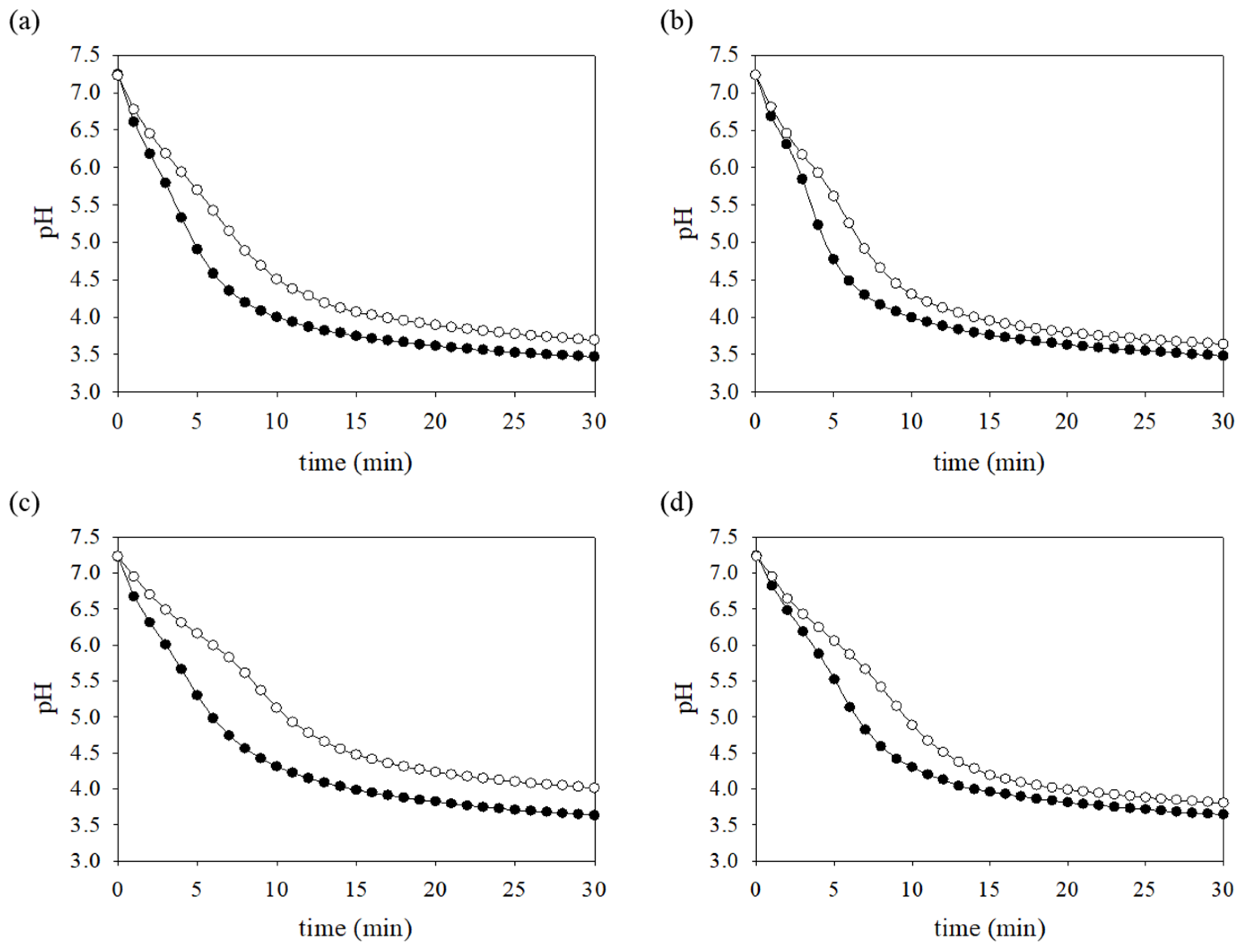
3.8. Production of Acid-End Products Is Reduced in the FR Strain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petersen, P.E. The World Oral Health Report 2003: Continuous improvement of oral health in the 21st century-the approach of the WHO Global Oral Health Programme. Community Dent. Oral Epidemiol. 2003, 31, 3–24. [Google Scholar] [CrossRef]
- Kassebaum, N.; Bernabe, E.; Dahiya, M.; Bhandari, B.; Murray, C.J.L.; Marcenes, W. Global Burden of Untreated Caries: A sys-tematic review and metaregression. J. Dent. Res. 2015, 94, 650–658. [Google Scholar] [CrossRef]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 1986, 50, 353–380. [Google Scholar] [CrossRef] [PubMed]
- Beighton, D. The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dent. Oral Epidemiol. 2005, 33, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Belli, W.A.; Marquis, R.E. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 1991, 57, 1134–1138. [Google Scholar] [CrossRef] [PubMed]
- Marquis, R.E.; Clock, S.A.; Mota-Meira, M. Fluoride and organic weak acids as modulators of microbial physiology. FEMS Microbiol. Rev. 2003, 26, 493–510. [Google Scholar] [CrossRef] [PubMed]
- Marinho, V.C.; Higgins, J.P.; Logan, S.; Deceased, A.S. Fluoride toothpastes for preventing dental caries in children and adolescents. Cochrane Database Syst. Rev. 2003, CD002278. [Google Scholar] [CrossRef] [PubMed]
- O’Mullane, D.M.; Baez, R.J.; Jones, S.; Lennon, M.A.; Petersen, P.E.; Rugg-Gunn, A.J.; Whelton, H.; Whitford, G.M. Fluoride and Oral Health. Community Dent Health 2016, 33, 69–99. [Google Scholar] [PubMed]
- Streckfuss, J.L.; Perkins, D.; Horton, I.M.; Brown, L.R.; Dreizen, S.; Graves, L. Fluoride Resistance and Adherence of Selected Strains of Streptococcus mutans to Smooth Surfaces After Exposure to Fluoride. J. Dent. Res. 1980, 59, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.; White, J.; Horton, I.; Dreizen, S.; Streckfuss, J. Effect of Continuous Fluoride Gel Use on Plaque Fluoride Retention and Microbial Activity. J. Dent. Res. 1983, 62, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Chen, J.; Brandt, B.W.; Zhu, Y.; Li, J.; Van Loveren, C.; Deng, D.M. Identification and Functional Analysis of Genome Mutations in a Fluoride-Resistant Streptococcus mutans Strain. PLoS ONE 2015, 10, e0122630. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tian, J.; Liu, L.; Zhu, T.; Yu, X.; Chu, X.; Yao, B.; Wu, N.; Fan, Y. Identification of an operon involved in fluoride resistance in Enterobacter cloacae FRM. Sci. Rep. 2017, 7, 6786. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhang, Z.; Liang, J. Fatty-acid profiles and expression of the fabM gene in a fluoride-resistant strain of Streptococcus mutans. Arch. Oral Biol. 2012, 57, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Liao, Y.; Brandt, B.W.; Wei, X.; Liu, H.; Crielaard, W.; Van Loveren, C.; Deng, D.M. The Fitness Cost of Fluoride Resistance for Different Streptococcus mutans Strains in Biofilms. Front. Microbiol. 2017, 8, 1630. [Google Scholar] [CrossRef] [PubMed]
- Hoelscher, G.L.; Hudson, M.C. Characterization of an Unusual Fluoride-Resistant Streptococcus mutans Isolate. Curr. Microbiol. 1996, 32, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, I. Effects of Fluoride on Enzymatic Regulation of Bacterial Carbohydrate Metabolism. Caries Res. 1977, 11, 262–291. [Google Scholar] [CrossRef]
- Bender, G.R.; Sutton, S.V.; Marquis, R.E. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect. Immun. 1986, 53, 331–338. [Google Scholar] [CrossRef]
- Sutton, S.V.; Bender, G.R.; Marquis, R.E. Fluoride inhibition of proton-translocating ATPases of oral bacteria. Infect. Immun. 1987, 55, 2597–2603. [Google Scholar] [CrossRef] [PubMed]
- Bunick, F.J.; Kashket, S. Enolases from fluoride-sensitive and fluoride-resistant streptococci. Infect. Immun. 1981, 34, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Van Loveren, C.; Hoogenkamp, M.; Deng, D.; Cate, J.T. Effects of Different Kinds of Fluorides on Enolase and ATPase Activity of a Fluoride-Sensitive and Fluoride-Resistant Streptococcus mutans Strain. Caries Res. 2008, 42, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Yang, J.; Brandt, B.W.; Li, J.; Crielaard, W.; Van Loveren, C.; Deng, D.M. Genetic Loci Associated With Fluoride Resistance in Streptococcus mutans. Front. Microbiol. 2018, 9, 3093. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.L.; Sudarsan, N.; Weinberg, Z.; Roth, A.; Stockbridge, R.B.; Breaker, R.R. Widespread Genetic Switches and Toxicity Resistance Proteins for Fluoride. Science 2011, 335, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R. New Insight on the Response of Bacteria to Fluoride. Caries Res. 2012, 46, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Men, X.; Shibata, Y.; Takeshita, T.; Yamashita, Y. Identification of Anion Channels Responsible for Fluoride Resistance in Oral Streptococci. PLoS ONE 2016, 11, e0165900. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Hanada, N. Contribution of chloride channel permease to fluoride resistance inStreptococcus mutans. FEMS Microbiol. Lett. 2016, 363, fnw101. [Google Scholar] [CrossRef]
- Liao, S.; Klein, M.I.; Heim, K.P.; Fan, Y.; Bitoun, J.P.; Ahn, S.-J.; Burne, R.A.; Koo, H.; Brady, L.J.; Wen, Z.T. Streptococcus mutans Extracellular DNA Is Upregulated during Growth in Biofilms, Actively Released via Membrane Vesicles, and Influenced by Components of the Protein Secretion Machinery. J. Bacteriol. 2014, 196, 2355–2366. [Google Scholar] [CrossRef] [PubMed]
- Loo, C.Y.; Corliss, D.A.; Ganeshkumar, N. Streptococcus gordonii Biofilm Formation: Identification of Genes that Code for Biofilm Phenotypes. J. Bacteriol. 2000, 182, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, H.L.; Reeves, R.E. Inducible phosphoenolpyruvate-dependent hexose phosphotransferase activities in Escherichia coli. Biochem. J. 1972, 128, 1339–1344. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Burne, R.A.; Zeng, L. Uptake and Metabolism of N-Acetylglucosamine and Glucosamine by Streptococcus mutans. Appl. Environ. Microbiol. 2014, 80, 5053–5067. [Google Scholar] [CrossRef] [PubMed]
- Burne, R.A.; Wen, Z.T.; Chen, Y.-Y.M.; Penders, J.E.C. Regulation of Expression of the Fructan Hydrolase Gene of Streptococcus mutans GS-5 by Induction and Carbon Catabolite Repression. J. Bacteriol. 1999, 181, 2863–2871. [Google Scholar] [CrossRef] [PubMed]
- Abranches, J.; Nascimento, M.M.; Zeng, L.; Browngardt, C.M.; Wen, Z.T.; Rivera, M.F.; Burne, R.A. CcpA Regulates Central Metabolism and Virulence Gene Expression in Streptococcus mutans. J. Bacteriol. 2008, 190, 2340–2349. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.A.; Burne, R.A. A model of efficiency: Stress tolerance by Streptococcus mutans. Microbiology 2008, 154, 3247–3255. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.A.C.; Abranches, J.; Burne, R.A. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 2005, 7, 95–107. [Google Scholar] [PubMed]
- Sargent, M.G. Control of cell length in Bacillus subtilis. J. Bacteriol. 1975, 123, 7–19. [Google Scholar] [CrossRef]
- Kreth, J.; Zhu, L.; Merritt, J.; Shi, W.; Qi, F. Role of sucrose in the fitness of Streptococcus mutans. Oral Microbiol. Immunol. 2008, 23, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Webb, A.J.; Homer, K.A.; Hosie, A.H.F. A Phosphoenolpyruvate-Dependent Phosphotransferase System Is the Principal Maltose Transporter in Streptococcus mutans. J. Bacteriol. 2007, 189, 3322–3327. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Webb, A.J.; Homer, K.A.; Hosie, A.H.F. Two Closely Related ABC Transporters in Streptococcus mutans Are Involved in Disaccharide and/or Oligosaccharide Uptake. J. Bacteriol. 2007, 190, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Sutcliffe, I.; Russell, R.; Ferretti, J. Transport of Sugars, Including Sucrose, by the msm Transport System of Streptococcus mutans. J. Dent. Res. 1993, 72, 1386–1390. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.; Aduse-Opoku, J.; Sutcliffe, I.; Tao, L.; Ferretti, J. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J. Biol. Chem. 1992, 267, 4631–4637. [Google Scholar] [CrossRef]
- Sato, Y.; Okamoto-Shibayama, K.; Azuma, T. The malQ gene is essential for starch metabolism in Streptococcus mutans. J. Oral Microbiol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Schäffer, C.; Messner, P. The structure of secondary cell wall polymers: How Gram-positive bacteria stick their cell walls together. Microbiolohy 2005, 151, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Dobrogosz, W.J. N-Acetylglucosamine Assimilation in Escherichia coli and Its Relation to Catabolite Repression1. J. Bacteriol. 1968, 95, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Trent, M.S. Biosynthesis, transport, and modification of lipid A. Biochem. Cell Biol. 2004, 82, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-AñorveL, I.; Bustos-Jaimes, I.; Calcagno, M.L.; Plumbridge, J. Allosteric Regulation of Glucosamine-6-Phosphate Deaminase (NagB) and Growth of Escherichia coli on Glucosamine. J. Bacteriol. 2009, 191, 6401–6407. [Google Scholar] [CrossRef] [PubMed]
- White, R.J. The role of the phosphoenolpyruvate phosphotransferase system in the transport of N-acetyl-d-glucosamine by Escherichia coli. Biochem. J. 1970, 118, 89–92. [Google Scholar] [CrossRef]
- Ajdić, D.; McShan, W.M.; Laughlin, R.E.; Savić, G.; Chang, J.; Carson, M.B.; Primeaux, C.; Tian, R.; Kenton, S.; Jia, H.; et al. Genome sequence of Streptococcus mutans UA159, acariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 2002, 99, 14434–14439. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.; Palmer, S.; Zeng, L.; Wen, Z.; Kajfasz, J.; Freires, I.; Abranches, J.; Brady, L. The Biology of Streptococcus mutans. Microbiol. Spectr. 2019, 7, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Heller, K.B.; Lin, E.C.; Wilson, T.H. Substrate specificity and transport properties of the glycerol facilitator of Escherichia coli. J. Bacteriol. 1980, 144, 274–278. [Google Scholar] [CrossRef]
- Bender, G.; Thibodeau, E.; Marquis, R. Reduction of Acidurance of Streptococcal Growth and Glycolysis by Fluoride and Gramicidin. J. Dent. Res. 1985, 64, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Brandt, B.W.; Zhang, M.; Li, J.; Crielaard, W.; Van Loveren, C.; Deng, D.M. A single nucleotide change in the promotermutpenhances fluoride resistance of Streptococcus mutans. Antimicrob. Agents Chemother. 2016, 60, 7509–7512. [Google Scholar] [CrossRef]
- Doi, Y. Glycerol metabolism and its regulation in lactic acid bacteria. Appl. Microbiol. Biotechnol. 2019, 103, 5079–5093. [Google Scholar] [CrossRef]
- Brussock, S.; Kral, T. Effects of pH on Expression of Sodium Fluoride Resistance in Streptococcus mutans. J. Dent. Res. 1987, 66, 1594–1596. [Google Scholar] [CrossRef]
- Li, Y.-H.; Lau, P.C.Y.; Lee, J.H.; Ellen, R.P.; Cvitkovitch, D.G. Natural Genetic Transformation of Streptococcus mutans Growing in Biofilms. J. Bacteriol. 2001, 183, 897–908. [Google Scholar] [CrossRef]
- Sheng, J.; Marquis, R.E. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol. Lett. 2007, 272, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Lemme, A.; Sztajer, H.; Wagner-Döbler, I. Characterization of mleR, a positive regulator of malolactic fermentation and part of the acid tolerance response in Streptococcus mutans. BMC Microbiol. 2010, 10, 58. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Baldeck, J.D.; Nguyen, P.T.; Quivey, R.G.; Marquis, R.E. Alkali production associated with malolactic fermentation by oral streptococci and protection against acid, oxidative, or starvation damage. Can. J. Microbiol. 2010, 56, 539–547. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Description | Mutation and Location | Effect |
|---|---|---|---|
| hsdS | Putative restriction endonuclease | 563 A→ACAGAATTGACCT | Disruptive inframe insertion |
| glpF | Glycerol uptake facilitator protein | 464 C→A | Stop codon |
| pykF | Pyruvate kinase | 1156 G→A | Val→Ile |
| murC2 | Putative UDP-N-acetylmuramyl tripeptide synthetase | 289 G→T | Ala→Ser |
| SMU.2059c | Putative integral membrane protein | 707 A→T | Tyr→Phe |
| SMU.1289c SMU.1290c | Chloride channel permease | A→G (Intergenic region of two genes) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-J.; Song, J.; Kim, J.N. Genetic Mutations That Confer Fluoride Resistance Modify Gene Expression and Virulence Traits of Streptococcus mutans. Microorganisms 2021, 9, 849. https://doi.org/10.3390/microorganisms9040849
Lee H-J, Song J, Kim JN. Genetic Mutations That Confer Fluoride Resistance Modify Gene Expression and Virulence Traits of Streptococcus mutans. Microorganisms. 2021; 9(4):849. https://doi.org/10.3390/microorganisms9040849
Chicago/Turabian StyleLee, Hyeon-Jeong, Jihee Song, and Jeong Nam Kim. 2021. "Genetic Mutations That Confer Fluoride Resistance Modify Gene Expression and Virulence Traits of Streptococcus mutans" Microorganisms 9, no. 4: 849. https://doi.org/10.3390/microorganisms9040849
APA StyleLee, H.-J., Song, J., & Kim, J. N. (2021). Genetic Mutations That Confer Fluoride Resistance Modify Gene Expression and Virulence Traits of Streptococcus mutans. Microorganisms, 9(4), 849. https://doi.org/10.3390/microorganisms9040849






