Probiotic Propionibacterium freudenreichii MJ2 Enhances Osteoblast Differentiation and Mineralization by Increasing the OPG/RANKL Ratio
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Isolation of Propionibacterium freudenreichii MJ2 and Preparation of Heat-Killed P. freudenreichii MJ2
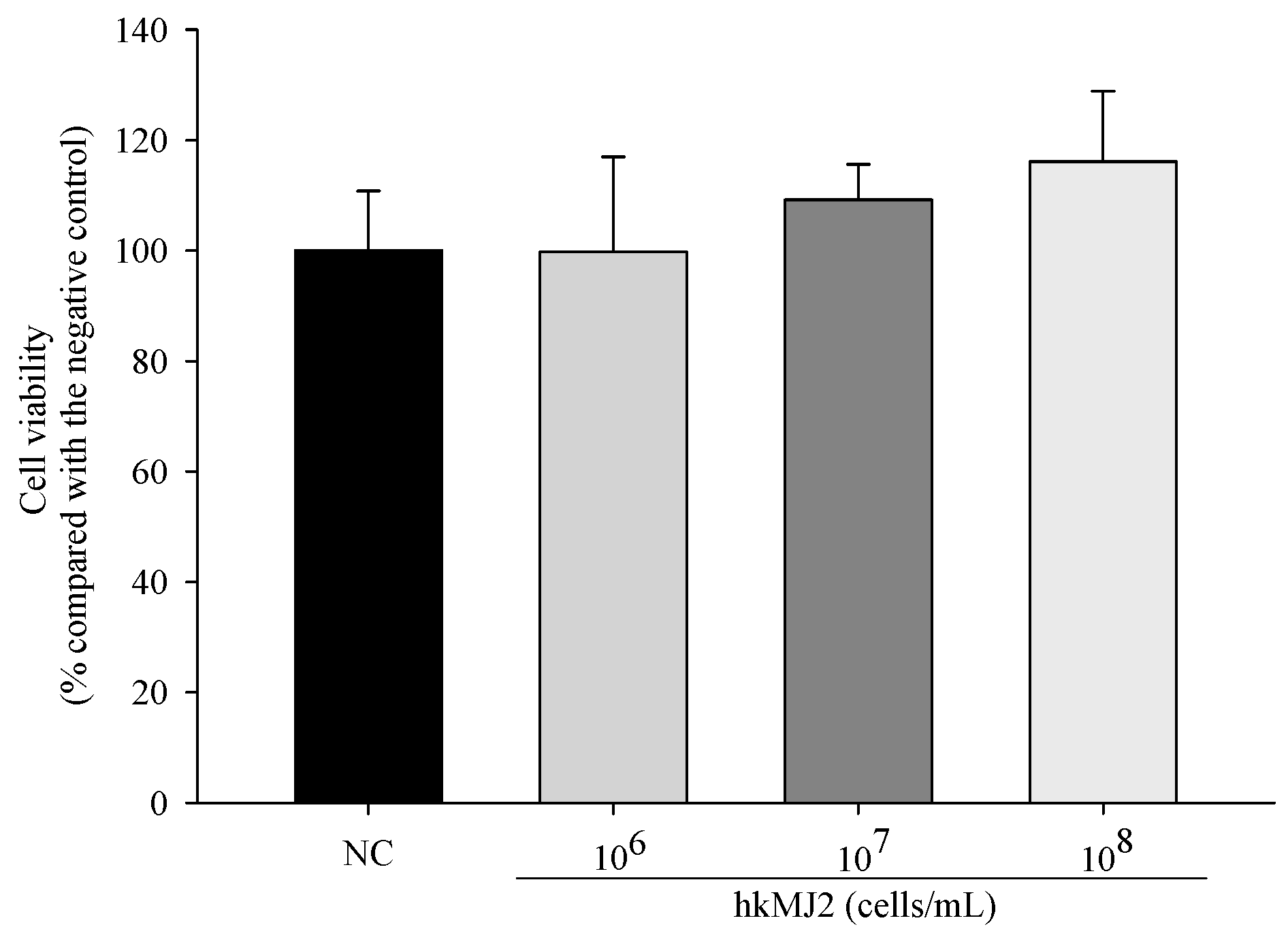
2.4. Measurement of Cell Viability
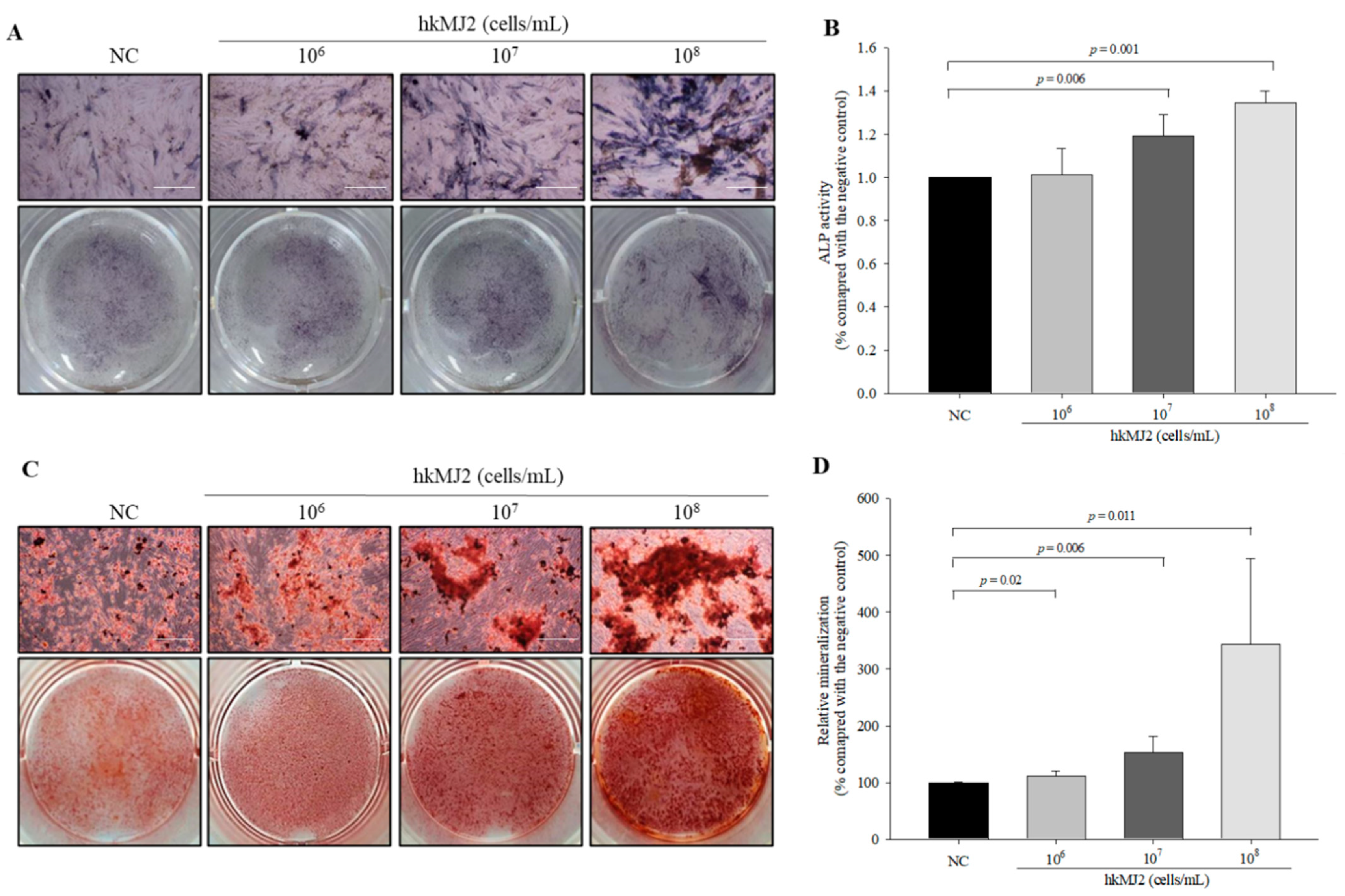
2.5. Alkaline Phosphatase (ALP) Activity Assay
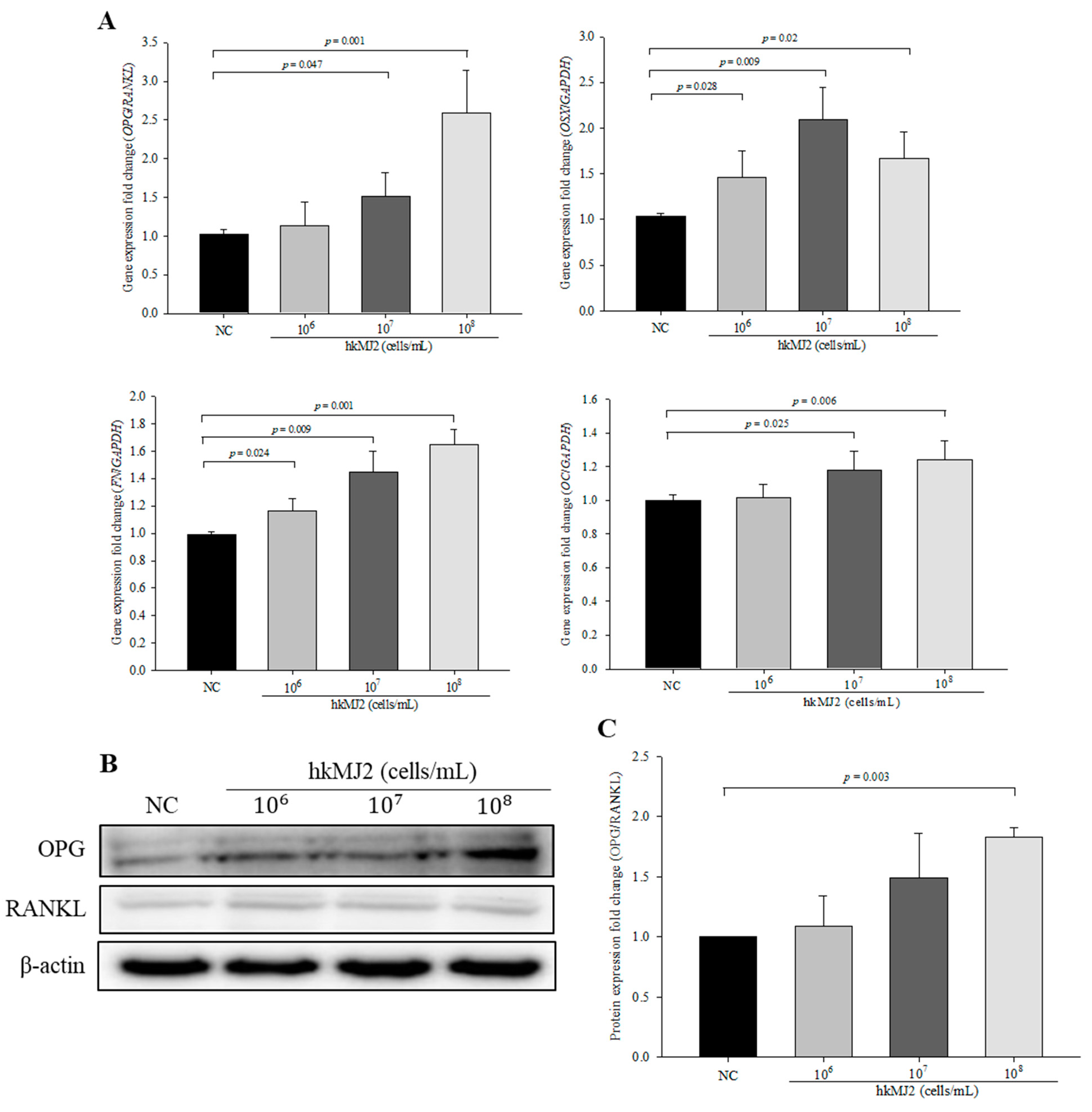
2.6. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.7. Western Blotting
2.8. Two-Dimensional Electrophoresis (2-DE) and LC-MS/MS
2.9. Immunocytochemistry (ICC)
2.10. Alizarin Red S Staining
2.11. Animal Model
2.12. Measurement of Bone Mineral Density (BMD)
2.13. Histological Analysis
2.14. Extraction of Surface Proteins from P. freudenreichii MJ2
2.15. Statistical Analysis
3. Results
3.1. Cytotoxicity of hkMJ2 in hFOB 1.19 Cells
3.2. Increase in ALP Activity and Osteoblast Mineralization Induced by hkMJ2
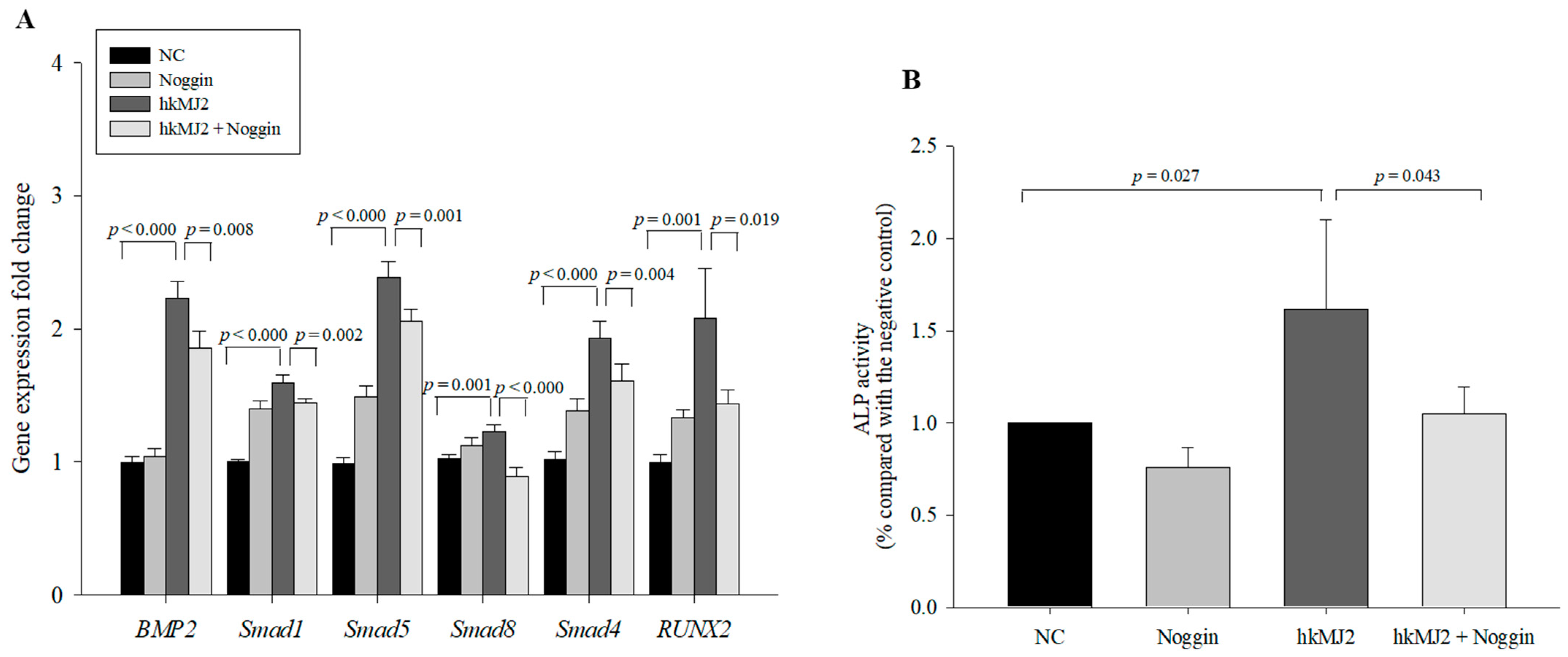
3.3. Expression of Osteoblast Differentiation-Related Genes and Proteins Induced by hkMJ2 Treatment
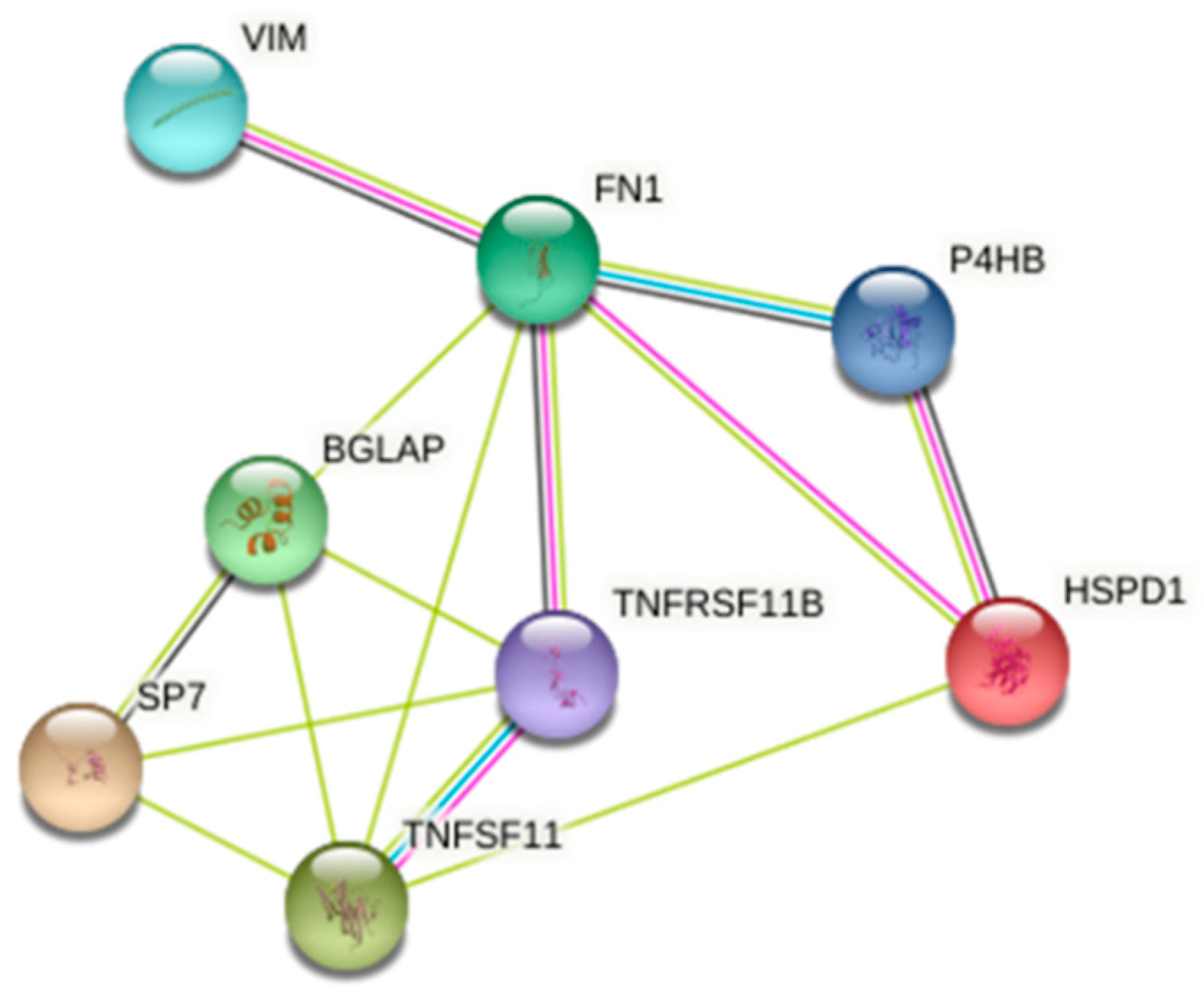
3.4. 2-DE and Protein Identification
3.5. hkMJ2 Treatment Activates the BMP2/RUNX2 Signaling Pathway
3.6. Effect of P. freudenreichii MJ2 (MJ2) and/or L. plantarum (LP) Administration to Ovariectomized (OVX) Rats on Body Weight and Hepatotoxicity
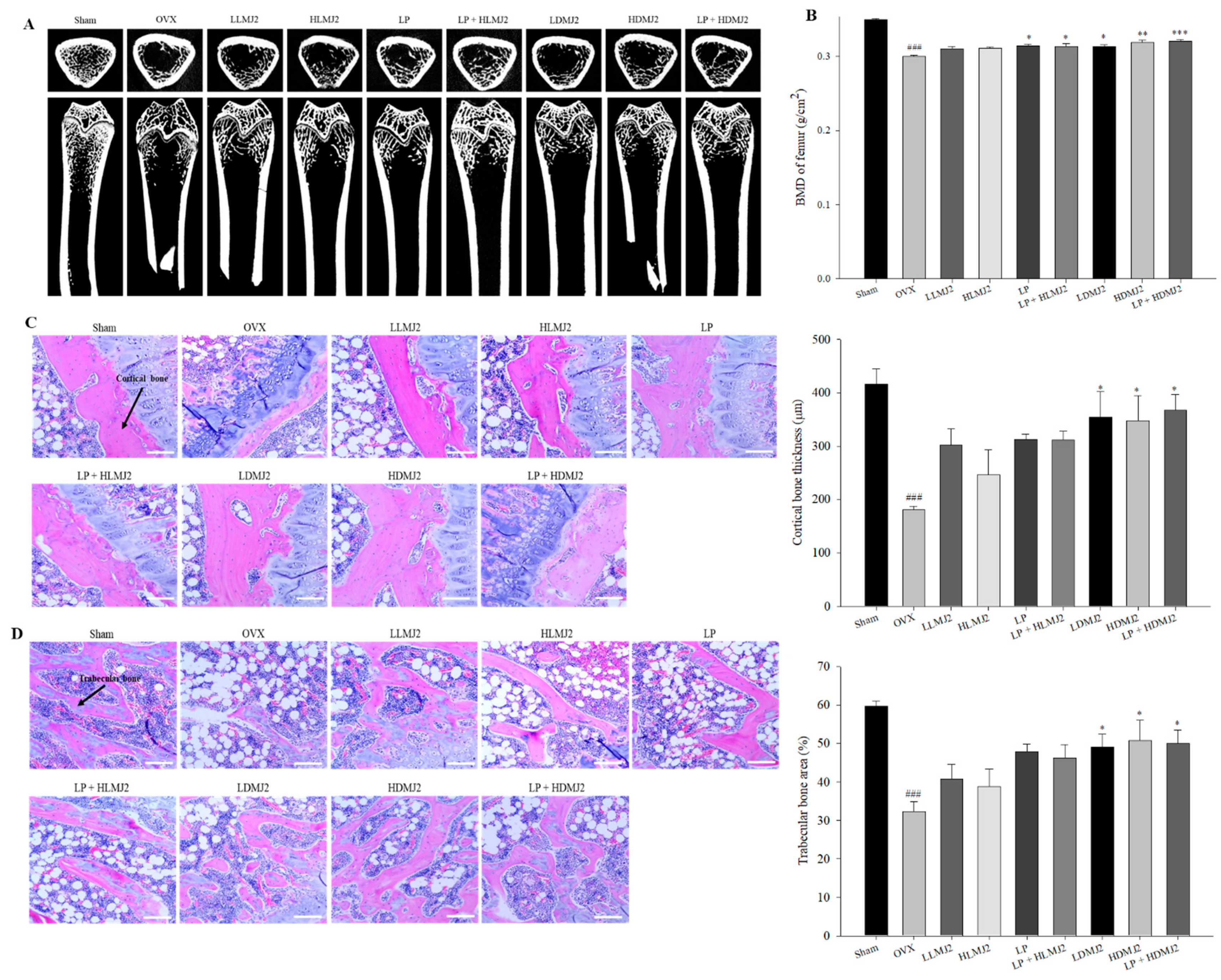
3.7. Improvement of Femoral BMD and Inhibitory Effect on Bone Loss in MJ2-Treated OVX Rats
3.8. Inhibitory Effect on Bone Loss in MJ2-Treated OVX Rats
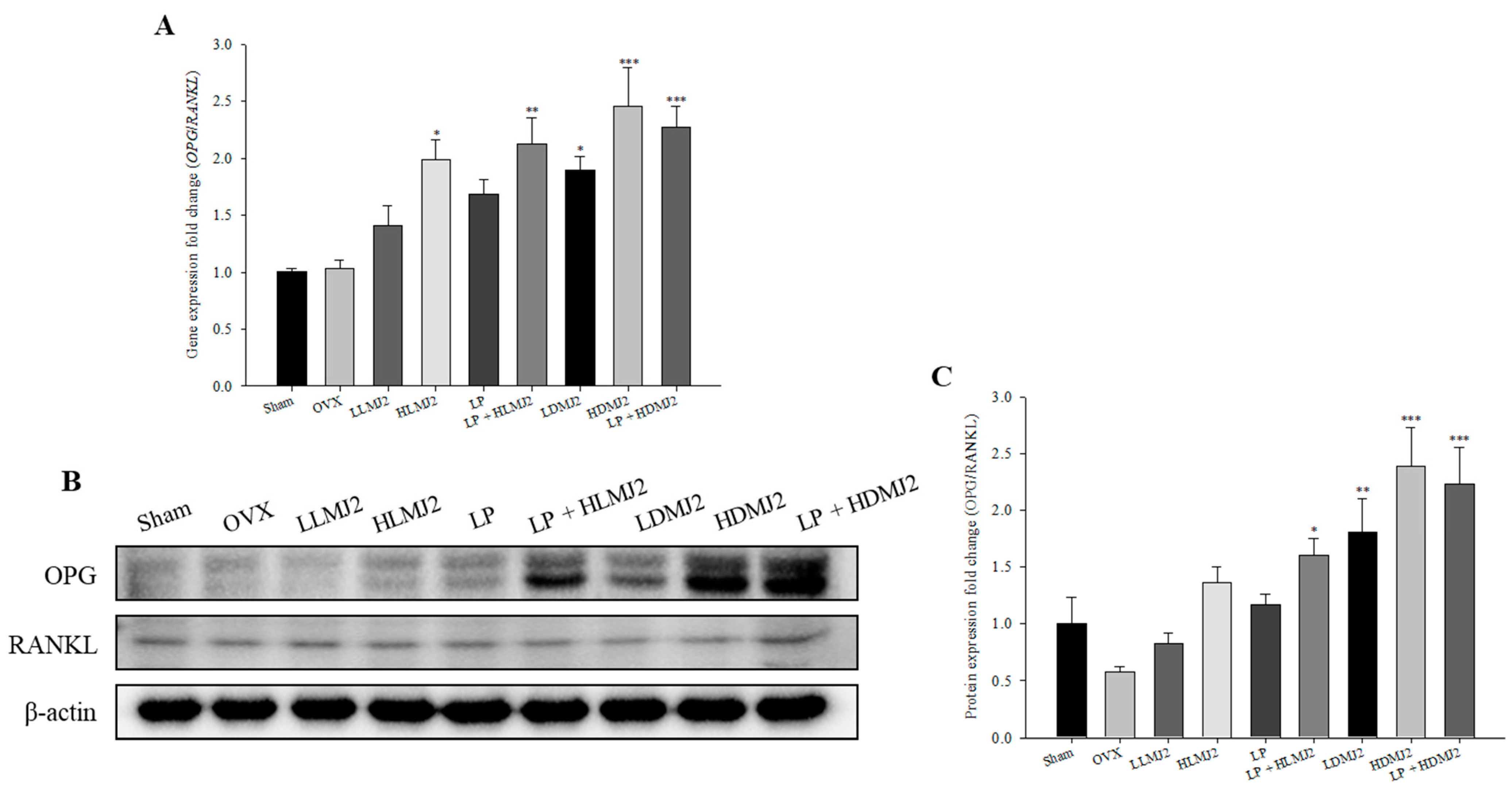
3.9. MJ2-Treated OVX Rats Show an Increase in the OPG/RANKL Ratio
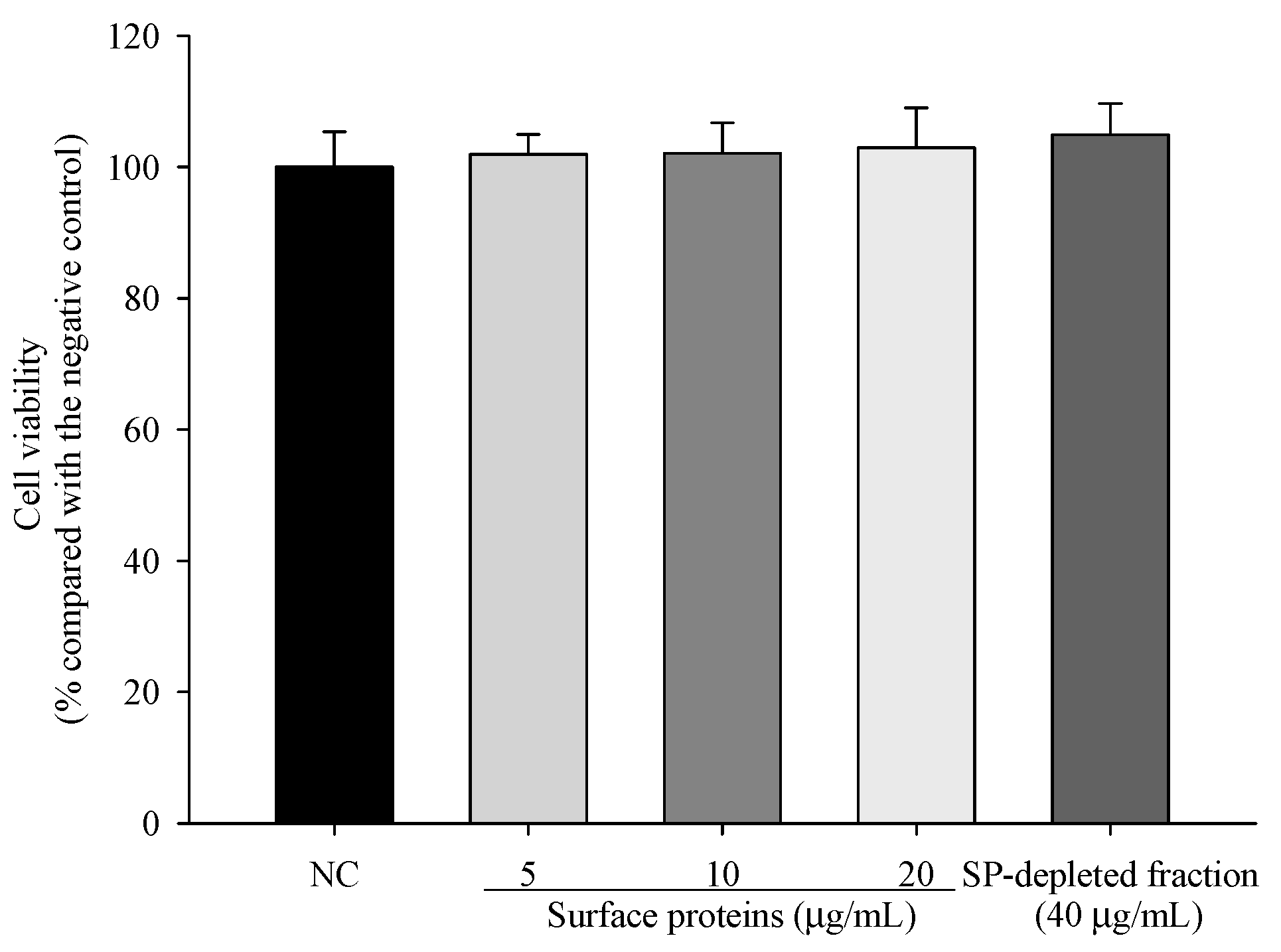
3.10. Viability of Cells Treated with Surface Proteins (SP) Extracted from MJ2
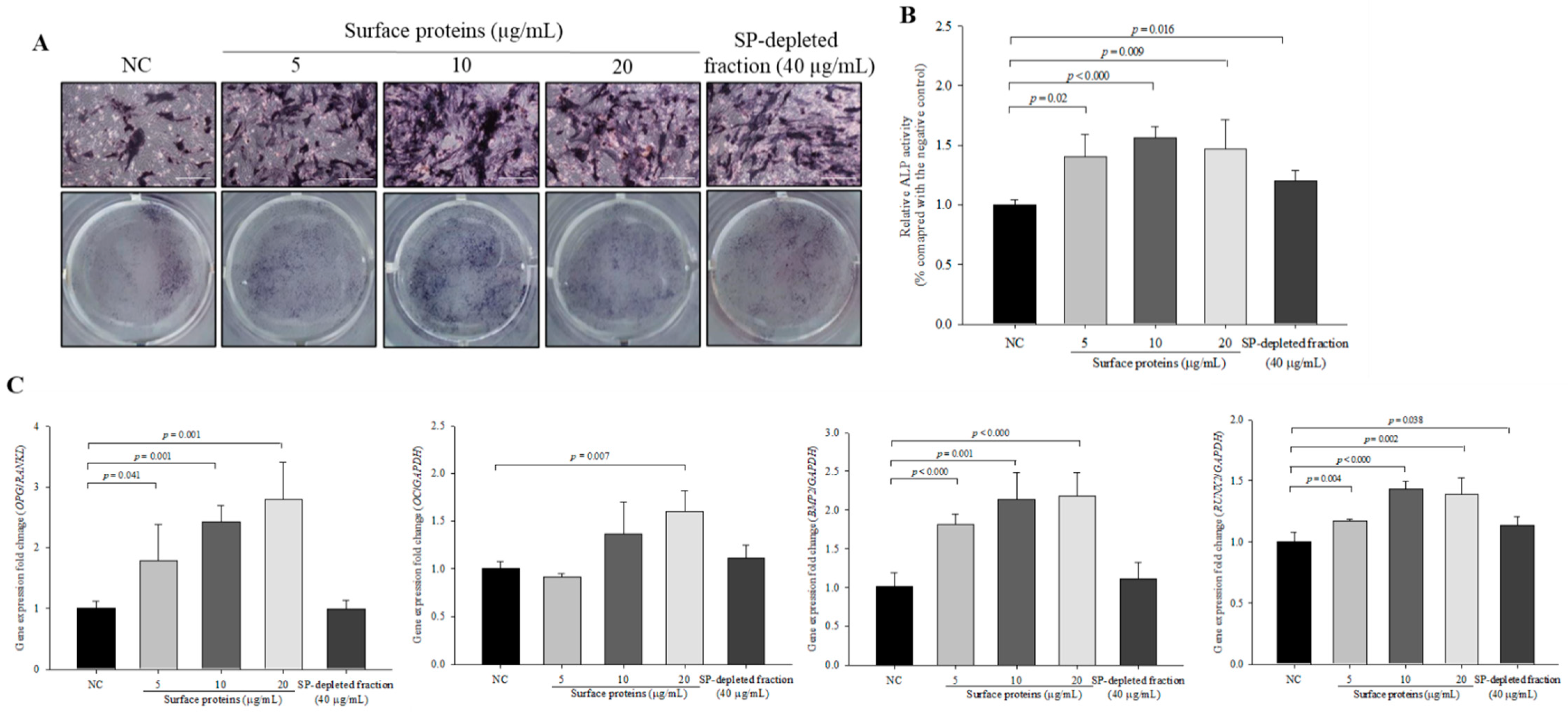
3.11. ALP Activity and the Expression Levels of Osteoblast Differentiation-Related Genes in Cells Treated with SPs Extracted from MJ2
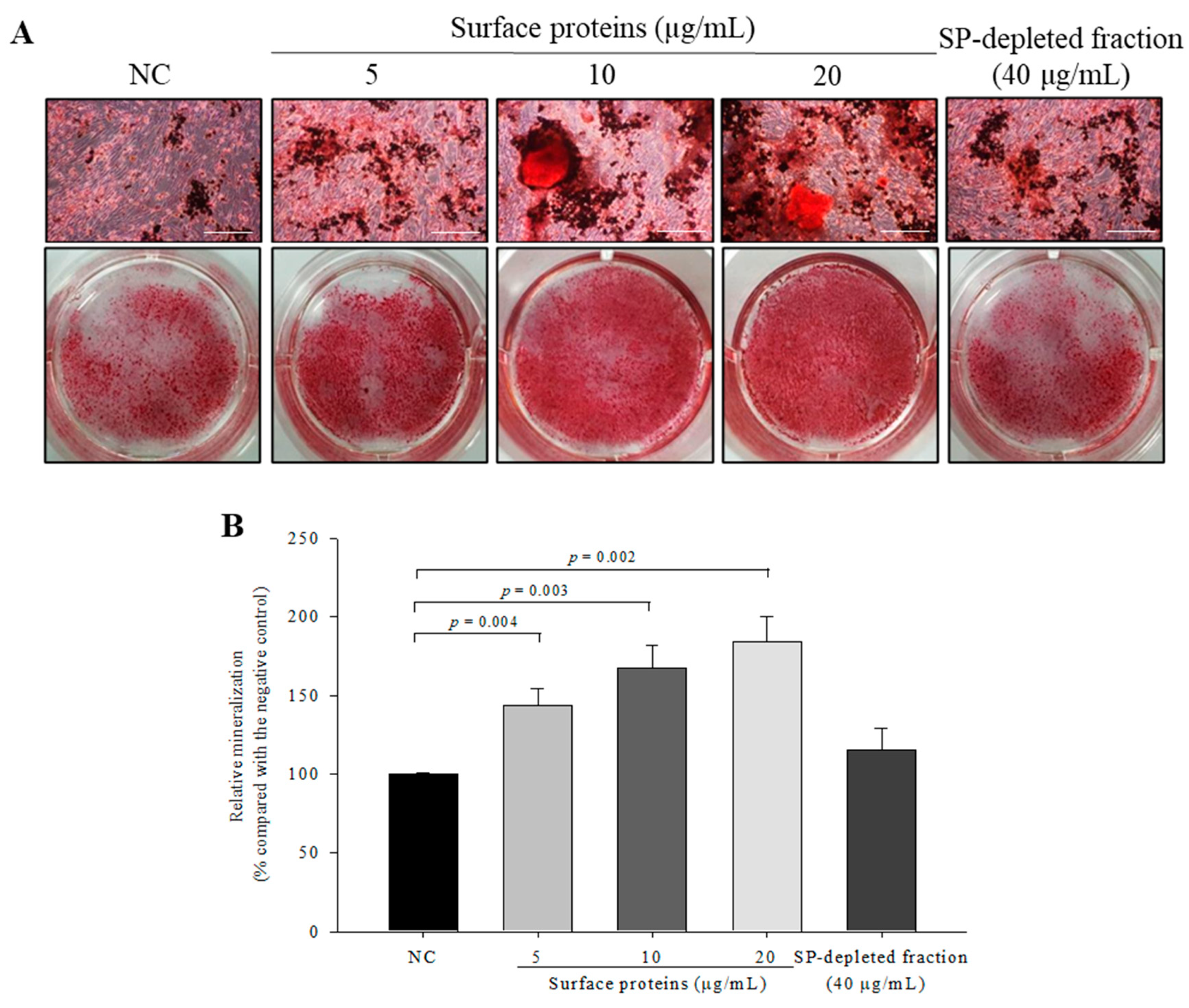
3.12. Osteoblast Mineralization in Cells Treated with SPs Extracted from MJ2
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Su, N.; Yang, J.; Xie, Y.; Du, X.; Chen, H.; Zhou, H.; Chen, L. Bone function, dysfunction and its role in diseases including critical illness. Int. J. Biol. Sci. 2019, 15, 776–787. [Google Scholar] [CrossRef]
- Safadi, F.F.; Barbe, M.F.; Abdelmagid, S.M.; Rico, M.C.; Aswad, R.A.; Litvin, J.; Popoff, S.N. Bone structure, development and bone biology. In Bone Pathology; Khurana, J.S., Ed.; Humana Press: New York, NY, USA, 2009; pp. 1–50. [Google Scholar]
- Atsuta, Y.A.-O.; Takahashi, Y.A.-O. Early formation of the Müllerian duct is regulated by sequential actions of BMP/Pax2 and FGF/Lim1 signaling. Development 2016, 143, 3549–3559. [Google Scholar] [CrossRef]
- Kirkham, G.; Cartmell, S. Genes and proteins involved in the regulation of osteogenesis. In Topics in Tissue Engineering; Ashammakhi, N., Reis, R., Chiellini, E., Eds.; 2007; Volume 3, pp. 1–22. Available online: https://www.researchgate.net/profile/Sarah-Cartmell-2/publication/267368490_Genes_and_Proteins_Involved_in_the_Regulation_of_Osteogenesis/links/548034770cf2ccc7f8bb2589/Genes-and-Proteins-Involved-in-the-Regulation-of-Osteogenesis.pdf (accessed on 12 January 2021).
- Yu, H.; de Vos, P.; Ren, Y. Overexpression of osteoprotegerin promotes preosteoblast differentiation to mature osteoblasts. Angle Orthod. 2011, 81, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Nakayamada, S.; Okada, Y. Osteoblasts and osteoclasts in bone remodeling and inflammation. Curr. Drug Targets Inflamm. Allergy 2005, 4, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Danks, L.; Sabokbar, A.; Athanasou, N. Osteoclast formation and activity in the pathogenesis of osteoporosis in rheumatoid arthritis. Rheumatology 2002, 41, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Raisz, L.G. Pathogenesis of osteoporosis: Concepts, conflicts, and prospects. J. Clin. Investig. 2005, 115, 3318–3325. [Google Scholar] [CrossRef]
- Riggs, B.L. The mechanisms of estrogen regulation of bone resorption. J. Clin. Investig. 2000, 106, 1203–1204. [Google Scholar] [CrossRef]
- Kennel, K.A.; Drake, M.T. Adverse effects of bisphosphonates: Implications for osteoporosis management. Mayo Clin. Proc. 2009, 84, 632–638. [Google Scholar] [CrossRef]
- Skjødt, M.K.; Frost, M.; Abrahamsen, B. Side effects of drugs for osteoporosis and metastatic bone disease. Br. J. Clin. Pharmacol. 2019, 85, 1063–1071. [Google Scholar] [CrossRef]
- Wentzensen, N.; Trabert, B. Hormone therapy: Short-term relief, long-term consequences. Lancet 2015, 385, 1806–1808. [Google Scholar] [CrossRef]
- Brouns, F.; Vermeer, C. Functional food ingredients for reducing the risks of osteoporosis. Trends Food Sci. Technol. 2000, 11, 22–33. [Google Scholar] [CrossRef]
- Rajput, R.; Wairkar, S.; Gaud, R. Nutraceuticals for better management of osteoporosis: An overview. J. Funct. Foods 2018, 47, 480–490. [Google Scholar] [CrossRef]
- Collins, F.L.; Rios-Arce, N.D.; Schepper, J.D.; Parameswaran, N.; Mccabe, L.R. The potential of probiotics as a therapy for osteoporosis. Microbiol. Spectr. 2017, 5, BAD-0015-2016. [Google Scholar]
- Jansson, P.-A.; Curiac, D.; Ahrén, I.L.; Hansson, F.; Niskanen, T.M.; Sjögren, K.; Ohlsson, C. Probiotic treatment using a mix of three Lactobacillus strains for lumbar spine bone loss in postmenopausal women: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet Rheumatol. 2019, 1, e154–e162. [Google Scholar] [CrossRef]
- Zárate, G. Dairy Propionibacteria: Less conventional probiotics to improve the human and animal health. In Probiotic in Animals; InTech: London, UK, 2012; pp. 153–202. [Google Scholar]
- Ma, S.; Yeom, J.; Lim, Y.-H. Dairy Propionibacterium freudenreichii ameliorates acute colitis by stimulating MUC2 expression in intestinal goblet cell in a DSS-induced colitis rat model. Sci. Rep. 2020, 10, 5523. [Google Scholar] [CrossRef] [PubMed]
- Kwon, G.; Lee, J.; Lim, Y.-H. Dairy Propionibacterium extends the mean lifespan of Caenorhabditis elegans via activation of the innate immune system. Sci. Rep. 2016, 6, 31713. [Google Scholar] [CrossRef]
- Savage-Dunn, C. TGF-beta signaling. WormBook 2005, 1–12. [Google Scholar] [CrossRef]
- Subramaniam, M.; Jalal, S.M.; Rickard, D.J.; Harris, S.A.; Bolander, M.E.; Spelsberg, T.C. Further characterization of human fetal osteoblastic hFOB 1.19 and hFOB/ERα cells: Bone formation in vivo and karyotype analysis using multicolor fluorescent in situ hybridization. J. Cell. Biochem. 2002, 87, 9–15. [Google Scholar] [CrossRef] [PubMed]
- De Souza Malaspina, T.S.; Dos Santos, C.X.; Campanelli, A.P.; Laurindo, F.R.M.; Sogayar, M.C.; Granjeiro, J.M. Tartrate-resistant acid phosphatase activity and glutathione levels are modulated during hFOB 1.19 osteoblastic differentiation. J. Mol. Histol. 2008, 39, 627–634. [Google Scholar] [CrossRef]
- Al Mamun, M.A.; Hosen, M.J.; Islam, K.; Khatun, A.; Alam, M.M.; Al-Bari, M.A.A. Tridax procumbens flavonoids promote osteoblast differentiation and bone formation. Biol. Res. 2015, 48, 1–8. [Google Scholar] [CrossRef]
- Thierry, A.; Madec, M. Enumeration of propionibacteria in raw milk using a new selective medium. Le Lait 1995, 75, 315–323. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-S.; Kim, H.; Kim, N.-G.; Cho, S.Y.; Choi, K.-H.; Seong, J.K.; Paik, Y.-K. Proteomic analysis and molecular characterization of tissue ferritin light chain in hepatocellular carcinoma. Hepatology 2002, 35, 1459–1466. [Google Scholar] [CrossRef]
- Kim, D.-E.; Kim, J.-K.; Han, S.-K.; Jang, S.-E.; Han, M.J.; Kim, D.-H. Lactobacillus plantarum NK3 and Bifidobacterium longum NK49 Alleviate Bacterial Vaginosis and Osteoporosis in Mice by Suppressing NF-κ B-Linked TNF-α Expression. J. Med. Food 2019, 22, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Do Carmo, F.L.R.; Rabah, H.; Huang, S.; Gaucher, F.; Deplanche, M.; Dutertre, S.; Jardin, J.; Le Loir, Y.; Azevedo, V.; Jan, G. Propionibacterium freudenreichii Surface Protein SlpB Is Involved in Adhesion to Intestinal HT-29 Cells. Front. Microbiol. 2017, 8, 1033. [Google Scholar] [CrossRef]
- Le Maréchal, C.; Peton, V.; Plé, C.; Vroland, C.; Jardin, J.; Briard-Bion, V.; Durant, G.; Chuat, V.; Loux, V.; Foligné, B. Surface proteins of Propionibacterium freudenreichii are involved in its anti-inflammatory properties. J. Proteom. 2015, 113, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.-G.; Kim, E.-J.; Kim, D.-K.; Ryoo, H.-M.; Lee, K.-B.; Kim, S.-H.; Choi, H.-S.; Koh, J.-T. BMP2 protein regulates osteocalcin expression via Runx2-mediated Atf6 gene transcription. J. Biol. Chem. 2012, 287, 905–915. [Google Scholar] [CrossRef]
- Murakami, T.; Saito, A.; Hino, S.-I.; Kondo, S.; Kanemoto, S.; Chihara, K.; Sekiya, H.; Tsumagari, K.; Ochiai, K.; Yoshinaga, K. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat. Cell Biol. 2009, 11, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Kim, J.; Cheng, C.; Rawlins, B.A.; Boachie-Adjei, O.; Crystal, R.G.; Hidaka, C. Noggin regulation of bone morphogenetic protein (BMP) 2/7 heterodimer activity in vitro. Bone 2006, 39, 61–71. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Z.; Duan, N.; Zhu, G.; Schwarz, E.M.; Xie, C. Osteoblast–osteoclast interactions. Connect. Tissue Res. 2018, 59, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Parvaneh, K.; Jamaluddin, R.; Karimi, G.; Erfani, R. Effect of probiotics supplementation on bone mineral content and bone mass density. Sci. World J. 2014, 2014, 565962. [Google Scholar] [CrossRef] [PubMed]
- Parvaneh, K.; Ebrahimi, M.; Sabran, M.R.; Karimi, G.; Hwei, A.N.M.; Abdul-Majeed, S.; Ahmad, Z.; Ibrahim, Z.; Jamaluddin, R. Probiotics (Bifidobacterium longum) increase bone mass density and upregulate Sparc and Bmp-2 genes in rats with bone loss resulting from ovariectomy. BioMed. Res. Int. 2015, 2015, 897639. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.M.; Yu, M.; Darby, T.M.; Vaccaro, C.; Li, J.-Y.; Owens, J.A.; Hsu, E.; Adams, J.; Weitzmann, M.N.; Jones, R.M. The microbial metabolite butyrate stimulates bone formation via T regulatory cell-mediated regulation of WNT10B expression. Immunity 2018, 49, 1116–1131. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Jia, X.; Mo, L.; Liu, C.; Zheng, L.; Yuan, Q.; Zhou, X. Intestinal microbiota: A potential target for the treatment of postmenopausal osteoporosis. Bone Res. 2017, 5, 17046. [Google Scholar] [CrossRef]
- Chen, M.; Weng, K.; Huang, S.; Liu, Y.; Tseng, S.; Ojcius, D.M.; Shih, S. Pretreatment with a heat-killed probiotic modulates monocyte chemoattractant protein-1 and reduces the pathogenicity of influenza and enterovirus 71 infections. Mucosal Immunol. 2017, 10, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Boyce, B.F.; Xing, L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch. Biochem. Biophys. 2008, 473, 139–146. [Google Scholar] [CrossRef]
- Long, F. Building strong bones: Molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 2012, 13, 27–38. [Google Scholar] [CrossRef]
- Yang, C.; Wang, C.; Zhou, J.; Liang, Q.; He, F.; Li, F.; Li, Y.; Chen, J.; Zhang, F.; Han, C. Fibronectin 1 activates WNT/β-catenin signaling to induce osteogenic differentiation via Integrin β1 interaction. Lab. Investig. 2020, 100, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Sila-Asna, M.; Bunyaratvej, A.; Maeda, S.; Kitaguchi, H.; Bunyaratavej, N. Osteoblast differentiation and bone formation gene expression in strontium-inducing bone marrow mesenchymal stem cell. Kobe J. Med. Sci. 2007, 53, 25–35. [Google Scholar] [PubMed]
- Lian, N.; Wang, W.; Li, L.; Elefteriou, F.; Yang, X. Vimentin inhibits ATF4-mediated osteocalcin transcription and osteoblast differentiation. J. Biol. Chem. 2009, 284, 30518–30525. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Watson, A.D.; Ji, S.; Boström, K.I. Heat shock protein 70 enhances vascular bone morphogenetic protein-4 signaling by binding matrix Gla protein. Circ. Res. 2009, 105, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Desvaux, M.; Dumas, E.; Chafsey, I.; Hébraud, M. Protein cell surface display in Gram-positive bacteria: From single protein to macromolecular protein structure. FEMS Microbiol. Lett. 2006, 256, 1–15. [Google Scholar] [CrossRef]
- Wang, W.; Jeffery, C.J. An analysis of surface proteomics results reveals novel candidates for intracellular/surface moonlighting proteins in bacteria. Mol. Biosyst. 2016, 12, 1420–1431. [Google Scholar] [CrossRef] [PubMed]

| Gene | Species | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|---|
| GAPDH | Human | GAG TCA ACG GAT TTG GTC GT | GAC AAG CTT CCC GTT CTC AG |
| BMP-2 | Human | CCC AGC GTG AAA AGA GAG AC | GAG ACCGCAGTC CGT CTA AG |
| Smad1 | Human | GGT TCC AAG ACA CAG CGA AT | TGG GAG AGT GAG GGT AGG TG |
| Smad5 | Human | AAC CTG AGC CAC AAT GAA CC | GTG GCA TAT AGG CAG GAG GA |
| Smad8 | Human | CCA CAG AAG CCT CTG AGA CC | CCC AAC TCG GTT GTT CAG TT |
| Smad4 | Human | AAA GGT GAA GGT GAT GTT TGG GTC | CTG GAG CTA TTC CAC CTA CTG ATC C |
| RUNX2 | Human | GCA GAC AGC TCA CAA AAC CA | GGA AAA GGG GAG AAG GAG TG |
| OSX | Human | GCT TAT CCA GCC CCC TTT AC | CAC TGG GCA GAC AGT CAG AA |
| FN | Human | AAT CCA AGC GGA GAG AGT CA | CAT CCT CAG GGC TCG AGT AG |
| OPG | Human | TGA GGA GGC ATT CTT CAG GT | CGC TGT TTT CAC AGA GGT CA |
| RANKL | Human | AGG CCT TTC AAG GAG CTG TG | TTG GAG ATC TTG GCC CAA CC |
| OC | Human | CCC GAA GGA GCT GAG GAC AC | CTT TGA CCC TGC TTC CAG AG |
| Β-actin | Rat | AGC CAT GTA CGT AGC CAT CC | CTC TCA GCT GTG GTG GTG AA |
| OPG | Rat | TGG GAA TGA AGA TCC TCC AG | GAG GAA GGA AAG GGC CTA TG |
| RANKL | Rat | CAT GAA ACC TCA GGG AGC GT | GTT GGA CAC CTG GAC GCT AA |
| Group | Ovariectomy (OVX) | Treatment | Abbreviation |
|---|---|---|---|
| 1 | Sham | Phosphate buffered saline (PBS) | Sham |
| 2 | OVX | PBS | OVX |
| 3 | OVX | Low-dose live P. freudenreichii MJ2 (1 × 107 CFU/mL) | LLMJ2 |
| 4 | OVX | High-dose live P. freudenreichii MJ2 (1 × 108 CFU/mL) | HLMJ2 |
| 5 | OVX | Live L. plantarum (1 × 108 CFU/mL) | LP |
| 6 | OVX | Live L. plantarum + high-dose live P. freudenreichii MJ2 | LP + HLMJ2 |
| 7 | OVX | Low-dose dead P. freudenreichii MJ2 (1 × 107 cells/mL) | LDMJ2 |
| 8 | OVX | High-dose dead P. freudenreichii MJ2 (1 × 108 cells/mL) | HDMJ2 |
| 9 | OVX | Live L. plantarum + high-dose dead P. freudenreichii MJ2 | LP + HDMJ2 |
| Protein Name | Expression Change (Fold) 1 | gi Number |
|---|---|---|
| Putative, partial | 0.125 | gi:553734 |
| Survival/evasion peptide (DCD) | 0.161 | gi:15375076 |
| 60 kDa heat shock protein (HSPD1) | 4.326 | gi:129379 |
| Chain A, protein disulfide isomerase (P4HB) | 7.752 | gi:159162689 |
| Vimentin (VIM) | 16.971 | gi:340219 |
| Mutant beta-actin (ACTB) | 4.096 | gi:283336 |
| Group | Body Weight (g) | GOT (U/L) | GPT (U/L) |
|---|---|---|---|
| Sham | 333.58 ± 5.73 | 87.0 ± 7.39 | 32.8 ± 3.86 |
| OVX | 401.03 ± 8.20 | 110.1 ± 8.69 | 52.5 ± 7.58 |
| LLMJ2 | 401.71 ± 5.25 | 89.6 ± 11.18 | 41.6 ± 6.81 |
| HLMJ2 | 388.79 ± 9.33 | 92.0 ± 14.13 | 33.8 ± 3.81 |
| LP | 391.67 ± 6.96 | 75.6 ± 5.20 | 30.4 ± 2.22 |
| LP + HLMJ2 | 373.54 ± 7.34 | 87.4 ± 15.94 | 36.5 ± 8.85 |
| LDMJ2 | 378.84 ± 5.37 | 73.4 ± 2.66 | 33.3 ± 1.82 |
| HDMJ2 | 372.51 ± 4.45 | 67.1 ± 3.81 | 30.5 ± 1.36 |
| LP + HDMJ2 | 396.98 ± 11.31 | 80.9 ± 14.14 | 39.8 ± 8.53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeom, J.; Ma, S.; Lim, Y.-H. Probiotic Propionibacterium freudenreichii MJ2 Enhances Osteoblast Differentiation and Mineralization by Increasing the OPG/RANKL Ratio. Microorganisms 2021, 9, 673. https://doi.org/10.3390/microorganisms9040673
Yeom J, Ma S, Lim Y-H. Probiotic Propionibacterium freudenreichii MJ2 Enhances Osteoblast Differentiation and Mineralization by Increasing the OPG/RANKL Ratio. Microorganisms. 2021; 9(4):673. https://doi.org/10.3390/microorganisms9040673
Chicago/Turabian StyleYeom, Jiah, Seongho Ma, and Young-Hee Lim. 2021. "Probiotic Propionibacterium freudenreichii MJ2 Enhances Osteoblast Differentiation and Mineralization by Increasing the OPG/RANKL Ratio" Microorganisms 9, no. 4: 673. https://doi.org/10.3390/microorganisms9040673
APA StyleYeom, J., Ma, S., & Lim, Y.-H. (2021). Probiotic Propionibacterium freudenreichii MJ2 Enhances Osteoblast Differentiation and Mineralization by Increasing the OPG/RANKL Ratio. Microorganisms, 9(4), 673. https://doi.org/10.3390/microorganisms9040673







