Arabidopsis thaliana Genes Associated with Cucumber mosaic virus Virulence and Their Link to Virus Seed Transmission
Abstract
1. Introduction
2. Materials and Methods
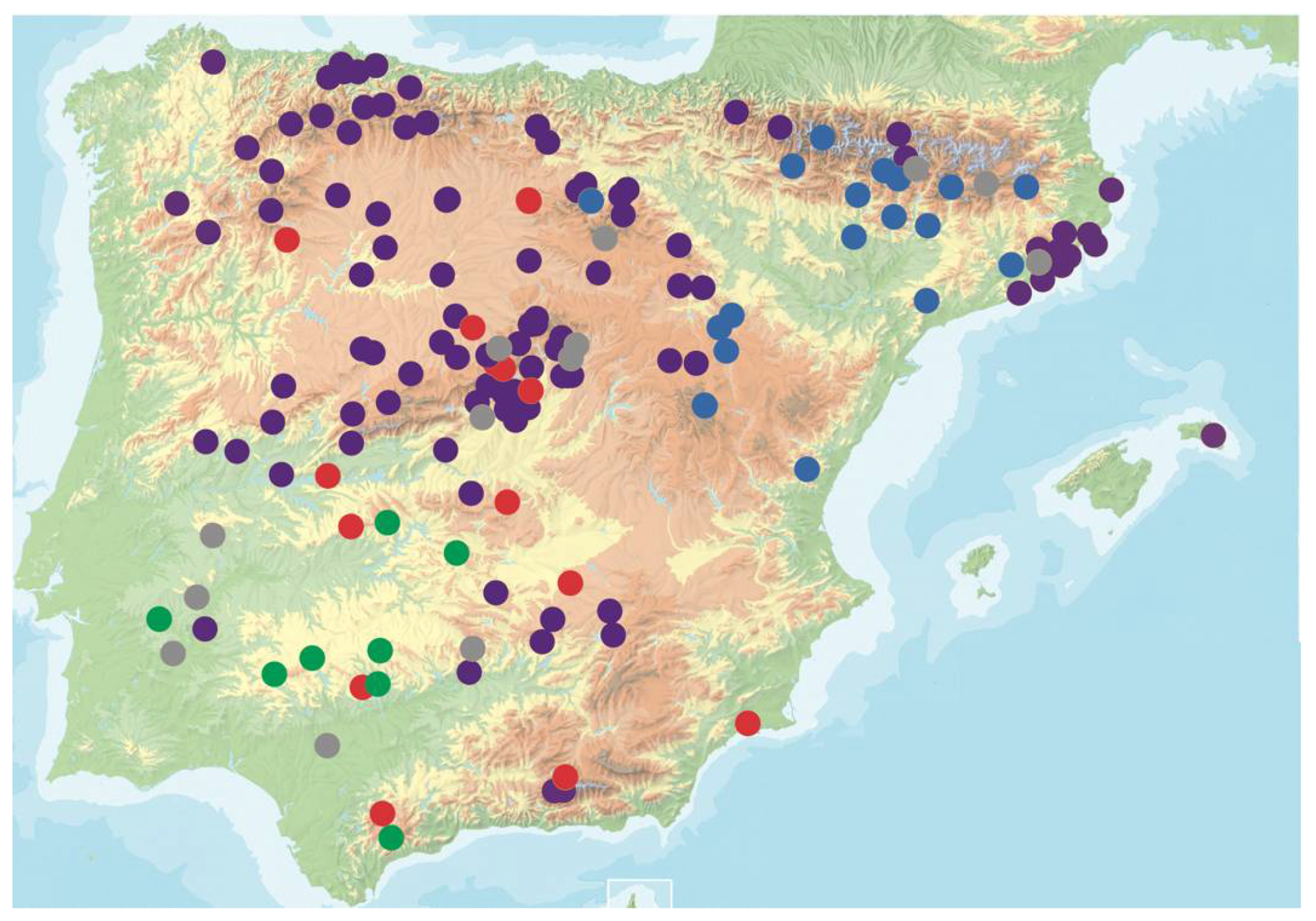
2.1. Plant Material
2.2. Virus Isolate and Inoculation
2.3. Quantification of Virulence
2.4. Efficiency of Virus Seed Transmission
2.5. Data Treatment and Statistical Analyses
2.6. Genome-Wide Association Study (GWAS)
2.7. Bayesian Sparse Linear Mixed Model (BSLMM)
2.8. Selection of SNPs Linking CMV Virulence and Seed Transmission Rate
3. Results
3.1. Natural Variation for CMV Virulence in Arabidopsis Genotypes
3.2. Genetic Architecture of Virulence in Arabidopsis
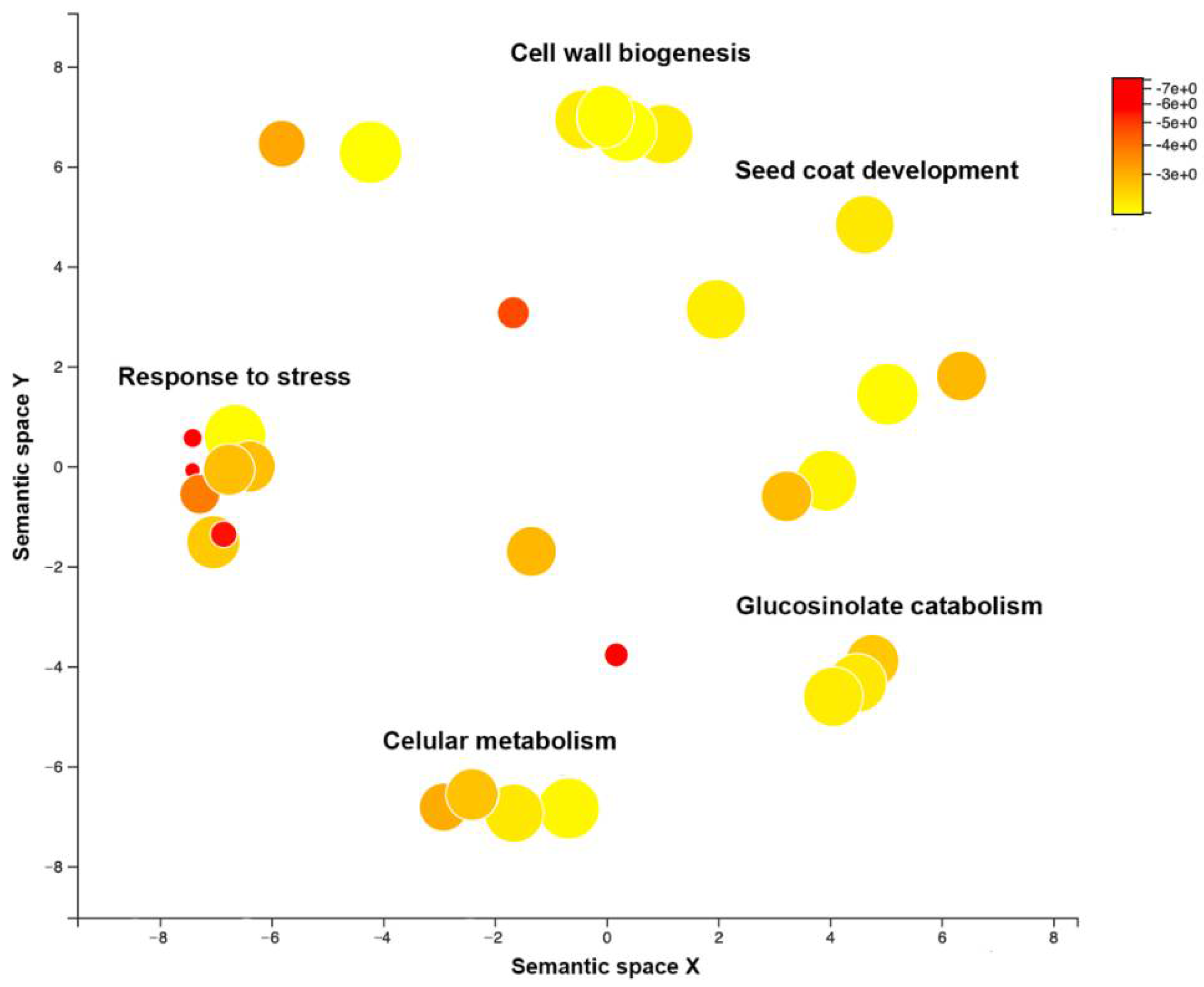
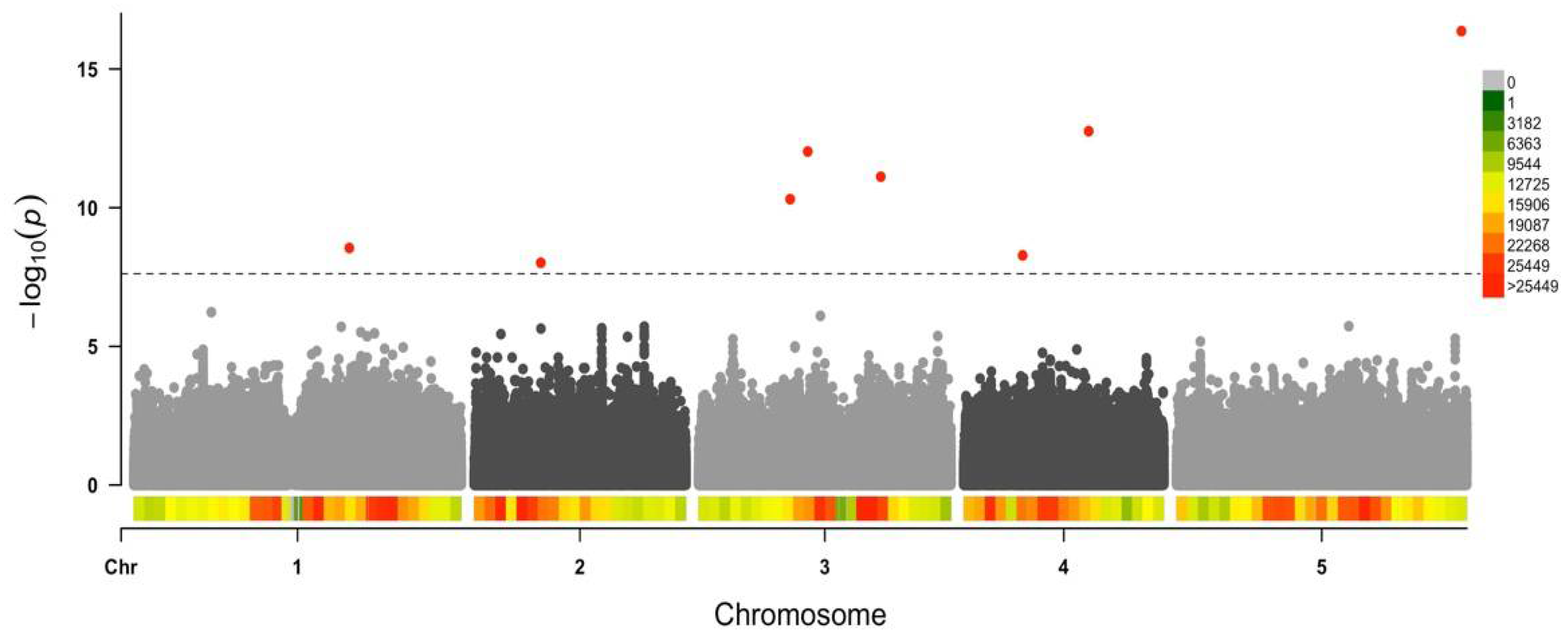
3.3. Arabidopsis Genetic Determinants of CMV Virulence Identified by GWAS
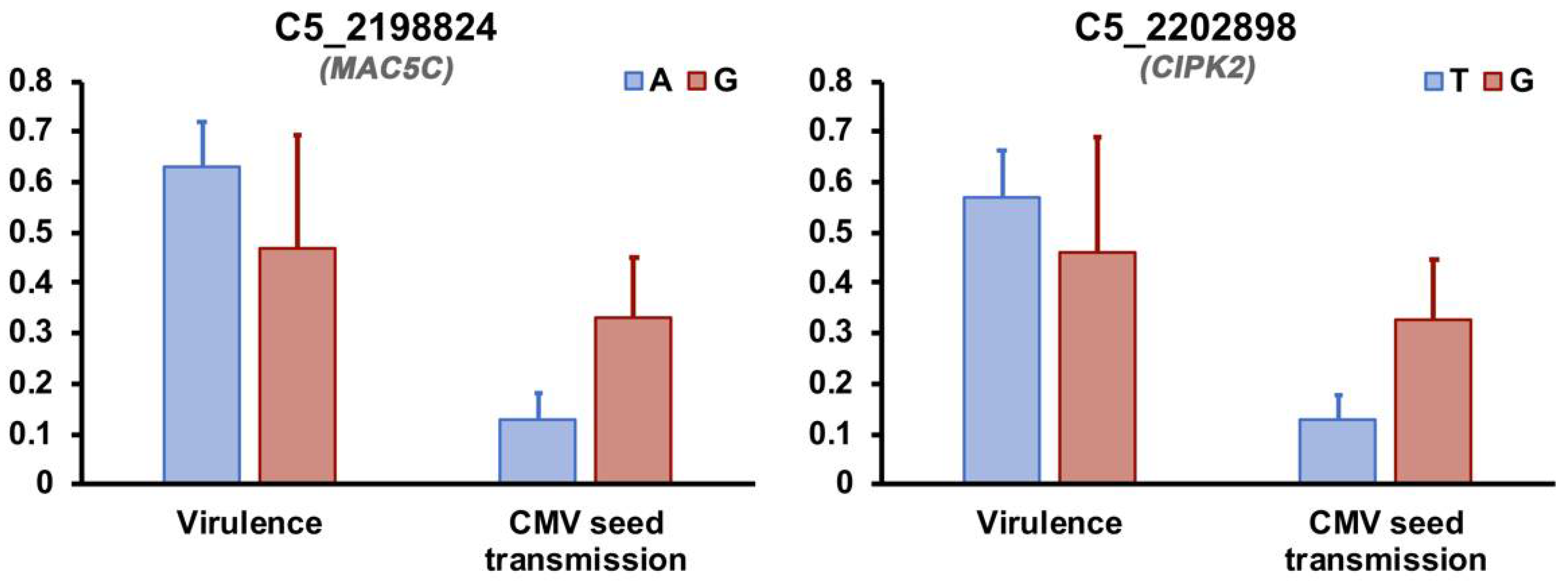
3.4. Link between Arabidopsis Genetic Determinants of CMV Virulence and Seed Transmission
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Read, A.F. The evolution of virulence. Trends Microbiol. 1994, 2, 73–76. [Google Scholar] [CrossRef]
- Oerke, E.-C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Pagán, I.; Alonso-Blanco, C.; García-Arenal, F. The relationship of within-host multiplication and virulence in a plant-virus system. PLoS ONE 2007, 2, e786. [Google Scholar] [CrossRef]
- Pallás, V.; García, J.A. How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 2011, 92, 2691–2705. [Google Scholar] [CrossRef]
- Paudel, D.B.; Sanfaçon, H. Exploring the diversity of mechanisms associated with plant tolerance to virus infection. Front. Plant Sci. 2018, 9, 1575. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Pagán, I.; García-Arenal, F. Effective tolerance based on resource reallocation is a virus-specific defence in Arabidopsis thaliana. Mol. Plant Pathol. 2018, 19, 1454–1465. [Google Scholar] [CrossRef]
- García, J.A.; Pallás, V. Viral factors involved in plant pathogenesis. Curr. Opt. Virol. 2015, 11, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P. Determinants of pathogenesis. In Cucumber Mosaic Virus; Palukaitis, P., García-Arenal, F., Eds.; APS Press: Washington, DC, USA, 2019; pp. 145–154. [Google Scholar]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed]
- García-Ruíz, H. Host factors against plant viruses. Mol. Plant Pathol. 2019, 20, 1588–1601. [Google Scholar] [CrossRef] [PubMed]
- Dieckmann, U.; Metz, J.A.; Sabelis, M.W.; Sigmund, K. Adaptive dynamics of infectious diseases. In Pursuit of Virulence Management; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Kang, B.-C.; Yeam, I.; Jahn, M.M. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 2005, 43, 581–621. [Google Scholar] [CrossRef]
- de Ronde, D.; Butterbach, P.; Kormelink, R. Dominant resistance against plant viruses. Front. Plant Sci. 2014, 5, 307. [Google Scholar] [CrossRef] [PubMed]
- Faoro, F.; Gozzo, F. Is modulating virus virulence by induced systemic resistance realistic? Plant Sci. 2015, 234, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.P.; Murphy, A.M. Host responses: Susceptibility. In Cucumber Mosaic Virus; Palukaitis, P., García-Arenal, F., Eds.; APS Press: Washington, DC, USA, 2019; pp. 47–58. [Google Scholar]
- Drummond, D.A.; Raval, A.; Wilke, C.O. A single determinant dominates the rate of yeast protein evolution. Mol. Biol. Evol. 2006, 23, 327–3337. [Google Scholar] [CrossRef] [PubMed]
- Pagán, I.; García-Arenal, F. Tolerance to plant pathogens: Theory and experimental evidence. Int. J. Mol. Sci. 2018, 19, 810. [Google Scholar] [CrossRef]
- Malmstrom, C.M.; McCullough, A.J.; Johnson, H.A.; Newton, L.A.; Borer, E.T. Invasive annual grasses indirectly increase virus incidence in California native perennial bunchgrasses. Oecologia 2005, 145, 153–164. [Google Scholar] [CrossRef]
- Vijayan, V.; López-González, S.; Sánchez, F.; Ponz, F.; Pagán, I. Virulence evolution of a sterilizing plant virus: Tuning multiplication and resource exploitation. Virus Evol. 2017, 3, vex033. [Google Scholar] [CrossRef] [PubMed]
- Pagán, I.; Fraile, A.; Fernández-Fueyo, E.; Montes, N.; Alonso-Blanco, C.; García-Arenal, F. Arabidopsis thaliana as a model for the study of plant–virus co-evolution. Philos. Trans. R. Soc. Lond. B 2010, 365, 1983–1995. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Pérez, M.G.; Pagán, I.; Aragón-Caballero, L.; Cáceres, F.; Fraile, A.; García-Arenal, F. Ecological and genetic determinants of Pepino mosaic virus emergence. J. Virol. 2014, 88, 3359–43368. [Google Scholar] [CrossRef]
- Montes, N.; Alonso-Blanco, C.; García-Arenal, F. Cucumber mosaic virus infection as a potential selective pressure on Arabidopsis thaliana populations. PLoS Pathog. 2019, 15, e1007810. [Google Scholar] [CrossRef]
- Pagán, I.; García-Arenal, F. Tolerance of plants to plant pathogens: A unifying view. Annu Rev. Plant Pathol. 2020, 18, 9.1–9.20. [Google Scholar] [CrossRef]
- Pagán, I.; Alonso-Blanco, C.; García-Arenal, F. Host responses in life-history traits and tolerance to virus infection in Arabidopsis thaliana. PLoS Pathog. 2008, 4, e1000124. [Google Scholar] [CrossRef] [PubMed]
- Montes, N.; Vijayan, V.; Pagán, I. Trade-offs between host tolerances to different pathogens in plant-virus interactions. Virus Evol. 2020, 6, veaa019. [Google Scholar] [CrossRef] [PubMed]
- Sastry, K.S. Seed-Borne Plant Virus Diseases; Springer: New Delhi, India, 2013. [Google Scholar]
- Cobos, A.; Montes, N.; López-Herranz, M.; Gil-Valle, M.; Pagán, I. Within-host multiplication and speed of colonization as infection traits associated with plant virus vertical transmission. J. Virol. 2019, 93, e01078-19. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P.; García-Arenal, F. Cucumber Mosaic Virus; APS Press: Washington, DC, USA, 2019. [Google Scholar]
- Brennan, A.C.; Méndez-Vigo, B.; Haddioui, A.; Martínez-Zapater, J.M.; Picó, F.X.; Alonso-Blanco, C. The genetic structure of Arabidopsis thaliana in the south-western Mediterranean range reveals a shared history between North Africa and southern Europe. BMC Plant Biol. 2014, 14, 17. [Google Scholar] [CrossRef]
- Lee, C.R.; Svardal, H.; Farlow, A.; Exposito-Alonso, M.; Ding, W.; Novikova, P.; Alonso-Blanco, C.; Weigel, D.; Nordborg, M. On the post-glacial spread of human commensal Arabidopsis thaliana. Nat. Commun. 2017, 8, 14458. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, T.H.; Emerson, B.C. Functional variation in a disease resistance gene in populations of Arabidopsis thaliana. Mol. Ecol. 2008, 17, 4912–4923. [Google Scholar] [CrossRef]
- Huard-Chauveau, C.; Perchepied, L.; Debieu, M.; Rivas, S.; Kroj, T.; Kars, I.; Bergelson, J.; Roux, F.; Roby, D. An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PLoS Genet. 2013, 9, e1003766. [Google Scholar] [CrossRef] [PubMed]
- Karasov, T.L.; Kniskern, J.M.; Gao, L.; DeYoung, B.J.; Ding, J.; Dubiella, U.; Lastra, R.O.; Nallu, S.; Roux, F.; Innes, R.W.; et al. The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 2014, 512, 436–440. [Google Scholar] [CrossRef] [PubMed]
- The 1001 Genomes Consortium. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 2016, 166, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Picó, F.X.; Méndez-Vigo, B.; Martinez-Zapater, J.M.; Alonso-Blanco, C. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian Peninsula. Genetics 2008, 180, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Vigo, B.; Picó, F.X.; Ramiro, M.; Martinez-Zapater, J.M.; Alonso-Blanco, C. Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol. 2011, 157, 1942–1955. [Google Scholar] [CrossRef]
- Gomaa, N.H.; Montesinos-Navarro, A.; Alonso-Blanco, C.; Picó, F.X. Temporal variation in genetic diversity and effective population size of Mediterranean and subalpine Arabidopsis thaliana populations. Mol. Ecol. 2001, 20, 3540–3554. [Google Scholar] [CrossRef] [PubMed]
- Tabas-Madrid, D.; Méndez-Vigo, B.; Arteaga, N.; Marcer, A.; Pascual-Montano, A.; Weigel, D.; Picó, F.X.; Alonso-Blanco, C. Genome-wide signatures of flowering adaptation to climate temperature: Regional analyses in a highly diverse native range of Arabidopsis thaliana. Plant Cell Environ. 2018, 41, 1806–1820. [Google Scholar] [CrossRef] [PubMed]
- Manzano-Piedras, E.; Marcer, A.; Alonso-Blanco, C.; Picó, F.X. Deciphering the adjustment between environment and life history in annuals: Lessons from a geographically-explicit approach in Arabidopsis thaliana. PLoS ONE 2014, 9, e87836. [Google Scholar] [CrossRef] [PubMed]
- Vidigal, D.S.; Marques, A.C.; Willems, L.A.; Buijs, G.; Méndez-Vigo, B.; Hilhorst, H.W.; Bentsink, L.; Picó, F.X.; Alonso-Blanco, C. Altitudinal and climatic associations of seed dormancy and flowering traits evidence adaptation of annual life cycle timing in Arabidopsis thaliana. Plant Cell Environ. 2016, 39, 1737–1748. [Google Scholar] [CrossRef]
- Rizzo, T.M.; Palukaitis, P. Construction of full-length cDNA clones of cucumber mosaic virus RNAs 1, 2 and 3: Generation of infectious RNA transcripts. Mol. Gen. Genet. 1990, 222, 249–256. [Google Scholar] [CrossRef]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Görlach, J. Growth stage–based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer Associates Inc.: Sunderland, MA, USA, 1998. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models using lme4. J. Stat. Soft. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Tang, Y.; Liu, X.; Wang, J.; Li, M.; Wang, Q.; Tian, F.; Su, Z.; Pan, Y.; Liu, D.; Lipka, A.E.; et al. GAPIT Version 2: An enhanced integrated tool for genomic association and prediction. Plant Genome 2016, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, Z.; Li, X.; et al. rMVP: Memory-Efficient, Visualize-Enhanced, Parallel-Accelerated GWAS Tool. R Package Version 1.0.4. 2020. Available online: https://CRAN.R-project.org/package=rMVP (accessed on 26 February 2021).
- Van Rooijen, R.; Aarts, M.G.M.; Harbinson, J. Natural genetic variation for acclimation of photosynthetic light use efficiency to growth irradiance in Arabidopsis. Plant Physiol. 2015, 167, 1412–1484. [Google Scholar] [CrossRef] [PubMed]
- Thoen, M.P.M.; Olivas, N.H.D.; Kloth, K.J.; Coolen, S.; Huang, P.-P.; Aarts, M.G.M.; Bac-Molenaar, J.A.; Bakker, J.; Bouwmeester, H.J.; Broekgaarden, C.; et al. Genetic architecture of plant stress resistance: Multi-trait genome-wide association mapping. New Phytol. 2017, 213, 1346–1362. [Google Scholar] [CrossRef]
- Rubio, B.; Cosson, P.; Caballero, M.; Revers, F.; Bergelson, J.; Roux, F.; Schurdi-Levraud, V. Genome-wide association study reveals new loci involved in Arabidopsis thaliana and Turnip mosaic virus (TuMV) interactions in the field. New Phytol. 2019, 221, 2026–2038. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wangle, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucl. Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Carbonetto, P.; Stephens, M. Polygenic modeling with Bayesian Sparse Linear Mixed Models. PLoS Genet. 2013, 9, e1003264. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kuhn, M. Caret: Classification and Regression Training. R Package Version 6.0-86. 2020. Available online: https://CRAN.R-project.org/package=caret (accessed on 26 February 2021).
- Janitza, S.; Celik, E.; Boulesteix, A. A computationally fast variable importance test for random forests for high-dimensional data. Adv. Data Anal. Classif. 2018, 12, 885–915. [Google Scholar] [CrossRef]
- Szymczak, S.; Holzinger, E.; Dasgupta, A.; Malley, J.D.; Molloy, A.M.; Mills, J.L.; Brody, L.C.; Stambolian, D.; Bailey-Wilson, J.E. r2VIM: A new variable selection method for random forests in genome-wide association studies. BioData Min. 2016, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bonin, C.P.; Freshour, G.; Hahn, M.G.; Vanzin, G.F.; Reiter, W.-D. The GMD1 and GMD2 genes of Arabidopsis encode isoforms of GDP-D-mannose 4,6-dehydratase with cell type-specific expression patterns. Plant Physiol. 2003, 132, 883–892. [Google Scholar] [CrossRef]
- Sun, L.; van Nocker, S. Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis. BMC Plant Biol. 2010, 10, 152. [Google Scholar] [CrossRef]
- Ciftci-Yilmaza, S.; Mittler, R. The zinc finger network of plants. Cell Mol. Life Sci. 2008, 65, 1150–1160. [Google Scholar] [CrossRef]
- Waters, E.R.; Vierling, E. Plant small heat shock proteins—Evolutionary and functional diversity. New Phytol. 2020, 227, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Yin, M. Genetic Dissection of Nitric Oxide Signalling Network in Plant Defence Response. Ph.D. Thesis, The University of Edinburgh, Edinburgh, UK, 2014. Available online: http://hdl.handle.net/1842/10462 (accessed on 26 February 2021).
- Li, J.; Xiang, C.-Y.; Yang, J.; Chen, P.-P.; Zhang, H.-M. Interaction of HSP20 with a viral RdRp changes its sub-cellular localization and distribution pattern in plants. Sci. Rep. 2015, 5, 14016. [Google Scholar] [CrossRef] [PubMed]
- Knoth, C.; Eulgem, T. The oomycete response gene LURP1 is required for defense against Hyaloperonospora parasitica in Arabidopsis thaliana. Plant J. 2008, 55, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Davin, N.; Edger, P.P.; Hefer, C.A.; Mizrachi, E.; Schuetz, M.; Smets, E.; Myburg, A.A.; Douglas, C.J.; Schranz, M.E.; Lens, F. Functional network analysis of genes differentially expressed during xylogenesis in soc1ful woody Arabidopsis plants. Plant J. 2016, 86, 376–390. [Google Scholar] [CrossRef] [PubMed]
- Kraft, E.; Bostick, M.; Jacobsen, S.E.; Callis, J. ORTH/VIM proteins that regulate DNA methylation are functional ubiquitin E3 ligases. Plant J. 2008, 56, 704–715. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, T.; Li, T.; Wang, M.; Yang, G.; He, G. A CBL-interacting protein kinase TaCIPK2 confers drought tolerance in transgenic tobacco plants through regulating the stomatal movement. PLoS ONE 2016, 11, e0167962. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, C.; Roux, F. Genome-Wide Association Studies in plant pathosystems: Toward an ecological genomics approach. Front. Plant Sci. 2017, 8, 763. [Google Scholar] [CrossRef] [PubMed]
- Madden, L.V.; Jeger, M.J.; van den Bosch, F. A theoretical assessment of the effects of vector-virus transmission mechanism on plant virus disease epidemics. Phytopathology 2000, 90, 576–594. [Google Scholar] [CrossRef]
- Hily, J.M.; García, A.; Moreno, A.; Plaza, M.; Wilkinson, M.D.; Fereres, A.; Fraile, A.; García-Arenal, F. The relationship between host lifespan and pathogen reservoir potential: An analysis in the system Arabidopsis thaliana-Cucumber mosaic virus. PLoS Pathog. 2014, 10, e1004492. [Google Scholar] [CrossRef]
- Pagán, I. Movement between plants: Vertical transmission. In Cucumber Mosaic Virus; Palukaitis, P., García-Arenal, F., Eds.; APS Press: Washington, DC, USA, 2019; pp. 185–198. [Google Scholar]
- Yin, M.; Wang, Y.; Zhang, L.; Li, J.; Quan, W.; Yang, L.; Wang, Q.; Chan, Z. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J. Exp. Bot. 2017, 68, 2991–3005. [Google Scholar] [CrossRef] [PubMed]
- Sewelam, N.; Kazan, K.; Hüdig, M.; Maurino, V.G.; Schenk, P.M. The AtHSP17.4C1 gene expression is mediated by diverse signals that link biotic and abiotic stress factors with ROS and can be a useful molecular marker for oxidative stress. Int. J. Mol. Sci. 2019, 20, 3201. [Google Scholar] [CrossRef] [PubMed]
- Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-X.; Zhang, H.-X.; Wei, A.-M.; Gong, Z.-H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef]
- Senthil, G.; Liu, H.; Puram, V.G.; Clark, A.; Stromberg, A.; Goodin, M.M. Specific and common changes in Nicotiana benthamiana gene expression in response to infection by enveloped viruses. J. Gen. Virol. 2005, 86, 2615–2625. [Google Scholar] [CrossRef] [PubMed]
- Mitsuya, Y.; Takahashi, Y.; Berberich, T.; Miyazaki, A.; Matsumura, H.; Takahashi, H.; Terauchi, R.; Kusano, T. Spermine signaling plays a significant role in the defense response of Arabidopsis thaliana to cucumber mosaic virus. J. Plant Physiol. 2009, 166, 626–643. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Miller, J.; Nozaki, Y.; Takeda, M.; Shah, J.; Hase, S.; Ikegami, M.; Ehara, Y.; Dinesh-Kumar, S.P.; Sukamto. RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J. 2002, 32, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Herlihy, J.; Ludwig, N.R.; van den Ackerveken, G.; McDowell, J.M. Oomycetes used in Arabidopsis research. In The Arabidopsis Book; BioOne: Washington, DC, USA, 2019; pp. 1–26. [Google Scholar]
- Bellincampi, D.; Cervone, F.; Lionetti, V. Plant cell wall dynamics and wall-related susceptibility in plant–pathogen interactions. Front. Plant Sci. 2014, 5, 228. [Google Scholar] [CrossRef] [PubMed]
- Bethke, G.; Thao, A.; Xiong, G.; Li, B.; Soltis, N.E.; Hatsugai, N.; Hillmer, R.A.; Katagiri, F.; Kliebenstein, D.J.; Pauly, M.; et al. Pectin biosynthesis is critical for cell wall integrity and immunity in Arabidopsis thaliana. Plant Cell 2016, 28, 537–556. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-H.; Sheng, J.; Hind, G.; Handa, A.K.; Citovsky, V. Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 2000, 5, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Raiola, A.; Cervone, F.; Bellincampi, D. Transgenic expression of pectin methylesterase inhibitors limits tobamovirus spread in tobacco and Arabidopsis. Mol. Plant. Pathol. 2014, 15, 265–274. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Lockhart, B.E.L. Plant cell wall dynamics incompatible and incompatible potato response to infection caused by Potato Virus Y (PVYNTN). Int. J. Mol. Sci. 2018, 19, 862. [Google Scholar] [CrossRef]
- Vlad, F.; Spano, T.; Vlad, D.; Bou Daher, F.; Ouelhadj, A.; Kalaitzis, P. Arabidopsis prolyl 4-hydroxylases are differentially expressed in response to hypoxia, anoxia and mechanical wounding. Physiol. Plant. 2007, 130, 471–483. [Google Scholar] [CrossRef]
- Hily, J.M.; Poulicard, N.; Mora, M.A.; Pagán, I.; García-Arenal, F. Environment and host genotype determine the outcome of a plant-virus interaction: From antagonism to mutualism. New Phytol. 2016, 209, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yu, Y.; Ruan, Y.; Meyer, D.; Wolff, M.; Xu, L.; Wang, N.; Steinmetz, A.; Shen, W.-H. Plant SET- and RING-associated domain proteins in heterochromatinization. Plant J. 2007, 52, 914–926. [Google Scholar] [CrossRef] [PubMed]
- Baulcome, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Du, P.; Wang, X.; Yu, Y.Q.; Qiu, Y.H.; Li, W.; Gal-On, A.; Zhou, C.; Li, Y.; Ding, S.W. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14613. [Google Scholar] [CrossRef]
- Sardar, A.; Nandi, A.K.; Chattopadhyay, D. CBL-interacting protein kinase 6 negatively regulates immune response to Pseudomonas syringae in Arabidopsis. J. Exp. Bot. 2017, 68, 3573–3584. [Google Scholar] [CrossRef] [PubMed]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high-resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, B.J.; Pearce, S.; van Bolderen-Veldkamp, R.P.; Marshall, A.; Widera, P.; Gilbert, J.; Drost, H.G.; Bassel, G.W.; Müller, K.; King, J.R.; et al. Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant. Physiol. 2013, 163, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.; Zhang, B.; You, C.; Zhang, Y.; Zeng, L.; Li, S.; Johnson, K.C.M.; Yu, B.; Li, X.; Chen, X. The Arabidopsis MOS4-Associated Complex promotes microRNA biogenesis and precursor messenger RNA splicing. Plant Cell 2017, 29, 2626–2643. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, J.; Xu, F.; Xu, S.; Zhang, Y.; Li, X. Two putative RNA-binding proteins function with unequal genetic redundancy in the MOS4-associated complex. Plant. Physiol. 2010, 154, 1783–1793. [Google Scholar] [CrossRef] [PubMed]
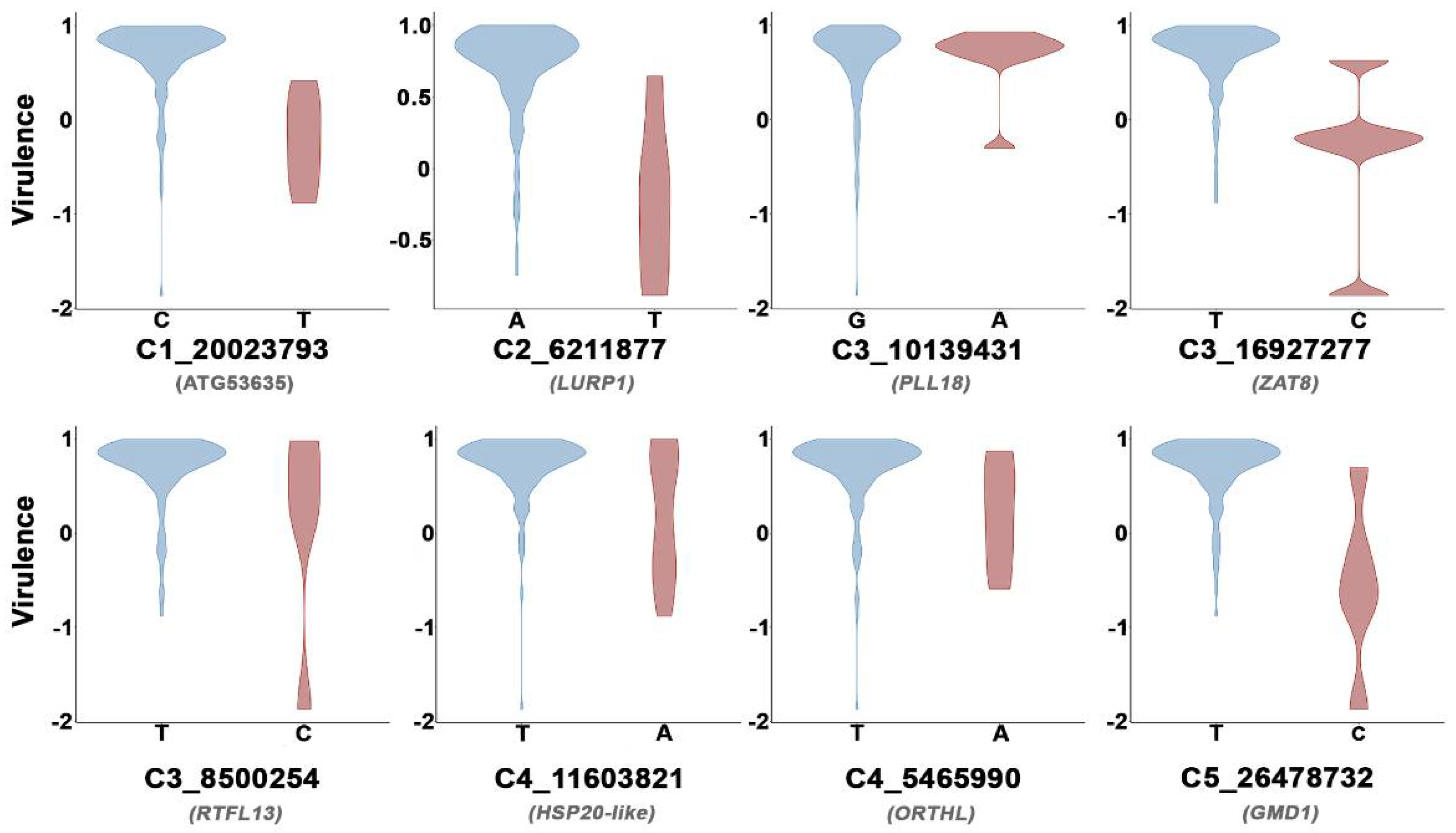
| SNP a | Protein | Description | FDR | Effect |
|---|---|---|---|---|
| C5_26478732 | GMD1 | Cell wall metabolism | 8.92 × 10−11 | −0.42 |
| C3_10139431 | PLL18 | Cell wall metabolism | 6.53 × 10−7 | −0.40 |
| C4_11603821 | HSP20-like | Abiotic stress response | 1.81 × 10−7 | −0.22 |
| C3_16927277 | ZAT8 | Biotic and abiotic stress response | 3.96 × 10−6 | −0.30 |
| C3_8500254 | RTFL13 | Flowering time | 2.05 × 10−5 | −0.23 |
| C1_20023793 | AT1G53635 | Unknown | 9.81 × 10−4 | −0.23 |
| C4_5465990 | ORTHL | DNA methylation | 1.56 × 10−3 | −0.21 |
| C2_6211877 | LURP1 | Biotic stress response | 2.51 × 10−3 | −0.22 |
| SNP a | Protein | Description | p-Value | % Inc. MSE b |
|---|---|---|---|---|
| C5_2202898 | CIPK2 | Response to abiotic stress | 0.012 | 8.11 |
| C5_2198824 | MAC5C | Cell wall metabolism | 0.012 | 2.77 |
| C5_11777493 | AT5G31963 | Unknown | 2 × 10−16 | 0.83 |
| C2_7038732 | MEG | Embryogenesis | 2 × 10−16 | 0.73 |
| C3_15811438 | AT3G44030 | Unknown | 0.041 | 0.72 |
| C1_12253373 | JAL4 | Response to biotic stress | 0.041 | 0.53 |
| C1_16528990 | AT1G43750 | Unknown | 0.033 | 0.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montes, N.; Cobos, A.; Gil-Valle, M.; Caro, E.; Pagán, I. Arabidopsis thaliana Genes Associated with Cucumber mosaic virus Virulence and Their Link to Virus Seed Transmission. Microorganisms 2021, 9, 692. https://doi.org/10.3390/microorganisms9040692
Montes N, Cobos A, Gil-Valle M, Caro E, Pagán I. Arabidopsis thaliana Genes Associated with Cucumber mosaic virus Virulence and Their Link to Virus Seed Transmission. Microorganisms. 2021; 9(4):692. https://doi.org/10.3390/microorganisms9040692
Chicago/Turabian StyleMontes, Nuria, Alberto Cobos, Miriam Gil-Valle, Elena Caro, and Israel Pagán. 2021. "Arabidopsis thaliana Genes Associated with Cucumber mosaic virus Virulence and Their Link to Virus Seed Transmission" Microorganisms 9, no. 4: 692. https://doi.org/10.3390/microorganisms9040692
APA StyleMontes, N., Cobos, A., Gil-Valle, M., Caro, E., & Pagán, I. (2021). Arabidopsis thaliana Genes Associated with Cucumber mosaic virus Virulence and Their Link to Virus Seed Transmission. Microorganisms, 9(4), 692. https://doi.org/10.3390/microorganisms9040692







