Critical Role of 3′-Downstream Region of pmrB in Polymyxin Resistance in Escherichia coli BL21(DE3)
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Culture Conditions
2.2. Transformation of TOP10 with Putative Colistin-Resistant Genes and Different Lengths of pmrB from BL21
2.3. Functional Cloning of Colistin-Resistant Elements from BL21
2.4. Site-Directed Mutagenesis of the Genes in BL21
2.5. Antimicrobial Susceptibility Test
2.6. Quantitative Real-Time RT-PCR (qRT-PCR) Analysis of pmrB mRNA Level at Steady Phase
2.7. Determination of Decay Rate of pmrB mRNA
2.8. Lipid A Profile Analysis
2.9. Statistical Analysis
3. Results
3.1. The Genes from BL21 Implicated in Polymyxin Resistance Failed to Confer Colistin Resistance in TOP10
3.2. Discovery of Colistin-Resistant Determinants Using Functional Cloning
3.3. PmrB but Not PhoQ Is Required for Colistin Resistance in BL21
3.4. Identification of Critical 3′-Downstream Region of pmrB Required for Colistin Resistance
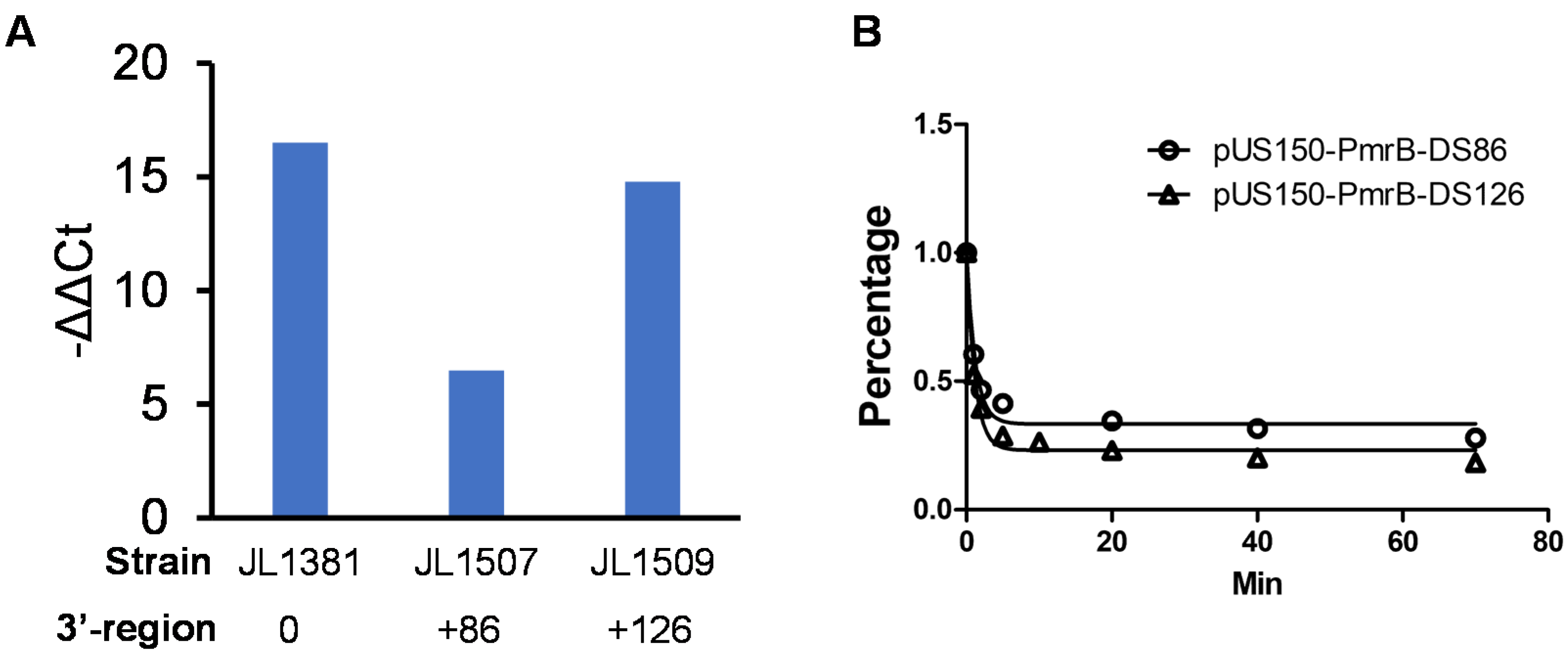
3.5. 3′-Downstream Region of pmrB Modulates the Expression Level of pmrB
3.6. 3′-Downstream Region Did Not Affect mRNA Stability of pmrB
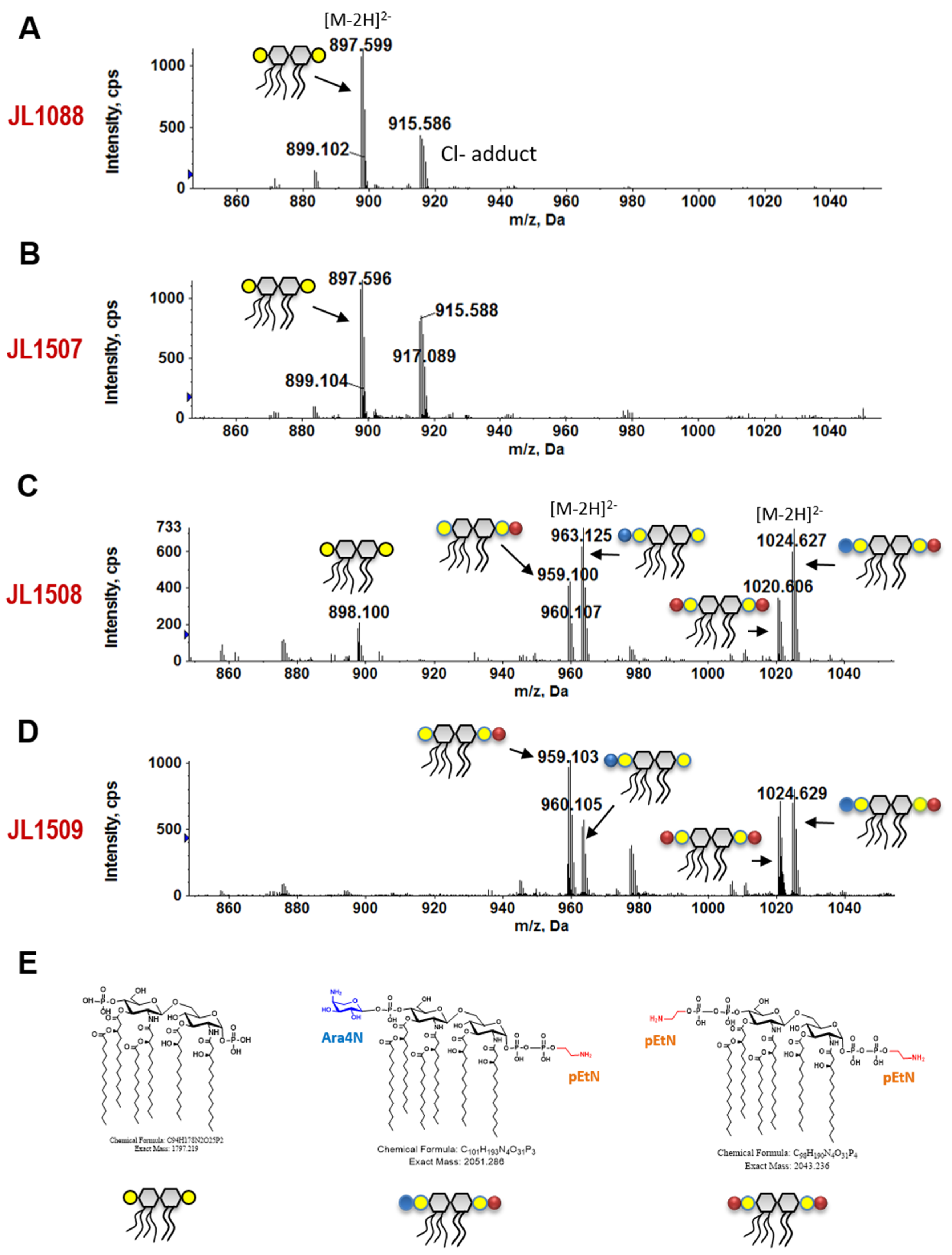
3.7. 3′-Downstream Region of pmrB Modulates Lipid A Modification
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; Nation, R.L.; Milne, R.W.; Turnidge, J.D.; Coulthard, K. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int. J. Antimicrob. Agents 2005, 25, 11–25. [Google Scholar] [CrossRef]
- Li, J.; Nation, R.L.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: The re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006, 6, 589–601. [Google Scholar] [CrossRef]
- Srinivas, P.; Rivard, K. Polymyxin Resistance in Gram-negative Pathogens. Curr. Infect. Dis. Rep. 2017, 19, 38. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E. Peptide antibiotics. Lancet 1997, 349, 418–422. [Google Scholar] [CrossRef]
- Ahmed, M.A.E.E.-S.; Zhong, L.-L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.-B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Gunn, J.S.; Lim, K.B.; Krueger, J.; Kim, K.; Guo, L.; Hackett, M.; Miller, S.I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 1998, 27, 1171–1182. [Google Scholar] [CrossRef]
- Wösten, M.M.; Kox, L.F.; Chamnongpol, S.; Soncini, F.C.; Groisman, E.A. A Signal Transduction System that Responds to Extracellular Iron. Cell 2000, 103, 113–125. [Google Scholar] [CrossRef]
- Phan, M.-D.; Nhu, N.T.K.; Achard, M.E.S.; Forde, B.M.; Hong, K.W.; Chong, T.M.; Yin, W.-F.; Chan, K.-G.; West, N.P.; Walker, M.J.; et al. Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J. Antimicrob. Chemother. 2017, 72, 2729–2736. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.C.; Groisman, E.A. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol. Microbiol. 2007, 63, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.M.; Hankins, J.V.; Trent, M.S. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol. Microbiol. 2010, 76, 1444–1460. [Google Scholar] [CrossRef]
- Kato, A. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004, 18, 2302–2313. [Google Scholar] [CrossRef]
- Lee, H.; Hsu, F.-F.; Turk, J.; Groisman, E.A. The PmrA-Regulated pmrC Gene Mediates Phosphoethanolamine Modification of Lipid A and Polymyxin Resistance in Salmonella enterica. J. Bacteriol. 2004, 186, 4124–4133. [Google Scholar] [CrossRef]
- Véscovi, E.G.; Soncini, F.C.; Groisman, E.A. Mg2+ as an Extracellular Signal: Environmental Regulation of Salmonella Virulence. Cell 1996, 84, 165–174. [Google Scholar] [CrossRef]
- Bader, M.W.; Sanowar, S.; Daley, M.E.; Schneider, A.R.; Cho, U.; Xu, W.; Klevit, R.E.; Le Moual, H.; Miller, S.I. Recognition of Antimicrobial Peptides by a Bacterial Sensor Kinase. Cell 2005, 122, 461–472. [Google Scholar] [CrossRef]
- Gunn, J.S.; Richards, S.M. Recognition and Integration of Multiple Environmental Signals by the Bacterial Sensor Kinase PhoQ. Cell Host Microbe 2007, 1, 163–165. [Google Scholar] [CrossRef]
- Gunn, J.S.; Ryan, S.S.; Van Velkinburgh, J.C.; Ernst, R.K.; Miller, S.I. Genetic and Functional Analysis of a PmrA-PmrB-Regulated Locus Necessary for Lipopolysaccharide Modification, Antimicrobial Peptide Resistance, and Oral Virulence of Salmonella entericaSerovar Typhimurium. Infect. Immun. 2000, 68, 6139–6146. [Google Scholar] [CrossRef]
- Kox, L.F.; Wösten, M.M.; Groisman, E.A. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 2000, 19, 1861–1872. [Google Scholar] [CrossRef]
- Xu, F.; Zeng, X.; Hinenoya, A.; Lin, J. MCR-1 Confers Cross-Resistance to Bacitracin, a Widely Used In-Feed Antibiotic. mSphere 2018, 3, e00411-18. [Google Scholar] [CrossRef]
- Lutz, R. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997, 25, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Datta, S.; Costantino, N.; Court, D.L. A set of recombineering plasmids for gram-negative bacteria. Gene 2006, 379, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, Y.; Lin, J. Functional Cloning and Characterization of Antibiotic Resistance Genes from the Chicken Gut Microbiome. Appl. Environ. Microbiol. 2012, 78, 3028–3032. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Michel, L.O.; Zhang, Q. CmeABC Functions as a Multidrug Efflux System in Campylobacter jejuni. Antimicrob. Agents Chemother. 2002, 46, 2124–2131. [Google Scholar] [CrossRef]
- Zeng, X.; Brown, S.; Gillespie, B.; Lin, J. A single nucleotide in the promoter region modulates the expression of the -lactamase OXA-61 in Campylobacter jejuni. J. Antimicrob. Chemother. 2014, 69, 1215–1223. [Google Scholar] [CrossRef]
- Ye, S.; Dhillon, S.; Ke, X.; Collins, A.R.; Day, I.N.M. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001, 29, e88. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Wu, Z.; Xu, C.; Sahin, O.; Yaeger, M.; Plummer, P.J.; Zhang, Q. The Rho-Independent Transcription Terminator for theporAGene Enhances Expression of the Major Outer Membrane Protein andCampylobacter jejuniVirulence in Abortion Induction. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Henderson, J.C.; O’Brien, J.P.; Brodbelt, J.S.; Trent, M.S. Isolation and Chemical Characterization of Lipid A from Gram-negative Bacteria. J. Vis. Exp. 2013, 10, e50623. [Google Scholar] [CrossRef] [PubMed]
- Joyce, L.R.; Guan, Z.; Palmer, K.L. Phosphatidylcholine Biosynthesis in Mitis Group Streptococci via Host Metabolite Scavenging. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.K.; Bogdanov, M.; Zhao, J.; Dowhan, W.; Raetz, C.R.H.; Guan, Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. USA 2012, 109, 16504–16509. [Google Scholar] [CrossRef]
- Pinske, C.; Bönn, M.; Krüger, S.; Lindenstrauß, U.; Sawers, R.G. Metabolic Deficiences Revealed in the Biotechnologically Important Model Bacterium Escherichia coli BL21(DE3). PLoS ONE 2011, 6, e22830. [Google Scholar] [CrossRef] [PubMed]
- Cannatelli, A.; D’Andrea, M.M.; Giani, T.; Di Pilato, V.; Arena, F.; Ambretti, S.; Gaibani, P.; Rossolini, G.M. In VivoEmergence of Colistin Resistance in Klebsiella pneumoniae Producing KPC-Type Carbapenemases Mediated by Insertional Inactivation of the PhoQ/PhoPmgrBRegulator. Antimicrob. Agents Chemother. 2013, 57, 5521–5526. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.T.; Lin, A.S.; Beckett, A.C.; McClain, M.S.; Cover, T.L. Role of a Stem-Loop Structure in Helicobacter pylori cagA Transcript Stability. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.F.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.D.; Jewett, M.W.; Groisman, E.A. Ancestral Genes Can Control the Ability of Horizontally Acquired Loci to Confer New Traits. PLoS Genet. 2011, 7, e1002184. [Google Scholar] [CrossRef][Green Version]
- Rubin, E.J.; Herrera, C.M.; Crofts, A.A.; Trent, M.S. PmrD Is Required for Modifications to Escherichia coli Endotoxin That Promote Antimicrobial Resistance. Antimicrob. Agents Chemother. 2015, 59, 2051–2061. [Google Scholar] [CrossRef] [PubMed]
- De Los Mozos, I.R.; Vergara-Irigaray, M.; Segura, V.; Villanueva, M.; Bitarte, N.; Saramago, M.; Domingues, S.; Arraiano, C.M.; Fechter, P.; Romby, P.; et al. Base Pairing Interaction between 5′- and 3′-UTRs Controls icaR mRNA Translation in Staphylococcus aureus. PLoS Genet. 2013, 9, e1004001. [Google Scholar] [CrossRef] [PubMed]
- Miyakoshi, M.; Chao, Y.; Vogel, J. Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr. Opin. Microbiol. 2015, 24, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, E.; Li, L.; Bischler, T.; Barquist, L.; Vogel, J. Global Maps of ProQ Binding In Vivo Reveal Target Recognition via RNA Structure and Stability Control at mRNA 3′ Ends. Mol. Cell 2018, 70, 971–982. [Google Scholar] [CrossRef]
- Kato, A.; Chen, H.D.; Latifi, T.; Groisman, E.A. Reciprocal Control between a Bacterium’s Regulatory System and the Modification Status of Its Lipopolysaccharide. Mol. Cell 2012, 47, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Winfield, M.D.; Groisman, E.A. Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc. Natl. Acad. Sci. USA 2004, 101, 17162–17167. [Google Scholar] [CrossRef] [PubMed]

| Plasmids or Strains | Description | Source or Reference |
|---|---|---|
| Plasmids | ||
| pZE21 | Cloning and expression vector; kanamycin resistant (Kanr) | [22] |
| pUC19 | Clone vector, ampicillin resistant (Ampr) | Invitrogen |
| pKD13 | Template plasmid of kanamycin resistant cassette for gene disruption. | [23] |
| pSIM6 | Ampicillin resistant. Heat inducible red recombinase expression plasmid, with a temperature-sensitive origin of replication | [24] |
| pZE21-EptA | pZE21 derivative containing eptA ORF | This study |
| pZE21-EptB | pZE21 derivative containing eptB ORF | This study |
| pZE21-CptA | pZE21 derivative containing cptA ORF | This study |
| pZE21-OpgEA | pZE21 derivative containing opgE ORF | This study |
| pZE21-PmrAB | pZE21 derivative containing pmrA and pmrB ORFs | This study |
| pZE21-PmrB | pZE21 derivative containing pmrB ORF | This study |
| pZE21-PmrD | pZE21 derivative containing pmrD ORF | This study |
| pPmrA-PmrB-ProP | pZE21 derivative containing PmrA(partial)-PmrB-ProP(partial) | This study |
| pUS150-PmrB-DS34 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 34 bp | This study |
| pUS150-PmrB-DS86 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 86 bp | This study |
| pUS150-PmrB-DS103 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 103 bp | This study |
| pUS150-PmrB-DS126 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 126 bp | This study |
| pUS150-PmrB-DS134 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 134 bp | This study |
| pUS150-PmrB-DS176 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 176 bp | This study |
| pUS150-PmrBMG1655-DS176 | pZE21 derivative containing pmrBMG1655 region from upstream 150 bp to downstream 176 bp | This study |
| Strains | ||
| BL21(DE3) | F− ompT hsdSB (rB−, mB−) gal dcm (DE3) | Stratagene/Novagen |
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| JL1374 | TOP10/pZE21-EptA | This study |
| JL1375 | TOP10/pZE21-EptB | This study |
| JL1376 | TOP10/pZE21-CptA | This study |
| JL1377 | TOP10/pZE21-OpgE | This study |
| JL1371 | TOP10/pZE21-PmrAB | This study |
| JL1373 | TOP10/pZE21-PmrD | This study |
| JL1365 | Transformant #1 from functional cloning, ColR | This study |
| JL1366 | Transformant #2 from functional cloning, ColR | This study |
| JL1367 | Transformant #3 from functional cloning, ColR | This study |
| JL1368 | Transformant #4 from functional cloning, ColR | This study |
| JL1369 | Transformant #5 from functional cloning, ColR | This study |
| JL1397 | TOP10/pPmrA-PmrB-ProP | This study |
| JL1088 | TOP10/pZE21 | This study |
| JL1381 | TOP10/pPmrB, containing pmrB ORF only | This study |
| JL1431 | TOP10/pUS150-PmrB-DS176 | This study |
| JL1432 | TOP10/pUS150-PmrBMG1655-DS176 | This study |
| JL1611 | TOP10/pUS150-PmrB-DS34 | This study |
| JL1507 | TOP10/pUS150-PmrB-DS86 | This study |
| JL1508 | TOP10/pUS150-PmrB-DS103 | This study |
| JL1509 | TOP10/pUS150-PmrB-DS126 | This study |
| JL1444 | TOP10/pUS150-PmrB-DS134 | This study |
| JL1435 | BL21(DE3), phoQ::kan | This study |
| JL1436 | BL21(DE3), pmrB::kan | This study |
| Primer | DNA Sequence (5′-3′) a | Product Size (bp) b | Target Gene/Region and Function |
|---|---|---|---|
| EptA_F | ATGTTGAAGCGCCTACTAAAAAGAC | 1644 | eptA ORF |
| EptA_R | CGCGGATCCTCATTCACTCACTCTCCT (BamHI) | ||
| EptB_F | ATGAGATACATCAAATCGATTACAC | 1692 | eptB ORF |
| EptB_R | CGCGGATCCTTAGTTAGCCGCTGCCTC (BamHI) | ||
| CptA_F | ATGCATTCCACAGAAGTCCAGGCT | 1734 | cptA ORF |
| CptA_R | CGCGGATCCTTACTGATTACCCACCTG (BamHI) | ||
| OpgE_F | ATGAATTTAACCCTCAAAGAATCGC | 1584 | opgE ORF |
| OpgE_R | CGCGGATCCTTAAGGTTTCGGGTCG (BamHI) | ||
| Prbas_F | AAATTCTGATTGTTGAAGACGATAC | 1766 | pmrA-pmrB ORF |
| PrEAbas_R | CGCGGATCCTTATATCTGGTTTGCCAC (BamHI) | ||
| PmrB-F1 | ATG CAT TTT CTG CGC CGA CCA ATA | 1092 | pmrB ORF |
| PmrB-R | ATATGGATCCTTATATCTGGTTTGCCACGT (BamHI) | ||
| p5Up-KpnI-F3 | ATATGGTACCGACGCTGAATATGGGTCGCC (KpnI) | 2897 | pmrA(partial)-pmrB-proP(partial) |
| p5Up-SalI-R3 | ATATGTCGACAAGTTTTTTTCCCGGGGGCTGA (SalI) | ||
| US150-PmrB-SalI-F1 | ATATGTCGACGACATCTATAACTGGGACAA (SalI) | ||
| DS34-PmrB-BamHI-R1 | ATATGGATCCCCGTGTTCAGCGTGCTGGTG (BamHI) | 1271 | pmrB ORF, with 150 bp upstream and 34 bp downstream |
| DS86-PmrB-BamHI-R1 | ATATGGATCCCAGGTTAACGGAGGAGAGTG (BamHI) | 1328 | pmrB ORF, with 150 bp upstream and 86 bp downstream |
| DS103-PmrB-BamHI-R1 | ATATGGATCCACGCGCATACTCTCCTCCAG (BamHI) | 1345 | pmrB ORF, with 150 bp upstream and 103 bp downstream |
| DS126-PmrB-BamHI-R1 | ATATGGATCCCTTAAGGTTCACTTAATCTC (BamHI) | 1368 | pmrB ORF, with 150 bp upstream and 126 bp downstream |
| DS134-PmrB-BamHI-R1 | ATATGGATCCATTGAACTCTTAAGGTTCACT (BamHI) | 1376 | pmrB ORF, with 150 bp upstream and 134 bp downstream |
| DS176-PmrB-BamHI-R1 | ATATGGATCCGCTGAAACGGATGGCCTGAT (BamHI) | 1418 | pmrB ORF, with 150 bp upstream and 176 bp downstream |
| pZE_F | GAATTCATTAAAGAGGAGAAAGGT | N/A | Forward sequencing primers for pZE21 derivatives |
| pZE_R | TTTCGTTTTATTTGATGCCTCTAG | N/A | Reverse sequencing primers for pZE21 derivatives |
| PmrB(BL21DE3)_pKD13_F3 | GCTTTGGCTATATGCTGGTCGCGAATGAGGAAAACTAATTGAATCTGATGTGTAGGCTGGAGCTGCTTCG | 1403 | Site-directed mutation of pmrB |
| PmrB(BL21DE3)_pKD13_R3 | TTCAGCGTGCTGGTGGTCAGCAGCTTTCTTTATATCTGGTTTGCCACGTAATTCCGGGGATCCGTCGACC | ||
| PmrB_F | AATGAACCCTCGACCAACAC | 1376 | Detect site-directed mutation of pmrB with k1 |
| PmrB_R | CGCTGTCTTATCAGGCCAAT | ||
| PhoQ(BL21DE3)_pKD13_F3 | GTGATTACCACCGTTCGCGGCCAGGGCTATCTGTTCGAATTGCGCTGATGTGTAGGCTGGAGCTGCTTCG | 1403 | Site-directed mutation of phoQ |
| PhoQ(BL21DE3)_pKD13_R3 | TTAACGTAATGCGTGAAGTATGGACATATTTATTCATCTTTCGGCGTAGAATTCCGGGGATCCGTCGACC | ||
| PhoQ_F | TAATGGCAAAGTGGTGAGCA | 1772 | Detect site-directed mutation of phoQ with K1 |
| PhoQ_R | TTCTGCCAGTGACGTTCAAG | ||
| K1 | CAGTCATAGCCGAATAGCCT | Common primer for detecting site-directed mutation | |
| RT-PmrB-BL21SNP-F3 | CATTGCCATTCACAGCGCCACCCGCA | 180 | RT-PCR detection of pmrBBL21(DE3) |
| RT-PmrB-BL21SNP-R3 | TGCGTTTTCGCCAGCAGTTCCAGATGCA | ||
| 16S-F | AAGTTAATACCTTTGCTCATTGAC | 118 | 16S rRNA internal control for RT-PCR |
| 16S-R | GCTTTACGCCCAGTAATTCC |
| Strain | Insert Size (bp) | Genome Location in BL21 | Annotated Genes and Organization a |
|---|---|---|---|
| JL1365 | 10,660 | 210,471–208,308 | dnaE |
| 3,818,298–3,821,491 | viaA-ravA-kup | ||
| 1,144,385–1,147,209 | rne-yceQ | ||
| 2,263,105–2,265,581 | arnF-pmrD-menE-menC | ||
| JL1366 | 3926 | 4,242,936–4,239,011 | proP-pmrB-pmrA-EptA-adiC |
| JL1367 | 6989 | 4,239,038–4,243,091 | proP-pmrB-pmrA-EptA-adiC |
| 1,256,348–1,259,815 | kdsA-ldrA-ldrB-ldrC-chaA | ||
| JL1368 | 3813 | 2,263,769–2,261,742 | arnT-arnE-arnF-pmrD |
| 2,759,470–2,757,690 | syd-queF-ygdH | ||
| JL1369 | 5269 | 1,879,745–1,877,377 | purT-eda-edd |
| 4,237,732–4,240,628 | proP-pmrB-pmrA |
| Strain | Colistin MIC (µg/mL) | Lipid A Modification | |
|---|---|---|---|
| pEtN | Ara4N | ||
| TOP10/pZE21 | 0.5 | - a | - |
| TOP10/pPmrB | 0.5 | ND b | ND |
| TOP10/pUS150-PmrB-DS34 | 0.5 | ND | ND |
| TOP10/pUS150-PmrB-DS86 | 0.5 | - | - |
| TOP10/pUS150-PmrB-DS103 | 4 | + | + |
| TOP10/pUS150-PmrB-DS126 | 16 | + | + |
| TOP10/pUS150-PmrB-DS134 | 16 | ND | ND |
| TOP10/pUS150-PmrB-DS176 | 16 | ND | ND |
| TOP10/pUS150-PmrBMG1655-DS176 | 0.5 | ND | ND |
| BL21, wild type | 16 | + | + |
| JL1435 (BL21, phoQ::kan) | 16 | ND | ND |
| JL1436 (BL21, pmrB::kan) | 0.5 | ND | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, F.; Hinenoya, A.; Zeng, X.; Li, X.-P.; Guan, Z.; Lin, J. Critical Role of 3′-Downstream Region of pmrB in Polymyxin Resistance in Escherichia coli BL21(DE3). Microorganisms 2021, 9, 655. https://doi.org/10.3390/microorganisms9030655
Xu F, Hinenoya A, Zeng X, Li X-P, Guan Z, Lin J. Critical Role of 3′-Downstream Region of pmrB in Polymyxin Resistance in Escherichia coli BL21(DE3). Microorganisms. 2021; 9(3):655. https://doi.org/10.3390/microorganisms9030655
Chicago/Turabian StyleXu, Fuzhou, Atsushi Hinenoya, Ximin Zeng, Xing-Ping Li, Ziqiang Guan, and Jun Lin. 2021. "Critical Role of 3′-Downstream Region of pmrB in Polymyxin Resistance in Escherichia coli BL21(DE3)" Microorganisms 9, no. 3: 655. https://doi.org/10.3390/microorganisms9030655
APA StyleXu, F., Hinenoya, A., Zeng, X., Li, X.-P., Guan, Z., & Lin, J. (2021). Critical Role of 3′-Downstream Region of pmrB in Polymyxin Resistance in Escherichia coli BL21(DE3). Microorganisms, 9(3), 655. https://doi.org/10.3390/microorganisms9030655






