Candida albicans Hexokinase 2 Challenges the Saccharomyces cerevisiae Moonlight Protein Model
Abstract
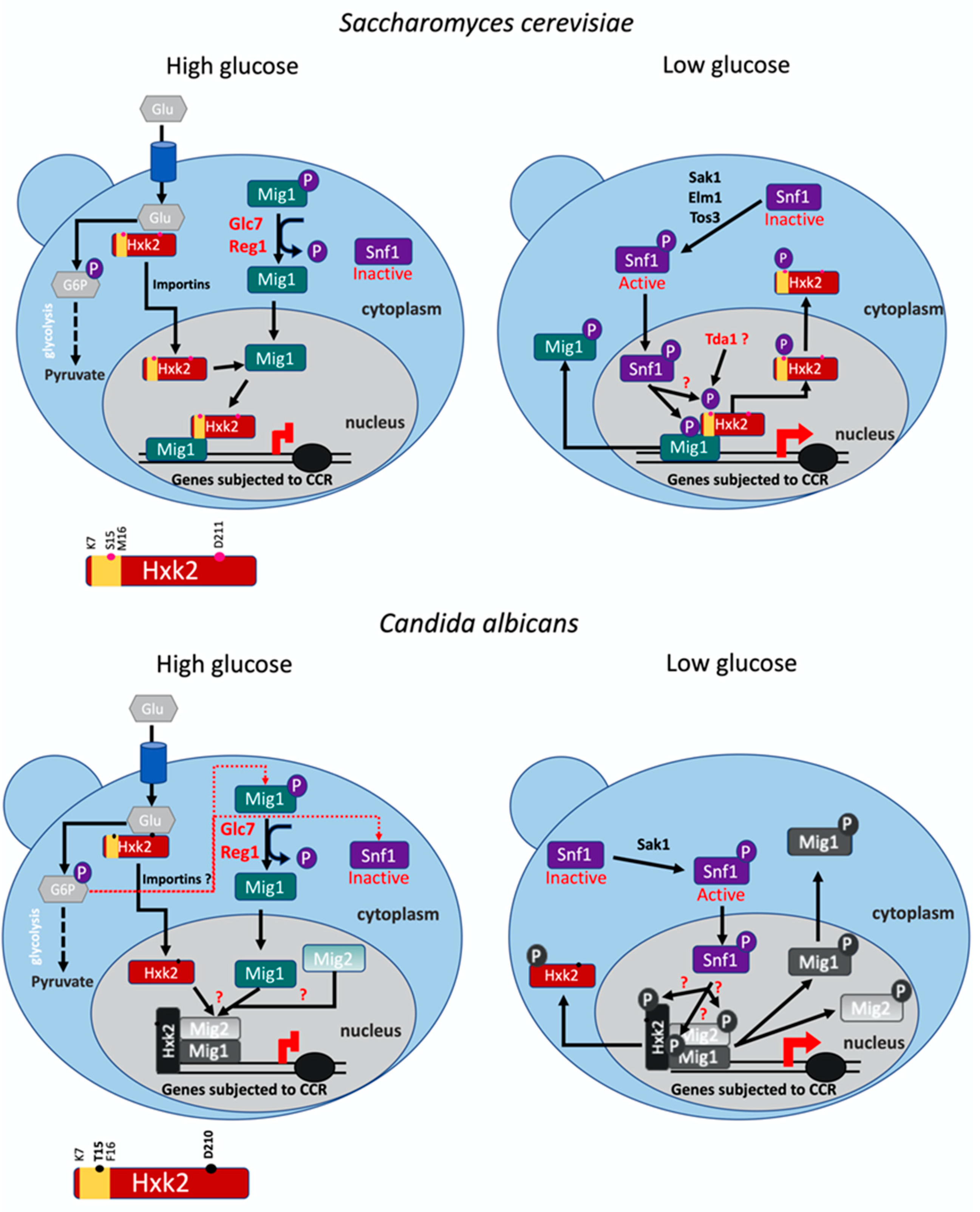
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Growth Conditions
2.2. Construction of Mutant Strains
2.3. Preparation of Yeast Cell Extract
2.4. Determination of Glucose Kinase Activity
2.5. Determination of Glucose Concentration
2.6. RNA Extraction and RT-qPCR Analysis
2.7. Infection of Phagocytes with Yeasts
2.8. Flow Cytometry Analysis
3. Results
3.1. The Regulatory Domains and Catalytic Residues Are Conserved in CaHxk2
3.2. The Aspartic Acid Residue D210 Is Implicated in Glucose Phosphorylation in C. albicans
3.3. The CaHxk2 Catalytic Residue D210 Is Involved in Glucose Repression in C. albicans But Neither the N-terminal Decapeptide K7F16 Nor the Threonine T15
3.4. Filamentation Is Affected in Cahxk2D210A
3.5. Virulence in a Macrophage Model Is Affected in Cahxk2D210A Mutant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Odds, F.C. Candida and Candidosis, A Review and Bibliography (Second Edition). X + 468 S., 97 Abb., 92 Table u. 22 Farbtafeln. London—Philadelphia—Toronto—Sydney—Tokyo 1988. Baillière Tindall (W. B. Saunders). ISBN: 0–7020–1265–3. J. Basic Microbiol. 1988, 30, 382–383. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Bender, J.A.; Fink, G.R. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 2004, 3, 1076–1087. [Google Scholar] [CrossRef]
- Barelle, C.J.; Priest, C.L.; Maccallum, D.M.; Gow, N.A.R.; Odds, F.C.; Brown, A.J.P. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 2006, 8, 961–971. [Google Scholar] [CrossRef] [PubMed]
- Zeman, I.; Neboháčová, M.; Gérecová, G.; Katonová, K.; Jánošíková, E.; Jakúbková, M.; Centárová, I.; Dunčková, I.; Tomáška, L.; Pryszcz, L.P.; et al. Mitochondrial carriers link the catabolism of hydroxyaromatic compounds to the central metabolism in Candida parapsilosis. G3 2016, 6, 4047–4058. [Google Scholar] [CrossRef]
- Miramón, P.; Lorenz, M.C. A Feast for Candida: Metabolic plasticity confers an edge for virulence. PLoS Pathog. 2017, 13, e1006144. [Google Scholar] [CrossRef]
- Tejima, K.; Ishiai, M.; Murayama, S.O.; Iwatani, S.; Kajiwara, S. Candida albicans Fatty acyl-CoA synthetase, CaFaa4p, is involved in the uptake of exogenous long-chain fatty acids and cell activity in the biofilm. Curr. Genet. 2018, 64, 429–441. [Google Scholar] [CrossRef]
- Laurian, R.; Jacot-des-Combes, C.; Bastian, F.; Dementhon, K.; Cotton, P. Carbon metabolism snapshot by ddPCR during the early step of Candida albicans phagocytosis by macrophages. Pathog. Dis. 2020, 78, ftaa014. [Google Scholar] [CrossRef]
- Williams, R.B.; Lorenz, M.C. Multiple alternative carbon pathways combine to promote Candida albicans stress resistance, immune interactions, and virulence. mbio 2020, 11, e03070-19. [Google Scholar] [CrossRef]
- Brown, A.J.P.; Odds, F.C.; Gow, N.A.R. Infection-related gene expression in Candida albicans. Curr. Opin. Microbiol. 2007, 10, 307–313. [Google Scholar] [CrossRef]
- Wilson, D.; Thewes, S.; Zakikhany, K.; Fradin, C.; Albrecht, A.; Almeida, R.; Brunke, S.; Grosse, K.; Martin, R.; Mayer, F.; et al. Identifying infection-associated genes of Candida albicans in the postgenomic era. FEMS Yeast Res. 2009, 9, 688–700. [Google Scholar] [CrossRef]
- Pérez, J.C.; Kumamoto, C.A.; Johnson, A.D. Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol. 2013, 11, e1001510. [Google Scholar] [CrossRef] [PubMed]
- Tucey, T.M.; Verma, J.; Harrison, P.F.; Snelgrove, S.L.; Lo, T.L.; Scherer, A.K.; Barugahare, A.A.; Powell, D.R.; Wheeler, R.T.; Hickey, M.J.; et al. Glucose homeostasis is important for immune cell viability during Candida challenge and host survival of systemic fungal infection. Cell Metab. 2018, 27, 988–1006. [Google Scholar] [CrossRef]
- Laurian, R.; Dementhon, K.; Doumèche, B.; Soulard, A.; Noel, T.; Lemaire, M.; Cotton, P. Hexokinase and Glucokinases are essential for fitness and virulence in the pathogenic yeast Candida albicans. Front. Microbiol. 2019, 10, 327. [Google Scholar] [CrossRef] [PubMed]
- Wijnants, S.; Riedelberger, M.; Penninger, P.; Kuchler, K.; Van Dijck, P. Sugar phosphorylation controls carbon source utilization and virulence of Candida albicans. Front. Microbiol. 2020, 11, 1274–1288. [Google Scholar] [CrossRef]
- Herrero, P.; Martínez-Campa, C.; Moreno, F. The Hexokinase 2 protein participates in regulatory DNA-Protein complexes necessary for glucose repression of the SUC2 gene in Saccharomyces cerevisiae. FEBS Lett. 1998, 434, 71–76. [Google Scholar] [CrossRef]
- Moreno, F.; Ahuatzi, D.; Riera, A.; Palomino, C.A.; Herrero, P. Glucose sensing through the Hxk2-dependent signalling pathway. Biochem. Soc. Trans. 2005, 33, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Gancedo, C.; Flores, C.-L. Moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 2008, 72, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Flores, C.-L.; Gancedo, C. Unraveling moonlighting functions with yeasts. IUBMB Life 2011, 63, 457–462. [Google Scholar] [CrossRef]
- Gancedo, C.; Flores, C.-L.; Gancedo, J.M. Evolution of moonlighting proteins: Insight from yeasts. Biochem. Soc. Trans. 2014, 42, 1715–1719. [Google Scholar] [CrossRef][Green Version]
- Kayikci, Ö.; Nielsen, J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov068. [Google Scholar] [CrossRef]
- Randez-Gil, F.; Herrero, P.; Sanz, P.; Prieto, J.A.; Moreno, F. Hexokinase PII has a double cytosolic-nuclear localisation in Saccharomyces cerevisiae. FEBS Lett. 1998, 425, 475–478. [Google Scholar] [CrossRef]
- Gancedo, J.M. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 1998, 62, 334–361. [Google Scholar] [CrossRef]
- Ahuatzi, D.; Herrero, P.; de la Cera, T.; Moreno, F. The glucose-regulated nuclear localization of Hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 2004, 279, 14440–14446. [Google Scholar] [CrossRef]
- Hedbacker, K.; Carlson, M. SNF1/AMPK Pathways in yeast. Front. Biosci. 2008, 13, 2408–2420. [Google Scholar] [CrossRef]
- Wilson, W.A.; Hawley, S.A.; Hardie, D.G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 1996, 6, 1426–1434. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef]
- Peláez, R.; Herrero, P.; Moreno, F. Functional domains of yeast Hexokinase 2. Biochem. J. 2010, 432, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Vega, M.; Riera, A.; Fernández-Cid, A.; Herrero, P.; Moreno, F. Hexokinase 2 is an intracellular glucose sensor of yeast cells that maintains the structure and activity of Mig1 protein repressor complex. J. Biol. Chem. 2016, 291, 7267–7285. [Google Scholar] [CrossRef] [PubMed]
- Ahuatzi, D.; Riera, A.; Peláez, R.; Herrero, P.; Moreno, F. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J. Biol. Chem. 2007, 282, 4485–4493. [Google Scholar] [CrossRef]
- Peláez, R.; Fernández-García, P.; Herrero, P.; Moreno, F. Nuclear import of the yeast Hexokinase 2 protein requires α/β-importin-dependent Pathway. J. Biol. Chem. 2012, 287, 3518–3529. [Google Scholar] [CrossRef] [PubMed]
- Fernández-García, P.; Peláez, R.; Herrero, P.; Moreno, F. Phosphorylation of yeast Hexokinase 2 regulates its nucleocytoplasmic shuttling. J. Biol. Chem. 2012, 287, 42151–42164. [Google Scholar] [CrossRef] [PubMed]
- Behlke, J.; Heidrich, K.; Naumann, M.; Müller, E.C.; Otto, A.; Reuter, R.; Kriegel, T. Hexokinase 2 from Saccharomyces cerevisiae: Regulation of oligomeric structure by in vivo phosphorylation at serine-14. Biochemistry 1998, 37, 11989–11995. [Google Scholar] [CrossRef] [PubMed]
- Kriegel, T.M.; Rush, J.; Vojtek, A.B.; Clifton, D.; Fraenkel, D.G. In vivo phosphorylation site of Hexokinase 2 in Saccharomyces cerevisiae. Biochemistry 1994, 33, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Kettner, K.; Krause, U.; Mosler, S.; Bodenstein, C.; Kriegel, T.M.; Rödel, G. Saccharomyces cerevisiae gene YMR291W/TDA1 mediates the in vivo phosphorylation of Hexokinase isoenzyme 2 at serine-15. FEBS Lett. 2012, 586, 455–458. [Google Scholar] [CrossRef]
- Kaps, S.; Kettner, K.; Migotti, R.; Kanashova, T.; Krause, U.; Rödel, G.; Dittmar, G.; Kriegel, T.M. Protein kinase Ymr291w/Tda1 is essential for glucose signaling in Saccharomyces cerevisiae on the level of hexokinase isoenzyme ScHxk2 phosphorylation. J. Biol. Chem. 2015, 290, 6243–6255. [Google Scholar] [CrossRef]
- Bennett, W.S.; Steitz, T.A. Structure of a complex between yeast Hexokinase A and glucose. II. Detailed comparisons of conformation and active site configuration with the native Hexokinase B monomer and dimer. J. Mol. Biol. 1980, 140, 211–230. [Google Scholar] [CrossRef]
- Arora, K.K.; Fanciulli, M.; Pedersen, P.L. Glucose phosphorylation in tumor cells. cloning, sequencing, and overexpression in active form of a full-length cDNA encoding a mitochondrial bindable form of Hexokinase. J. Biol. Chem. 1991, 265, 6481–6488. [Google Scholar] [CrossRef]
- Ramírez-Zavala, B.; Mottola, A.; Haubenreißer, J.; Schneider, S.; Allert, S.; Brunke, S.; Ohlsen, K.; Hube, B.; Morschhäuser, J. The Snf1-activating kinase Sak1 is a key regulator of metabolic adaptation and in vivo fitness of Candida albicans. Mol. Microbiol. 2017, 104, 989–1007. [Google Scholar] [CrossRef]
- Zaragoza, O.; Rodríguez, C.; Gancedo, C. Isolation of the MIG1 gene from Candida albicans and effects of its disruption on catabolite repression. J. Bacteriol. 2000, 182, 320–326. [Google Scholar] [CrossRef]
- Murad, A.M.; d’Enfert, C.; Gaillardin, C.; Tournu, H.; Tekaia, F.; Talibi, D.; Marechal, D.; Marchais, V.; Cottin, J.; Brown, A.J. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 2001, 42, 981–993. [Google Scholar] [CrossRef]
- Lagree, K.; Woolford, C.A.; Huang, M.Y.; May, G.; McManus, C.J.; Solis, N.V.; Filler, S.G.; Mitchell, A.P. Roles of Candida albicans Mig1 and Mig2 in glucose repression, pathogenicity traits, and SNF1 essentiality. PLoS Genet. 2020, 16, e1008582. [Google Scholar] [CrossRef]
- Mayordomo, I.; Sanz, P. Hexokinase PII: Structural analysis and glucose signalling in the yeast Saccharomyces cerevisiae. Yeast 2001, 18, 923–930. [Google Scholar] [CrossRef]
- Peláez, R.; Herrero, P.; Moreno, F. Nuclear export of the yeast Hexokinase 2 protein requires the Xpo1 (Crm1)-dependent pathway. J. Biol. Chem. 2009, 284, 20548–20555. [Google Scholar] [CrossRef] [PubMed]
- Reuss, O.; Vik, A.; Kolter, R.; Morschhäuser, J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 2004, 341, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Walther, A.; Wendland, J. An improved transformation protocol for the Human fungal pathogen Candida albicans. Curr. Genet. 2003, 42, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A.; Oliviero, S. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. 2001, 13. [Google Scholar] [CrossRef]
- Dementhon, K.; El-Kirat-Chatel, S.; Noël, T. Development of an in vitro model for the multi-parametric quantification of the cellular interactions between Candida yeasts and phagocytes. PLoS ONE 2012, 7, e32621. [Google Scholar] [CrossRef]
- Anderson, C.M.; Stenkamp, R.E.; McDonald, R.C.; Steitz, T.A. A refined model of the sugar binding site of yeast Hexokinase, B.J. Mol. Biol. 1978, 123, 207–219. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Fink, G.R. The glyoxylate cycle is required for fungal virulence. Nature 2001, 412, 83–86. [Google Scholar] [CrossRef]
- Ramírez, M.A.; Lorenz, M.C. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot. Cell 2007, 6, 280–290. [Google Scholar] [CrossRef]
- Chew, S.Y.; Chee, W.J.Y.; Than, L.T.L. The glyoxylate cycle and alternative carbon metabolism as metabolic adaptation strategies of Candida glabrata: Perspectives from Candida albicans and Saccharomyces cerevisiae. J. Biomed. Sci. 2019, 26, 52. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Chaturvedi, V.; Shen, S.-H. Identification and phylogenetic analysis of a glucose transporter gene family from the Human pathogenic yeast Candida albicans. J. Mol. Evol. 2002, 55, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Prigneau, O.; Porta, A.; Poudrier, J.A.; Colonna-Romano, S.; Noël, T.; Maresca, B. Genes involved in Beta-oxidation, energy metabolism and glyoxylate cycle are induced by Candida albicans during macrophage infection. Yeast 2003, 20, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Luongo, M.; Porta, A.; Maresca, B. Homology, disruption and phenotypic analysis of CaGS Candida albicans gene induced during macrophage infection. FEMS Immunol. Med. Microbiol. 2005, 45, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Moyes, D.L.; Runglall, M.; Murciano, C.; Shen, C.; Nayar, D.; Thavaraj, S.; Kohli, A.; Islam, A.; Mora-Montes, H.; Challacombe, S.J.; et al. A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe 2010, 8, 225–235. [Google Scholar] [CrossRef]
- Noble, S.M.; Gianetti, B.A.; Witchley, J.N. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol. 2017, 15, 96–108. [Google Scholar] [CrossRef]
- Rao, K.H.; Ruhela, D.; Ghosh, S.; Abdin, M.Z.; Datta, A. N-acetylglucosamine kinase, Hxk1 contributes to white-opaque morphological transition in Candida albicans. Biochem. Biophys. Res. Commun. 2014, 445, 138–144. [Google Scholar] [CrossRef]
- Niimi, K.; Niimi, M.; Shepherd, M.G.; Cannon, R.D. Regulation of N-acetylglucosaminidase production in Candida albicans. Arch. Microbiol. 1997, 168, 464–472. [Google Scholar] [CrossRef]
- Alvarez, F.J.; Konopka, J.B. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Mol. Biol. Cell 2007, 18, 965–975. [Google Scholar] [CrossRef]
- Vialás, V.; Perumal, P.; Gutierrez, D.; Ximénez-Embún, P.; Nombela, C.; Gil, C.; Chaffin, W.L. Cell surface shaving of Candida albicans biofilms, hyphae, and yeast form cells. Proteomics 2012, 12, 2331–2339. [Google Scholar] [CrossRef]
- Gil-Bona, A.; Parra-Giraldo, C.M.; Hernáez, M.L.; Reales-Calderon, J.A.; Solis, N.V.; Filler, S.G.; Monteoliva, L.; Gil, C. Candida albicans cell shaving uncovers new proteins involved in cell wall integrity, yeast to hypha transition, stress response and host-pathogen interaction. J. Proteom. 2015, 127, 340–351. [Google Scholar] [CrossRef]
- Willger, S.D.; Liu, Z.; Olarte, R.A.; Adamo, M.E.; Stajich, J.E.; Myers, L.C.; Kettenbach, A.N.; Hogan, D.A. Analysis of the Candida albicans phosphoproteome. Eukaryot. Cell 2015, 14, 474–485. [Google Scholar] [CrossRef]
- Ma, H.; Bloom, L.M.; Walsh, C.T.; Botstein, D. The residual enzymatic phosphorylation activity of Hexokinase II mutants is correlated with glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1989, 9, 5643–5649. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.; Albig, W.; Entian, K.D. Glucose repression in Saccharomyces cerevisiae is directly associated with hexose phosphorylation by Hexokinases PI and PII. Eur. J. Biochem. 1991, 199, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S.; Winderickx, J.; de Winde, J.H.; Valckx, D.; Cobbaert, P.; Luyten, K.; de Meirsman, C.; Ramos, J.; Thevelein, J.M. Novel alleles of yeast Hexokinase PII with distinct effects on catalytic activity and catabolite repression of SUC2. Microbiology 1999, 145, 703–714. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kraakman, L.; Lemaire, K.; Ma, P.; Teunissen, A.W.; Donaton, M.C.; Van Dijck, P.; Winderickx, J.; de Winde, J.H.; Thevelein, J.M. A Saccharomyces cerevisiae G-Protein Coupled Receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 1999, 32, 1002–1012. [Google Scholar] [CrossRef] [PubMed]
- Espinel, A.E.; Gómez-Toribio, V.; Peinado, J.M. The inactivation of hexokinase activity does not prevent glucose repression in Candida utilis. FEMS Microbiol. Lett. 1996, 135, 327–332. [Google Scholar] [CrossRef][Green Version]
- Lane, S.; Xu, H.; Oh, E.J.; Kim, H.; Lesmana, A.; Jeong, D.; Zhang, G.; Tsai, C.-S.; Jin, Y.-S.; Kim, S.R. Glucose repression can be alleviated by reducing glucose phosphorylation rate in Saccharomyces cerevisiae. Sci. Rep. 2018, 8, 2613. [Google Scholar] [CrossRef] [PubMed]
- Soncini, S.R.; Chandrashekarappa, D.G.; Augustine, D.A.; Callahan, K.P.; O’Donnell, A.F.; Schmidt, M.C. Spontaneous mutations that confer resistance to 2-deoxyglucose act through Hxk2 and Snf1 pathways to regulate gene expression and HXT Endocytosis. PLoS Genet. 2020, 16, e1008484. [Google Scholar] [CrossRef] [PubMed]
- Coccetti, P.; Nicastro, R.; Tripodi, F. Conventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae. Microb. Cell 2018, 5, 482–494. [Google Scholar] [CrossRef]
- Mayer, F.V.; Heath, R.; Underwood, E.; Sanders, M.J.; Carmena, D.; McCartney, R.R.; Leiper, F.C.; Xiao, B.; Jing, C.; Walker, P.A.; et al. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 2011, 14, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Bain, J.; Gow, N.A.R.; Erwig, L.-P. Novel insights into host-fungal pathogen interactions derived from live-cell imaging. Semin. Immunopathol. 2015, 37, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.C.; Fink, G.R. Life and death in a macrophage: Role of the glyoxylate cycle in virulence. Eukaryot. Cell 2002, 1, 657–662. [Google Scholar] [CrossRef]
- Ene, I.V.; Adya, A.K.; Wehmeier, S.; Brand, A.C.; MacCallum, D.M.; Gow, N.A.R.; Brown, A.J.P. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell. Microbiol. 2012, 14, 1319–1335. [Google Scholar] [CrossRef] [PubMed]
- Barelle, C.J.; Manson, C.L.; MacCallum, D.M.; Odds, F.C.; Gow, N.A.R.; Brown, A.J.P. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast 2004, 21, 333–340. [Google Scholar] [CrossRef]
- Brown, A.J.P.; Brown, G.D.; Netea, M.G.; Gow, N.A.R. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 2014, 22, 614–622. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laurian, R.; Ravent, J.; Dementhon, K.; Lemaire, M.; Soulard, A.; Cotton, P. Candida albicans Hexokinase 2 Challenges the Saccharomyces cerevisiae Moonlight Protein Model. Microorganisms 2021, 9, 848. https://doi.org/10.3390/microorganisms9040848
Laurian R, Ravent J, Dementhon K, Lemaire M, Soulard A, Cotton P. Candida albicans Hexokinase 2 Challenges the Saccharomyces cerevisiae Moonlight Protein Model. Microorganisms. 2021; 9(4):848. https://doi.org/10.3390/microorganisms9040848
Chicago/Turabian StyleLaurian, Romain, Jade Ravent, Karine Dementhon, Marc Lemaire, Alexandre Soulard, and Pascale Cotton. 2021. "Candida albicans Hexokinase 2 Challenges the Saccharomyces cerevisiae Moonlight Protein Model" Microorganisms 9, no. 4: 848. https://doi.org/10.3390/microorganisms9040848
APA StyleLaurian, R., Ravent, J., Dementhon, K., Lemaire, M., Soulard, A., & Cotton, P. (2021). Candida albicans Hexokinase 2 Challenges the Saccharomyces cerevisiae Moonlight Protein Model. Microorganisms, 9(4), 848. https://doi.org/10.3390/microorganisms9040848





