Critical Role of 3′-Downstream Region of pmrB in Polymyxin Resistance in Escherichia coli BL21(DE3)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Culture Conditions
2.2. Transformation of TOP10 with Putative Colistin-Resistant Genes and Different Lengths of pmrB from BL21
2.3. Functional Cloning of Colistin-Resistant Elements from BL21
2.4. Site-Directed Mutagenesis of the Genes in BL21
2.5. Antimicrobial Susceptibility Test
2.6. Quantitative Real-Time RT-PCR (qRT-PCR) Analysis of pmrB mRNA Level at Steady Phase
2.7. Determination of Decay Rate of pmrB mRNA
2.8. Lipid A Profile Analysis
2.9. Statistical Analysis
3. Results
3.1. The Genes from BL21 Implicated in Polymyxin Resistance Failed to Confer Colistin Resistance in TOP10
3.2. Discovery of Colistin-Resistant Determinants Using Functional Cloning
3.3. PmrB but Not PhoQ Is Required for Colistin Resistance in BL21
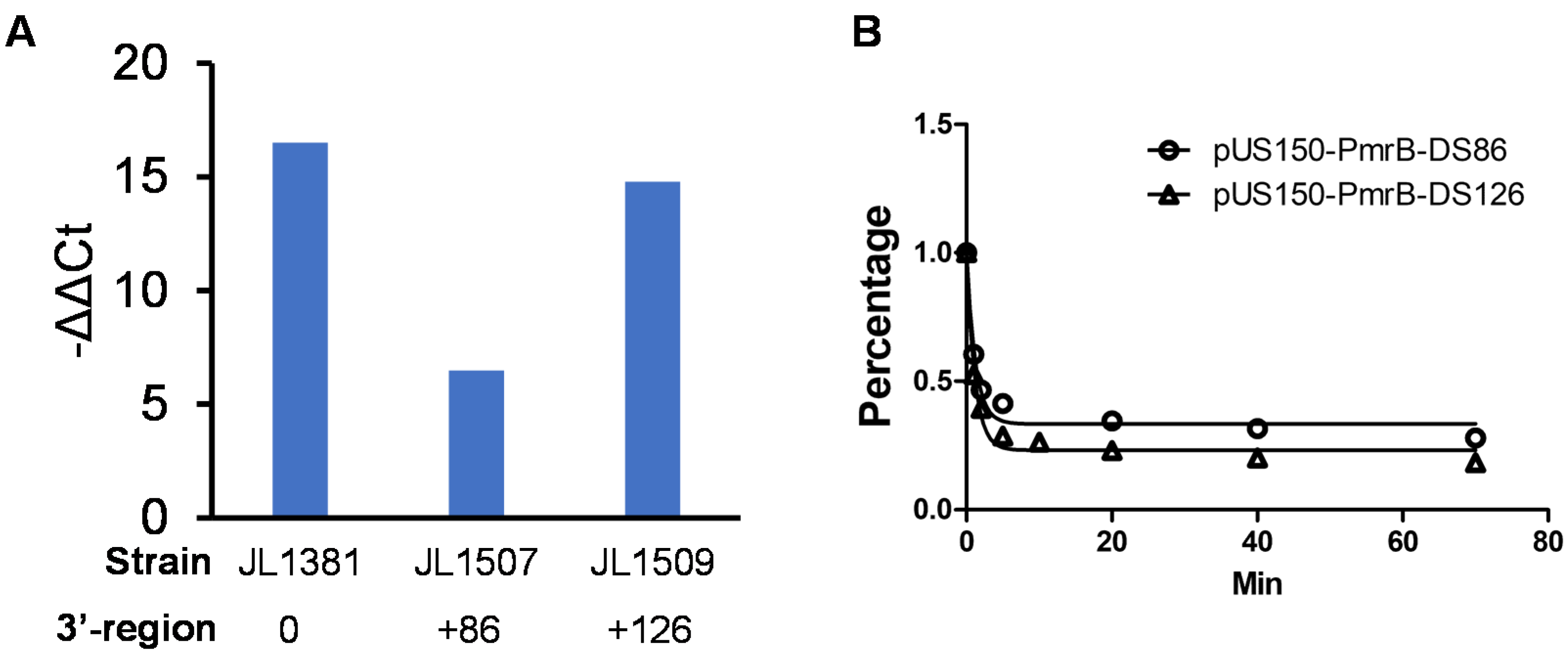
3.4. Identification of Critical 3′-Downstream Region of pmrB Required for Colistin Resistance
3.5. 3′-Downstream Region of pmrB Modulates the Expression Level of pmrB
3.6. 3′-Downstream Region Did Not Affect mRNA Stability of pmrB
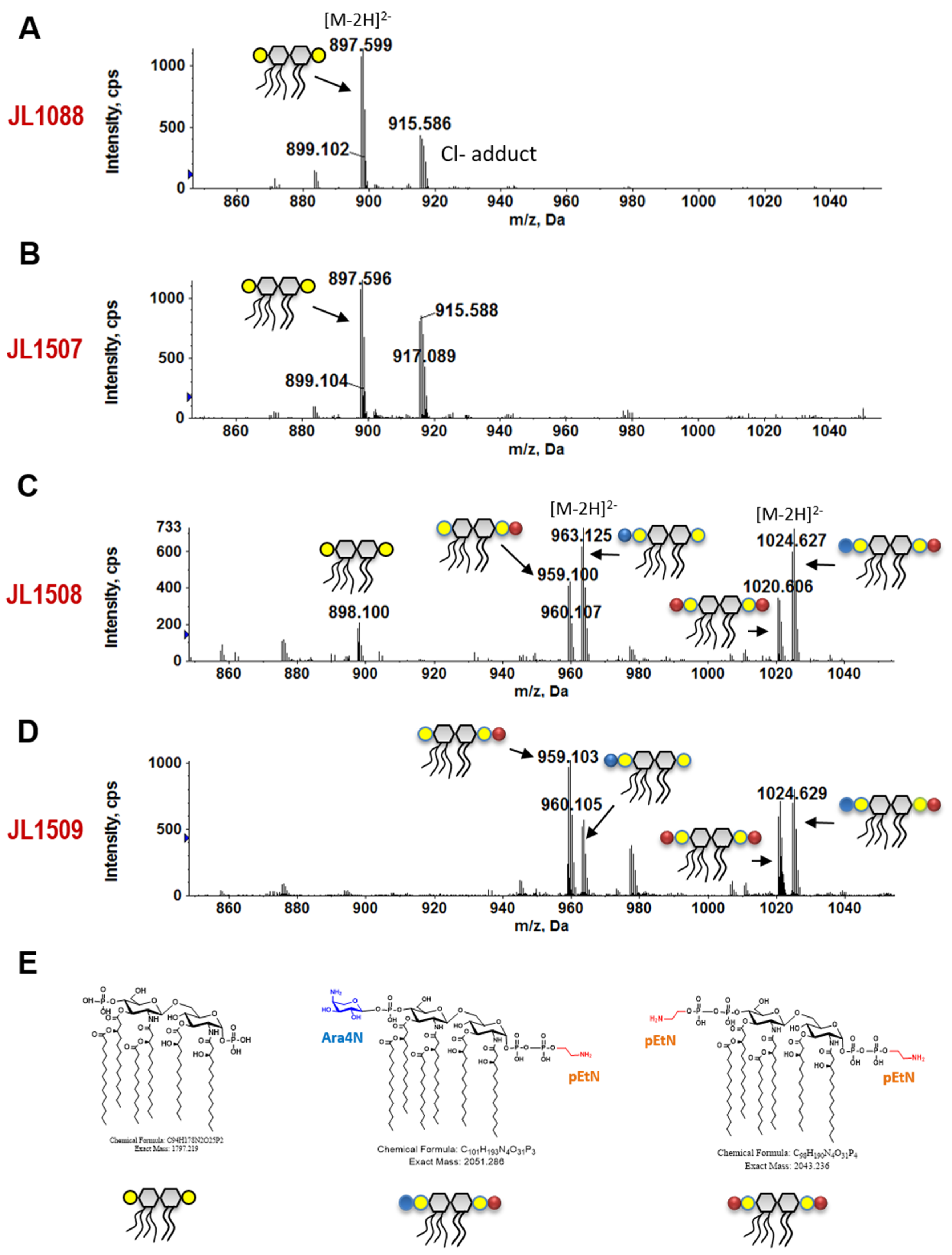
3.7. 3′-Downstream Region of pmrB Modulates Lipid A Modification
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; Nation, R.L.; Milne, R.W.; Turnidge, J.D.; Coulthard, K. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int. J. Antimicrob. Agents 2005, 25, 11–25. [Google Scholar] [CrossRef]
- Li, J.; Nation, R.L.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: The re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006, 6, 589–601. [Google Scholar] [CrossRef]
- Srinivas, P.; Rivard, K. Polymyxin Resistance in Gram-negative Pathogens. Curr. Infect. Dis. Rep. 2017, 19, 38. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, R.E. Peptide antibiotics. Lancet 1997, 349, 418–422. [Google Scholar] [CrossRef]
- Ahmed, M.A.E.E.-S.; Zhong, L.-L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.-B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [Green Version]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunn, J.S.; Lim, K.B.; Krueger, J.; Kim, K.; Guo, L.; Hackett, M.; Miller, S.I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 1998, 27, 1171–1182. [Google Scholar] [CrossRef]
- Wösten, M.M.; Kox, L.F.; Chamnongpol, S.; Soncini, F.C.; Groisman, E.A. A Signal Transduction System that Responds to Extracellular Iron. Cell 2000, 103, 113–125. [Google Scholar] [CrossRef] [Green Version]
- Phan, M.-D.; Nhu, N.T.K.; Achard, M.E.S.; Forde, B.M.; Hong, K.W.; Chong, T.M.; Yin, W.-F.; Chan, K.-G.; West, N.P.; Walker, M.J.; et al. Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J. Antimicrob. Chemother. 2017, 72, 2729–2736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, J.C.; Groisman, E.A. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol. Microbiol. 2007, 63, 283–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrera, C.M.; Hankins, J.V.; Trent, M.S. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol. Microbiol. 2010, 76, 1444–1460. [Google Scholar] [CrossRef]
- Kato, A. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004, 18, 2302–2313. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Hsu, F.-F.; Turk, J.; Groisman, E.A. The PmrA-Regulated pmrC Gene Mediates Phosphoethanolamine Modification of Lipid A and Polymyxin Resistance in Salmonella enterica. J. Bacteriol. 2004, 186, 4124–4133. [Google Scholar] [CrossRef] [Green Version]
- Véscovi, E.G.; Soncini, F.C.; Groisman, E.A. Mg2+ as an Extracellular Signal: Environmental Regulation of Salmonella Virulence. Cell 1996, 84, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Bader, M.W.; Sanowar, S.; Daley, M.E.; Schneider, A.R.; Cho, U.; Xu, W.; Klevit, R.E.; Le Moual, H.; Miller, S.I. Recognition of Antimicrobial Peptides by a Bacterial Sensor Kinase. Cell 2005, 122, 461–472. [Google Scholar] [CrossRef] [Green Version]
- Gunn, J.S.; Richards, S.M. Recognition and Integration of Multiple Environmental Signals by the Bacterial Sensor Kinase PhoQ. Cell Host Microbe 2007, 1, 163–165. [Google Scholar] [CrossRef] [Green Version]
- Gunn, J.S.; Ryan, S.S.; Van Velkinburgh, J.C.; Ernst, R.K.; Miller, S.I. Genetic and Functional Analysis of a PmrA-PmrB-Regulated Locus Necessary for Lipopolysaccharide Modification, Antimicrobial Peptide Resistance, and Oral Virulence of Salmonella entericaSerovar Typhimurium. Infect. Immun. 2000, 68, 6139–6146. [Google Scholar] [CrossRef]
- Kox, L.F.; Wösten, M.M.; Groisman, E.A. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 2000, 19, 1861–1872. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Zeng, X.; Hinenoya, A.; Lin, J. MCR-1 Confers Cross-Resistance to Bacitracin, a Widely Used In-Feed Antibiotic. mSphere 2018, 3, e00411-18. [Google Scholar] [CrossRef] [Green Version]
- Lutz, R. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997, 25, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [Green Version]
- Datta, S.; Costantino, N.; Court, D.L. A set of recombineering plasmids for gram-negative bacteria. Gene 2006, 379, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Wang, Y.; Lin, J. Functional Cloning and Characterization of Antibiotic Resistance Genes from the Chicken Gut Microbiome. Appl. Environ. Microbiol. 2012, 78, 3028–3032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Michel, L.O.; Zhang, Q. CmeABC Functions as a Multidrug Efflux System in Campylobacter jejuni. Antimicrob. Agents Chemother. 2002, 46, 2124–2131. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.; Brown, S.; Gillespie, B.; Lin, J. A single nucleotide in the promoter region modulates the expression of the -lactamase OXA-61 in Campylobacter jejuni. J. Antimicrob. Chemother. 2014, 69, 1215–1223. [Google Scholar] [CrossRef] [Green Version]
- Ye, S.; Dhillon, S.; Ke, X.; Collins, A.R.; Day, I.N.M. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001, 29, e88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, L.; Wu, Z.; Xu, C.; Sahin, O.; Yaeger, M.; Plummer, P.J.; Zhang, Q. The Rho-Independent Transcription Terminator for theporAGene Enhances Expression of the Major Outer Membrane Protein andCampylobacter jejuniVirulence in Abortion Induction. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [Green Version]
- Henderson, J.C.; O’Brien, J.P.; Brodbelt, J.S.; Trent, M.S. Isolation and Chemical Characterization of Lipid A from Gram-negative Bacteria. J. Vis. Exp. 2013, 10, e50623. [Google Scholar] [CrossRef] [PubMed]
- Joyce, L.R.; Guan, Z.; Palmer, K.L. Phosphatidylcholine Biosynthesis in Mitis Group Streptococci via Host Metabolite Scavenging. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, B.K.; Bogdanov, M.; Zhao, J.; Dowhan, W.; Raetz, C.R.H.; Guan, Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. USA 2012, 109, 16504–16509. [Google Scholar] [CrossRef] [Green Version]
- Pinske, C.; Bönn, M.; Krüger, S.; Lindenstrauß, U.; Sawers, R.G. Metabolic Deficiences Revealed in the Biotechnologically Important Model Bacterium Escherichia coli BL21(DE3). PLoS ONE 2011, 6, e22830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannatelli, A.; D’Andrea, M.M.; Giani, T.; Di Pilato, V.; Arena, F.; Ambretti, S.; Gaibani, P.; Rossolini, G.M. In VivoEmergence of Colistin Resistance in Klebsiella pneumoniae Producing KPC-Type Carbapenemases Mediated by Insertional Inactivation of the PhoQ/PhoPmgrBRegulator. Antimicrob. Agents Chemother. 2013, 57, 5521–5526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, J.T.; Lin, A.S.; Beckett, A.C.; McClain, M.S.; Cover, T.L. Role of a Stem-Loop Structure in Helicobacter pylori cagA Transcript Stability. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.F.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.D.; Jewett, M.W.; Groisman, E.A. Ancestral Genes Can Control the Ability of Horizontally Acquired Loci to Confer New Traits. PLoS Genet. 2011, 7, e1002184. [Google Scholar] [CrossRef] [Green Version]
- Rubin, E.J.; Herrera, C.M.; Crofts, A.A.; Trent, M.S. PmrD Is Required for Modifications to Escherichia coli Endotoxin That Promote Antimicrobial Resistance. Antimicrob. Agents Chemother. 2015, 59, 2051–2061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Los Mozos, I.R.; Vergara-Irigaray, M.; Segura, V.; Villanueva, M.; Bitarte, N.; Saramago, M.; Domingues, S.; Arraiano, C.M.; Fechter, P.; Romby, P.; et al. Base Pairing Interaction between 5′- and 3′-UTRs Controls icaR mRNA Translation in Staphylococcus aureus. PLoS Genet. 2013, 9, e1004001. [Google Scholar] [CrossRef] [PubMed]
- Miyakoshi, M.; Chao, Y.; Vogel, J. Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr. Opin. Microbiol. 2015, 24, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, E.; Li, L.; Bischler, T.; Barquist, L.; Vogel, J. Global Maps of ProQ Binding In Vivo Reveal Target Recognition via RNA Structure and Stability Control at mRNA 3′ Ends. Mol. Cell 2018, 70, 971–982. [Google Scholar] [CrossRef] [Green Version]
- Kato, A.; Chen, H.D.; Latifi, T.; Groisman, E.A. Reciprocal Control between a Bacterium’s Regulatory System and the Modification Status of Its Lipopolysaccharide. Mol. Cell 2012, 47, 897–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winfield, M.D.; Groisman, E.A. Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc. Natl. Acad. Sci. USA 2004, 101, 17162–17167. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Plasmids or Strains | Description | Source or Reference |
|---|---|---|
| Plasmids | ||
| pZE21 | Cloning and expression vector; kanamycin resistant (Kanr) | [22] |
| pUC19 | Clone vector, ampicillin resistant (Ampr) | Invitrogen |
| pKD13 | Template plasmid of kanamycin resistant cassette for gene disruption. | [23] |
| pSIM6 | Ampicillin resistant. Heat inducible red recombinase expression plasmid, with a temperature-sensitive origin of replication | [24] |
| pZE21-EptA | pZE21 derivative containing eptA ORF | This study |
| pZE21-EptB | pZE21 derivative containing eptB ORF | This study |
| pZE21-CptA | pZE21 derivative containing cptA ORF | This study |
| pZE21-OpgEA | pZE21 derivative containing opgE ORF | This study |
| pZE21-PmrAB | pZE21 derivative containing pmrA and pmrB ORFs | This study |
| pZE21-PmrB | pZE21 derivative containing pmrB ORF | This study |
| pZE21-PmrD | pZE21 derivative containing pmrD ORF | This study |
| pPmrA-PmrB-ProP | pZE21 derivative containing PmrA(partial)-PmrB-ProP(partial) | This study |
| pUS150-PmrB-DS34 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 34 bp | This study |
| pUS150-PmrB-DS86 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 86 bp | This study |
| pUS150-PmrB-DS103 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 103 bp | This study |
| pUS150-PmrB-DS126 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 126 bp | This study |
| pUS150-PmrB-DS134 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 134 bp | This study |
| pUS150-PmrB-DS176 | pZE21 derivative containing pmrB region from upstream 150 bp to downstream 176 bp | This study |
| pUS150-PmrBMG1655-DS176 | pZE21 derivative containing pmrBMG1655 region from upstream 150 bp to downstream 176 bp | This study |
| Strains | ||
| BL21(DE3) | F− ompT hsdSB (rB−, mB−) gal dcm (DE3) | Stratagene/Novagen |
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| JL1374 | TOP10/pZE21-EptA | This study |
| JL1375 | TOP10/pZE21-EptB | This study |
| JL1376 | TOP10/pZE21-CptA | This study |
| JL1377 | TOP10/pZE21-OpgE | This study |
| JL1371 | TOP10/pZE21-PmrAB | This study |
| JL1373 | TOP10/pZE21-PmrD | This study |
| JL1365 | Transformant #1 from functional cloning, ColR | This study |
| JL1366 | Transformant #2 from functional cloning, ColR | This study |
| JL1367 | Transformant #3 from functional cloning, ColR | This study |
| JL1368 | Transformant #4 from functional cloning, ColR | This study |
| JL1369 | Transformant #5 from functional cloning, ColR | This study |
| JL1397 | TOP10/pPmrA-PmrB-ProP | This study |
| JL1088 | TOP10/pZE21 | This study |
| JL1381 | TOP10/pPmrB, containing pmrB ORF only | This study |
| JL1431 | TOP10/pUS150-PmrB-DS176 | This study |
| JL1432 | TOP10/pUS150-PmrBMG1655-DS176 | This study |
| JL1611 | TOP10/pUS150-PmrB-DS34 | This study |
| JL1507 | TOP10/pUS150-PmrB-DS86 | This study |
| JL1508 | TOP10/pUS150-PmrB-DS103 | This study |
| JL1509 | TOP10/pUS150-PmrB-DS126 | This study |
| JL1444 | TOP10/pUS150-PmrB-DS134 | This study |
| JL1435 | BL21(DE3), phoQ::kan | This study |
| JL1436 | BL21(DE3), pmrB::kan | This study |
| Primer | DNA Sequence (5′-3′) a | Product Size (bp) b | Target Gene/Region and Function |
|---|---|---|---|
| EptA_F | ATGTTGAAGCGCCTACTAAAAAGAC | 1644 | eptA ORF |
| EptA_R | CGCGGATCCTCATTCACTCACTCTCCT (BamHI) | ||
| EptB_F | ATGAGATACATCAAATCGATTACAC | 1692 | eptB ORF |
| EptB_R | CGCGGATCCTTAGTTAGCCGCTGCCTC (BamHI) | ||
| CptA_F | ATGCATTCCACAGAAGTCCAGGCT | 1734 | cptA ORF |
| CptA_R | CGCGGATCCTTACTGATTACCCACCTG (BamHI) | ||
| OpgE_F | ATGAATTTAACCCTCAAAGAATCGC | 1584 | opgE ORF |
| OpgE_R | CGCGGATCCTTAAGGTTTCGGGTCG (BamHI) | ||
| Prbas_F | AAATTCTGATTGTTGAAGACGATAC | 1766 | pmrA-pmrB ORF |
| PrEAbas_R | CGCGGATCCTTATATCTGGTTTGCCAC (BamHI) | ||
| PmrB-F1 | ATG CAT TTT CTG CGC CGA CCA ATA | 1092 | pmrB ORF |
| PmrB-R | ATATGGATCCTTATATCTGGTTTGCCACGT (BamHI) | ||
| p5Up-KpnI-F3 | ATATGGTACCGACGCTGAATATGGGTCGCC (KpnI) | 2897 | pmrA(partial)-pmrB-proP(partial) |
| p5Up-SalI-R3 | ATATGTCGACAAGTTTTTTTCCCGGGGGCTGA (SalI) | ||
| US150-PmrB-SalI-F1 | ATATGTCGACGACATCTATAACTGGGACAA (SalI) | ||
| DS34-PmrB-BamHI-R1 | ATATGGATCCCCGTGTTCAGCGTGCTGGTG (BamHI) | 1271 | pmrB ORF, with 150 bp upstream and 34 bp downstream |
| DS86-PmrB-BamHI-R1 | ATATGGATCCCAGGTTAACGGAGGAGAGTG (BamHI) | 1328 | pmrB ORF, with 150 bp upstream and 86 bp downstream |
| DS103-PmrB-BamHI-R1 | ATATGGATCCACGCGCATACTCTCCTCCAG (BamHI) | 1345 | pmrB ORF, with 150 bp upstream and 103 bp downstream |
| DS126-PmrB-BamHI-R1 | ATATGGATCCCTTAAGGTTCACTTAATCTC (BamHI) | 1368 | pmrB ORF, with 150 bp upstream and 126 bp downstream |
| DS134-PmrB-BamHI-R1 | ATATGGATCCATTGAACTCTTAAGGTTCACT (BamHI) | 1376 | pmrB ORF, with 150 bp upstream and 134 bp downstream |
| DS176-PmrB-BamHI-R1 | ATATGGATCCGCTGAAACGGATGGCCTGAT (BamHI) | 1418 | pmrB ORF, with 150 bp upstream and 176 bp downstream |
| pZE_F | GAATTCATTAAAGAGGAGAAAGGT | N/A | Forward sequencing primers for pZE21 derivatives |
| pZE_R | TTTCGTTTTATTTGATGCCTCTAG | N/A | Reverse sequencing primers for pZE21 derivatives |
| PmrB(BL21DE3)_pKD13_F3 | GCTTTGGCTATATGCTGGTCGCGAATGAGGAAAACTAATTGAATCTGATGTGTAGGCTGGAGCTGCTTCG | 1403 | Site-directed mutation of pmrB |
| PmrB(BL21DE3)_pKD13_R3 | TTCAGCGTGCTGGTGGTCAGCAGCTTTCTTTATATCTGGTTTGCCACGTAATTCCGGGGATCCGTCGACC | ||
| PmrB_F | AATGAACCCTCGACCAACAC | 1376 | Detect site-directed mutation of pmrB with k1 |
| PmrB_R | CGCTGTCTTATCAGGCCAAT | ||
| PhoQ(BL21DE3)_pKD13_F3 | GTGATTACCACCGTTCGCGGCCAGGGCTATCTGTTCGAATTGCGCTGATGTGTAGGCTGGAGCTGCTTCG | 1403 | Site-directed mutation of phoQ |
| PhoQ(BL21DE3)_pKD13_R3 | TTAACGTAATGCGTGAAGTATGGACATATTTATTCATCTTTCGGCGTAGAATTCCGGGGATCCGTCGACC | ||
| PhoQ_F | TAATGGCAAAGTGGTGAGCA | 1772 | Detect site-directed mutation of phoQ with K1 |
| PhoQ_R | TTCTGCCAGTGACGTTCAAG | ||
| K1 | CAGTCATAGCCGAATAGCCT | Common primer for detecting site-directed mutation | |
| RT-PmrB-BL21SNP-F3 | CATTGCCATTCACAGCGCCACCCGCA | 180 | RT-PCR detection of pmrBBL21(DE3) |
| RT-PmrB-BL21SNP-R3 | TGCGTTTTCGCCAGCAGTTCCAGATGCA | ||
| 16S-F | AAGTTAATACCTTTGCTCATTGAC | 118 | 16S rRNA internal control for RT-PCR |
| 16S-R | GCTTTACGCCCAGTAATTCC |
| Strain | Insert Size (bp) | Genome Location in BL21 | Annotated Genes and Organization a |
|---|---|---|---|
| JL1365 | 10,660 | 210,471–208,308 | dnaE |
| 3,818,298–3,821,491 | viaA-ravA-kup | ||
| 1,144,385–1,147,209 | rne-yceQ | ||
| 2,263,105–2,265,581 | arnF-pmrD-menE-menC | ||
| JL1366 | 3926 | 4,242,936–4,239,011 | proP-pmrB-pmrA-EptA-adiC |
| JL1367 | 6989 | 4,239,038–4,243,091 | proP-pmrB-pmrA-EptA-adiC |
| 1,256,348–1,259,815 | kdsA-ldrA-ldrB-ldrC-chaA | ||
| JL1368 | 3813 | 2,263,769–2,261,742 | arnT-arnE-arnF-pmrD |
| 2,759,470–2,757,690 | syd-queF-ygdH | ||
| JL1369 | 5269 | 1,879,745–1,877,377 | purT-eda-edd |
| 4,237,732–4,240,628 | proP-pmrB-pmrA |
| Strain | Colistin MIC (µg/mL) | Lipid A Modification | |
|---|---|---|---|
| pEtN | Ara4N | ||
| TOP10/pZE21 | 0.5 | - a | - |
| TOP10/pPmrB | 0.5 | ND b | ND |
| TOP10/pUS150-PmrB-DS34 | 0.5 | ND | ND |
| TOP10/pUS150-PmrB-DS86 | 0.5 | - | - |
| TOP10/pUS150-PmrB-DS103 | 4 | + | + |
| TOP10/pUS150-PmrB-DS126 | 16 | + | + |
| TOP10/pUS150-PmrB-DS134 | 16 | ND | ND |
| TOP10/pUS150-PmrB-DS176 | 16 | ND | ND |
| TOP10/pUS150-PmrBMG1655-DS176 | 0.5 | ND | ND |
| BL21, wild type | 16 | + | + |
| JL1435 (BL21, phoQ::kan) | 16 | ND | ND |
| JL1436 (BL21, pmrB::kan) | 0.5 | ND | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, F.; Hinenoya, A.; Zeng, X.; Li, X.-P.; Guan, Z.; Lin, J. Critical Role of 3′-Downstream Region of pmrB in Polymyxin Resistance in Escherichia coli BL21(DE3). Microorganisms 2021, 9, 655. https://doi.org/10.3390/microorganisms9030655
Xu F, Hinenoya A, Zeng X, Li X-P, Guan Z, Lin J. Critical Role of 3′-Downstream Region of pmrB in Polymyxin Resistance in Escherichia coli BL21(DE3). Microorganisms. 2021; 9(3):655. https://doi.org/10.3390/microorganisms9030655
Chicago/Turabian StyleXu, Fuzhou, Atsushi Hinenoya, Ximin Zeng, Xing-Ping Li, Ziqiang Guan, and Jun Lin. 2021. "Critical Role of 3′-Downstream Region of pmrB in Polymyxin Resistance in Escherichia coli BL21(DE3)" Microorganisms 9, no. 3: 655. https://doi.org/10.3390/microorganisms9030655
APA StyleXu, F., Hinenoya, A., Zeng, X., Li, X.-P., Guan, Z., & Lin, J. (2021). Critical Role of 3′-Downstream Region of pmrB in Polymyxin Resistance in Escherichia coli BL21(DE3). Microorganisms, 9(3), 655. https://doi.org/10.3390/microorganisms9030655






