Comparative Analysis of Cytokine Expression in Oral Keratinocytes and THP-1 Macrophages in Response to the Most Prevalent Serotypes of Aggregatibacter actinomycetemcomitans
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria Stains
2.2. Oral Keratinocytes Culture
2.3. THP-1 Derived Macrophages Culture
2.4. Infection Assay
2.5. RNA Extraction and RT-PCR
2.6. qPCR
2.7. Statistical Analysis
3. Results
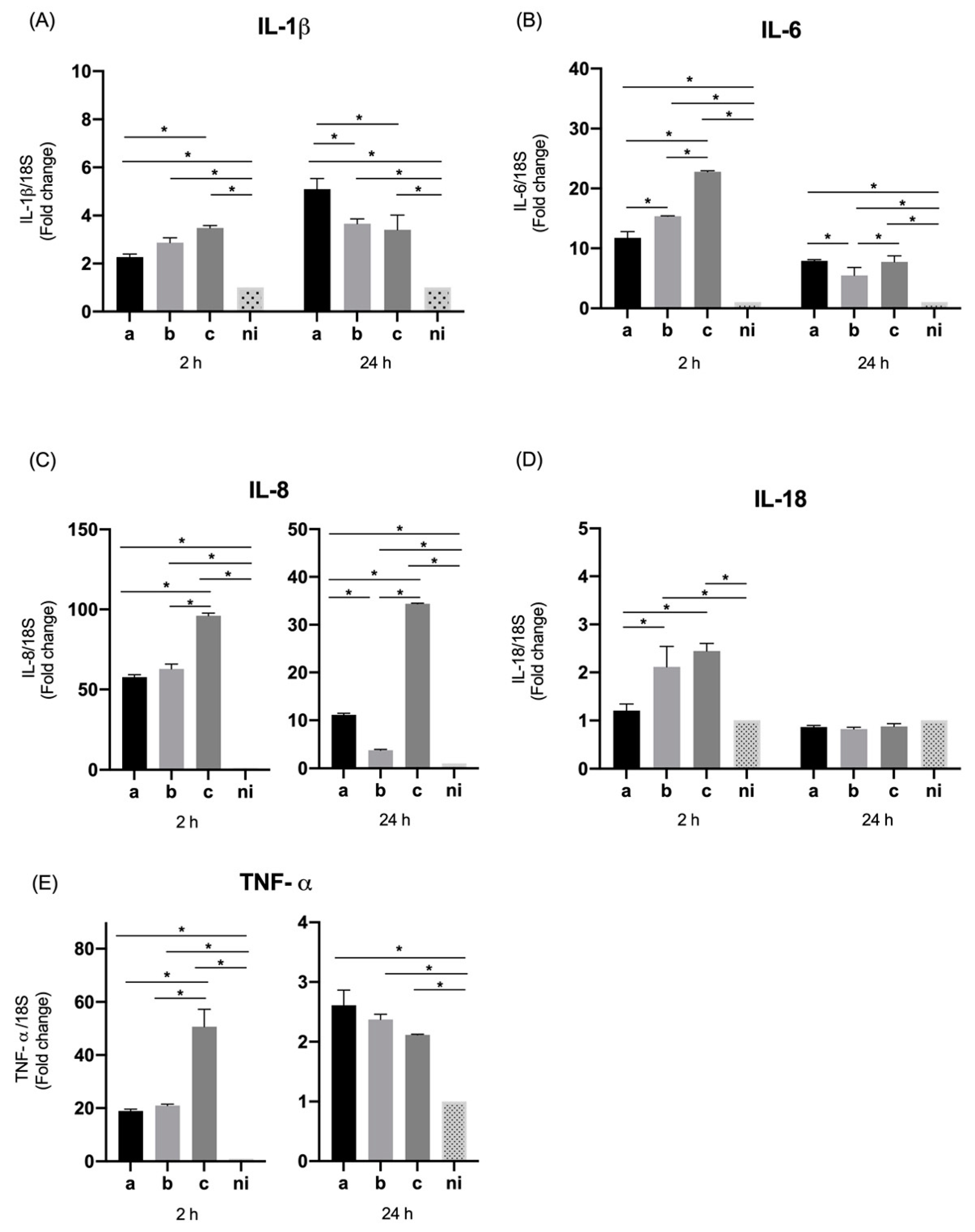
3.1. Expression of Pro-Inflammatory Cytokines and Chemokines by Oral Epithelial Cells
3.2. Expression of Molecules Associated with Tissue Destruction by Oral Epithelial Cells
3.3. Expression of TLR Receptors in Oral Epithelial Cells
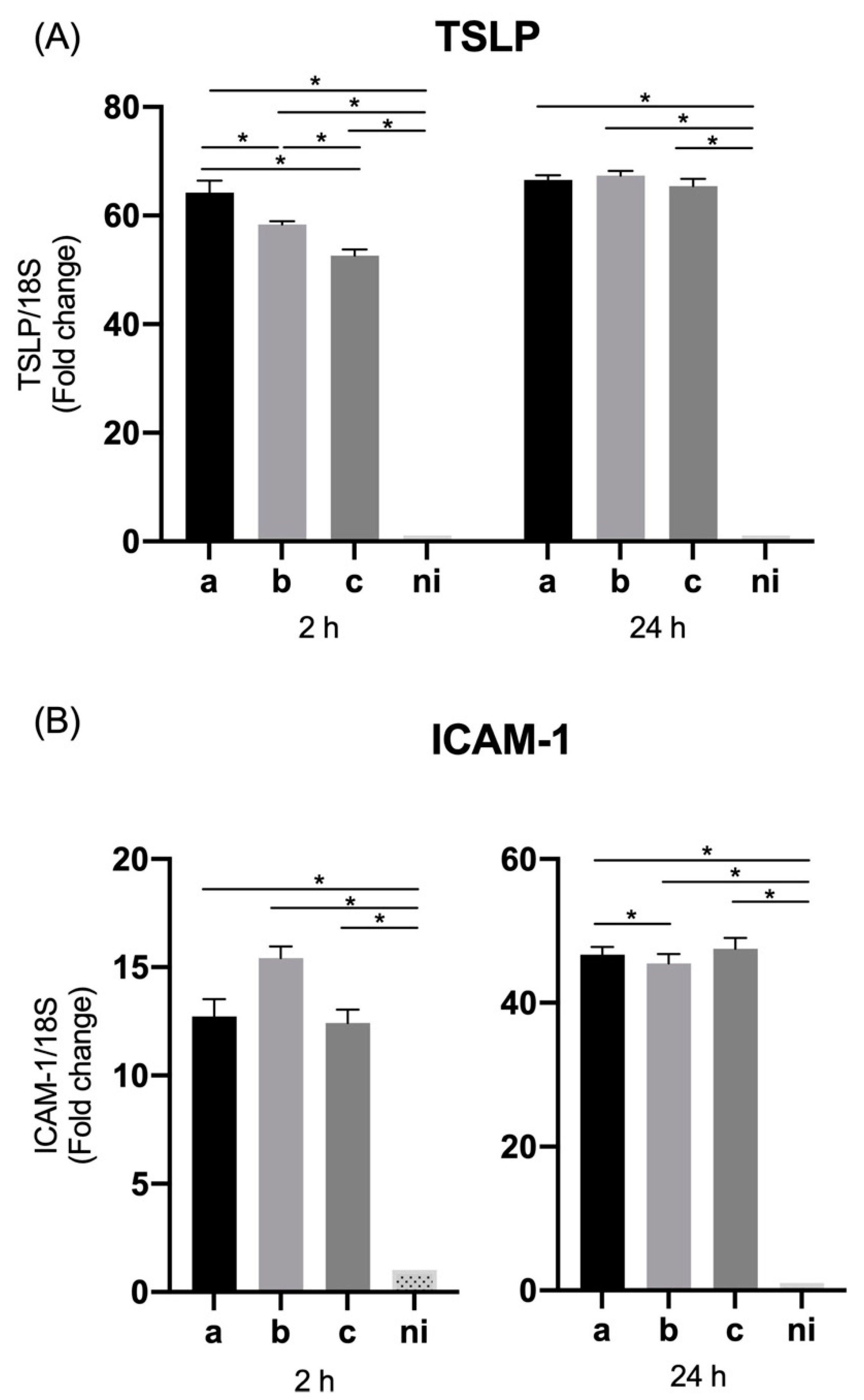
3.4. Expression of Thymic Stromal Lymphopoietin (TSLP) and ICAM-1 by Oral Epithelial Cells
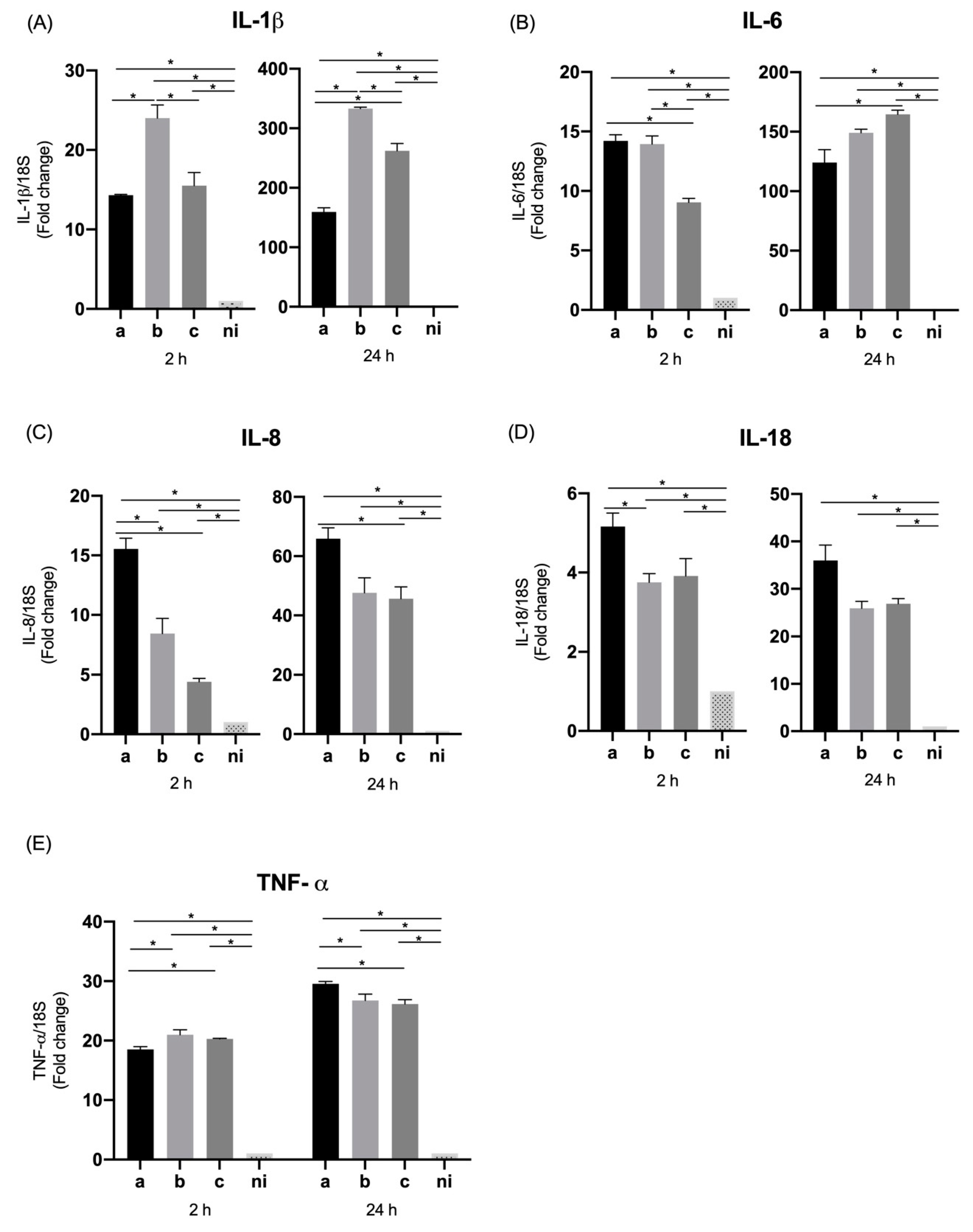
3.5. Expression of Pro-Inflammatory Cytokines and Chemokines by THP-1 Macrophage Cells
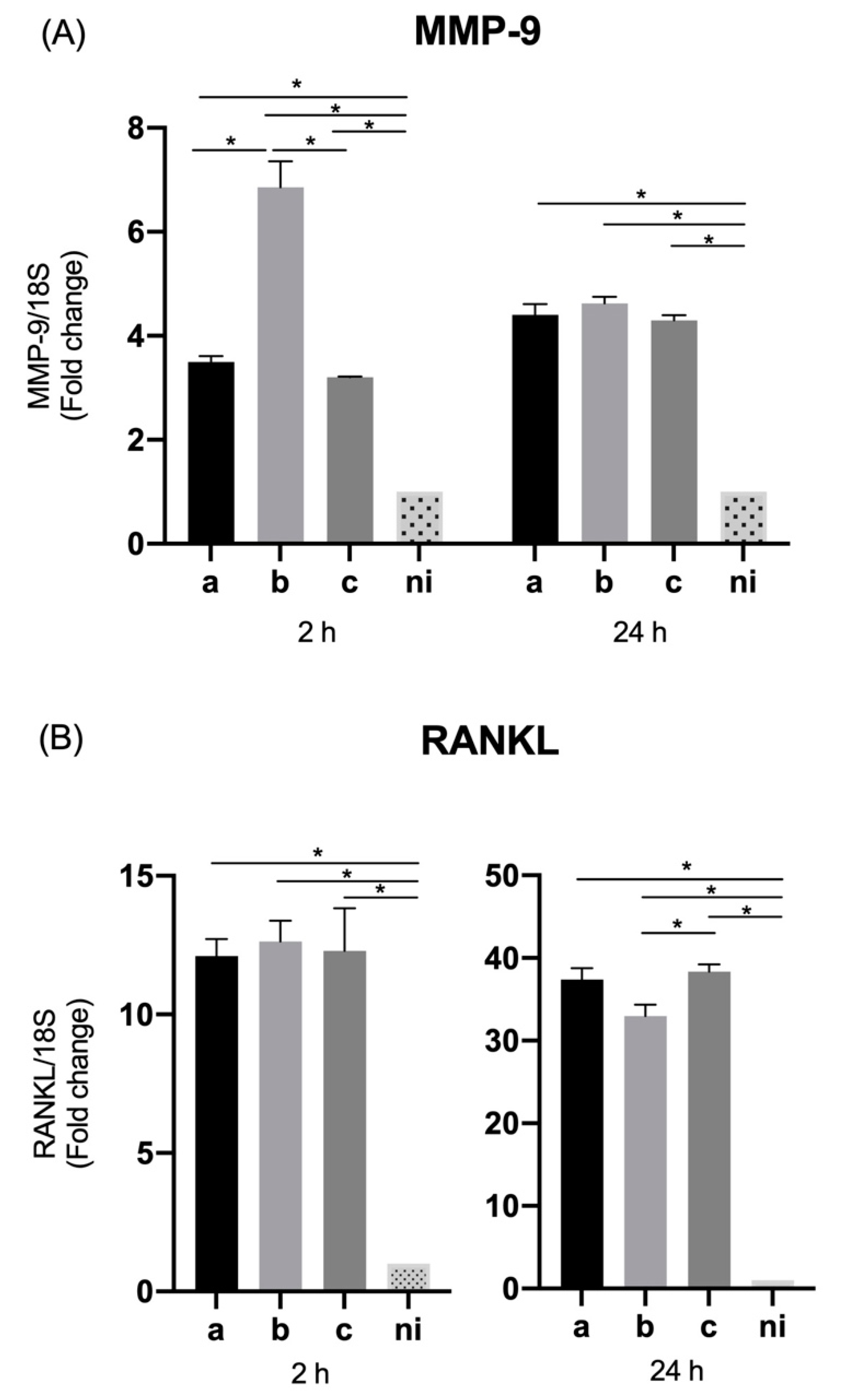
3.6. Expression of MMP-9 and RANKL by THP-1 Macrophage Cells
3.7. Expression of TLR Receptors in THP-1 Macrophage Cells
3.8. Expression of TSLP and ICAM-1 in THP-1 Macrophage Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89 (Suppl. 1), S173–S182. [Google Scholar] [CrossRef]
- Sanz, M.; Marco Del Castillo, A.; Jepsen, S.; Gonzalez-Juanatey, J.R.; D’Aiuto, F.; Bouchard, P.; Chapple, I.; Dietrich, T.; Gotsman, I.; Graziani, F.; et al. Periodontitis and cardiovascular diseases: Consensus report. J. Clin. Periodontol. 2020, 47, 268–288. [Google Scholar] [CrossRef]
- Khumaedi, A.I.; Purnamasari, D.; Wijaya, I.P.; Soeroso, Y. The relationship of diabetes, periodontitis and cardiovascular disease. Diabetes Metab. Syndr. 2019, 13, 1675–1678. [Google Scholar] [CrossRef]
- Polak, D.; Shapira, L. An update on the evidence for pathogenic mechanisms that may link periodontitis and diabetes. J. Clin. Periodontol. 2018, 45, 150–166. [Google Scholar] [CrossRef] [PubMed]
- Opacic, J.; Maldonado, A.; Ramseier, C.A.; Laugisch, O. Einfluss der Parodontitis auf Schwangerschaft und Geburt [Influence of periodontitis on pregnancy and childbirth]. Swiss Dent. J. 2019, 129, 581–589. [Google Scholar] [PubMed]
- Dominy, S.S.; Lynch, C.; Ermini, F.; Benedyk, M.; Marczyk, A.; Konradi, A.; Nguyen, M.; Haditsch, U.; Raha, D.; Griffin, C.; et al. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 2019, 5, eaau3333. [Google Scholar] [CrossRef]
- Nanayakkara, S.; Zhou, X. Periodontitis May Be Associated With Chronic Kidney Disease, but Evidence on Causal Association Is Limited. J. Evid. Based Dent. Pract. 2019, 19, 192–194. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Ferreira, R.; de Brito Silva, R.; Magno, M.B.; Carvalho Almeida, A.P.C.P.S.; Fagundes, N.C.F.; Maia, L.C.; Lima, R.R. Does periodontitis represent a risk factor for rheumatoid arthritis? A systematic review and meta-analysis. Ther. Adv. Musculoskelet. Dis. 2019, 11, 1759720X19858514. [Google Scholar] [CrossRef] [PubMed]
- Kalakonda, B.; Koppolu, P.; Baroudi, K.; Mishra, A. Periodontal Systemic Connections-Novel Associations-A Review of the Evidence with Implications for Medical Practitioners. Int. J. Health Sci. 2016, 10, 293–307. [Google Scholar] [CrossRef]
- Avila, M.; Ojcius, D.M.; Yilmaz, O. The oral microbiota: Living with a permanent guest. DNA Cell Biol. 2009, 28, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Mosaddad, S.A.; Tahmasebi, E.; Yazdanian, A.; Rezvani, M.B.; Seifalian, A.; Yazdanian, M.; Tebyanian, H. Oral microbial biofilms: An update. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2005–2019. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G. Immunomicrobial pathogenesis of periodontitis: Keystones, pathobionts, and host response. Trends Immunol. 2014, 35, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Meyle, J.; Chapple, I. Molecular aspects of the pathogenesis of periodontitis. Periodontology 2000 2015, 69, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Rosier, B.T.; Marsh, P.D.; Mira, A. Resilience of the Oral Microbiota in Health: Mechanisms that Prevent Dysbiosis. J. Dent. Res. 2018, 97, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Meuric, V.; Le Gall-David, S.; Boyer, E.; Acuña-Amador, L.; Martin, B.; Fong, S.B.; Barloy-Hubler, F.; Bonnaure-Mallet, M. Signature of Microbial Dysbiosis in Periodontitis. Appl. Environ. Microbiol. 2017, 83, e00462-17. [Google Scholar] [CrossRef]
- Deng, Z.L.; Szafrański, S.P.; Jarek, M.; Bhuju, S.; Wagner-Döbler, I. Dysbiosis in chronic periodontitis: Key microbial players and interactions with the human host. Sci. Rep. 2017, 7, 3703. [Google Scholar] [CrossRef]
- Slots, J.; Ting, M. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: Occurrence and treatment. Periodontology 2000 1999, 20, 82–121. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725. [Google Scholar] [CrossRef]
- Fine, D.H.; Patil, A.G.; Velusamy, S.K. Aggregatibacter actinomycetemcomitans (Aa) Under the Radar: Myths and Misunderstandings of Aa and Its Role in Aggressive Periodontitis. Front. Immunol. 2019, 10, 728. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Pormohammad, A.; Eslami, H.; Shokouhi, B.; Fakhrzadeh, V.; Kafil, H.S. Oral pathogenesis of Aggregatibacter actinomycetemcomitans. Microb Pathog. 2017, 113, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Raja, M.; Ummer, F.; Dhivakar, C.P. Aggregatibacter actinomycetemcomitans—A tooth killer? J. Clin. Diagn. Res. 2014, 8, ZE13–ZE16. [Google Scholar] [CrossRef] [PubMed]
- Belibasakis, G.N.; Maula, T.; Bao, K.; Lindholm, M.; Bostanci, N.; Oscarsson, J.; Ihalin, R.; Johansson, A. Virulence and Pathogenicity Properties of Aggregatibacter actinomycetemcomitans. Pathogens 2019, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Oscarsson, J.; Claesson, R.; Lindholm, M.; Höglund Åberg, C.; Johansson, A. Tools of Aggregatibacter actinomycetemcomitans to Evade the Host Response. J. Clin. Med. 2019, 8, 1079. [Google Scholar] [CrossRef]
- Tuuli, A.; Laura, K.; Terhi, M.; Jan, O.; Riikka, I. Aggregatibacter actinomycetemcomitans LPS binds human interleukin-8. J. Oral Microbiol. 2018, 11, 1549931. [Google Scholar] [CrossRef]
- Takada, K.; Saito, M.; Tsuzukibashi, O.; Kawashima, Y.; Ishida, S.; Hirasawa, M. Characterization of a new serotype g isolate of Aggregatibacter actinomycetemcomitans. Mol. Oral Microbiol. 2010, 25, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Melgar-Rodríguez, S.; Díaz-Zúñiga, J.; Alvarez, C.; Rojas, L.; Monasterio, G.; Carvajal, P.; Escobar, A.; Sanz, M.; Vernal, R. Serotype b of Aggregatibacter actinomycetemcomitans increases osteoclast and memory T-lymphocyte activation. Mol. Oral Microbiol. 2016, 31, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Zúñiga, J.; Monasterio, G.; Alvarez, C.; Melgar-Rodríguez, S.; Benítez, A.; Ciuchi, P.; García, M.; Arias, J.; Sanz, M.; Vernal, R. Variability of the dendritic cell response triggered by different serotypes of Aggregatibacter actinomycetemcomitans or Porphyromonas gingivalis is toll-like receptor 2 (TLR2) or TLR4 dependent. J. Periodontol. 2015, 86, 108–119. [Google Scholar] [CrossRef]
- Alvarez, C.; Benítez, A.; Rojas, L.; Pujol, M.; Carvajal, P.; Díaz-Zúñiga, J.; Vernal, R. Differential expression of CC chemokines (CCLs) and receptors (CCRs) by human T lymphocytes in response to different Aggregatibacter actinomycetemcomitans serotypes. J. Appl. Oral Sci. 2015, 23, 536–546. [Google Scholar] [CrossRef]
- Vernal, R.; Leon, R.; Herrera, D.; Garcia-Sanz, J.A.; Silva Sanz, M. Variability in the response of human dendritic cells stimulated with Porphyromonas gingivalis or Aggregatibacter actinomycetemcomitans. J. Periodontal Res. 2008, 43, 689–697. [Google Scholar] [CrossRef]
- Díaz-Zúñiga, J.; Yáñez, J.P.; Alvarez, C.; Melgar-Rodríguez, S.; Hernández, M.; Sanz, M.; Vernal, R. Serotype-dependent response of human dendritic cells stimulated with Aggregatibacter actinomycetemcomitans. J. Clin. Periodontol. 2014, 41, 242–251. [Google Scholar] [CrossRef]
- Henderson, B.; Nair, S.P.; Ward, J.M.; Wilson, M. Molecular pathogenicity of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans. Annu. Rev. Microbiol. 2003, 57, 29–55. [Google Scholar] [CrossRef]
- Herbert, B.A.; Novince, C.M.; Kirkwood, K.L. Aggregatibacter actinomycetemcomitans, a potent immunoregulator of the periodontal host defense system and alveolar bone homeostasis. Mol. Oral Microbiol. 2016, 31, 207–227. [Google Scholar] [CrossRef]
- Höglund Åberg, C.; Haubek, D.; Kwamin, F.; Johansson, A.; Claesson, R. Leukotoxic activity of Aggregatibacter actinomycetemcomitans and periodontal attachment loss. PLoS ONE 2014, 9, e104095. [Google Scholar] [CrossRef]
- Groeger, S.E.; Meyle, J. Epithelial barrier and oral bacterial infection. Periodontology 2000 2015, 69, 46–67. [Google Scholar] [CrossRef] [PubMed]
- Dale, B.A. Periodontal epithelium: A newly recognized role in health and disease. Periodontology 2000 2002, 30, 70–78. [Google Scholar] [CrossRef] [PubMed]
- McCormick, T.S.; Weinberg, A. Epithelial cell-derived antimicrobial peptides are multifunctional agents that bridge innate and adaptive immunity. Periodontology 2000 2010, 54, 195–206. [Google Scholar] [CrossRef]
- Ramage, G.; Lappin, D.F.; Millhouse, E.; Malcolm, J.; Jose, A.; Yang, J.; Bradshaw, D.J.; Pratten, J.R.; Culshaw, S. The epithelial cell response to health and disease associated oral biofilm models. J. Periodontal Res. 2017, 52, 325–333. [Google Scholar] [CrossRef]
- Kochi, S.; Yamashiro, K.; Hongo, S.; Yamamoto, T.; Ugawa, Y.; Shimoe, M.; Kawamura, M.; Hirata-Yoshihara, C.; Ideguchi, H.; Maeda, H.; et al. Aggregatibacter actinomycetemcomitans regulates the expression of integrins and reduces cell adhesion via integrin α5 in human gingival epithelial cells. Mol. Cell. Biochem. 2017, 436, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Suga, T.; Mitani, A.; Mogi, M.; Kikuchi, T.; Fujimura, T.; Takeda, H.; Hishikawa, T.; Yamamoto, G.; Hayashi, J.; Ishihara, Y.; et al. Aggregatibacter actinomycetemcomitans lipopolysaccharide stimulated epithelial cells produce interleukin-15 that regulates T cell activation. Arch. Oral Biol. 2013, 58, 1541–1548. [Google Scholar] [CrossRef]
- Park, E.K.; Jung, H.S.; Yang, H.I.; Yoo, M.C.; Kim, C.; Kim, K.S. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 2007, 56, 45–50. [Google Scholar] [CrossRef]
- Daigneault, M.; Preston, J.A.; Marriott, H.M.; Whyte, M.K.; Dockrell, D.H. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS ONE 2010, 5, e8668. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Darveau, R.P. Periodontitis: A polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 2010, 8, 481–490. [Google Scholar] [CrossRef]
- Gemmell, E.; Marshall, R.I.; Seymour, G.J. Cytokines and prostaglandins in immune homeostasis and tissue destruction in periodontal disease. Periodontology 2000 1997, 14, 112–143. [Google Scholar] [CrossRef]
- Kantrong, N.; To, T.T.; Darveau, R.P. Gingival Epithelial Cell Recognition of Lipopolysaccharide. Adv. Exp. Med. Biol. 2019, 1197, 55–67. [Google Scholar] [CrossRef]
- Yang, J.; Zhu, Y.; Duan, D.; Wang, P.; Xin, Y.; Bai, L.; Liu, Y.; Xu, Y. Enhanced activity of macrophage M1/M2 phenotypes in periodontitis. Arch. Oral Biol. 2018, 96, 234–242. [Google Scholar] [CrossRef]
- Pöllänen, M.T.; Salonen, J.I.; Uitto, V.J. Structure and function of the tooth-epithelial interface in health and disease. Periodontology 2000 2003, 31, 12–31. [Google Scholar] [CrossRef]
- Graves, D. Cytokines that promote periodontal tissue destruction. J. Periodontol. 2008, 79 (Suppl. 8), 1585–1591. [Google Scholar] [CrossRef]
- Pan, W.; Wang, Q.; Chen, Q. The cytokine network involved in the host immune response to periodontitis. Int. J. Oral Sci. 2019, 11, 30. [Google Scholar] [CrossRef]
- Dickinson, B.C.; Moffatt, C.E.; Hagerty, D.; Whitmore, S.E.; Brown, T.A.; Graves, D.T.; Lamont, R.J. Interaction of oral bacteria with gingival epithelial cell multilayers. Mol. Oral Microbiol. 2011, 26, 210–220. [Google Scholar] [CrossRef]
- Tanabe, S.I.; Grenier, D. Macrophage tolerance response to Aggregatibacter actinomycetemcomitans lipopolysaccharide induces differential regulation of tumor necrosis factor-alpha, interleukin-1 beta and matrix metalloproteinase 9 secretion. J. Periodontal Res. 2008, 43, 372–377. [Google Scholar] [CrossRef]
- Fine, D.H.; Schreiner, H.; Velusamy, S.K. Aggregatibacter, A Low Abundance Pathobiont That Influences Biogeography, Microbial Dysbiosis, and Host Defense Capabilities in Periodontitis: The History of A Bug, And Localization of Disease. Pathogens 2020, 9, 179. [Google Scholar] [CrossRef]
- Nowarski, R.; Jackson, R.; Gagliani, N.; de Zoete, M.R.; Palm, N.W.; Bailis, W.; Low, J.S.; Harman, C.C.; Graham, M.; Elinav, E.; et al. Epithelial IL-18 Equilibrium Controls Barrier Function in Colitis. Cell 2015, 163, 1444–1456. [Google Scholar] [CrossRef]
- Sugawara, S.; Uehara, A.; Nochi, T.; Yamaguchi, T.; Ueda, H.; Sugiyama, A.; Hanzawa, K.; Kumagai, K.; Okamura, H.; Takada, H. Neutrophil proteinase 3-mediated induction of bioactive IL-18 secretion by human oral epithelial cells. J. Immunol. 2001, 167, 6568–6575. [Google Scholar] [CrossRef]
- Vokurka, J.; Klapusová, L.; Pantuckova, P.; Kukletova, M.; Kukla, L.; Holla, L.I. The association of MMP-9 and IL-18 gene promoter polymorphisms with gingivitis in adolescents. Arch. Oral Biol. 2009, 54, 172–178. [Google Scholar] [CrossRef]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Dixit, V.M. Mechanisms and functions of inflammasomes. Cell 2014, 157, 1013–1022. [Google Scholar] [CrossRef]
- Xue, F.; Shu, R.; Xie, Y. The expression of NLRP3, NLRP1 and AIM2 in the gingival tissue of periodontitis patients: RT-PCR study and immunohistochemistry. Arch. Oral Biol. 2015, 60, 948–958. [Google Scholar] [CrossRef]
- Isaza-Guzman, D.M.; Medina-Piedrahita, V.M.; Gutierrez-Henao, C.; Tobon-Arroyave, S.I. Salivary Levels of NLRP3 Inflammasome-Related Proteins as Potential Biomarkers of Periodontal Clinical Status. J. Periodontol. 2017, 88, 1329–1338. [Google Scholar] [CrossRef]
- Kim, S.; Park, M.H.; Song, Y.R.; Na, H.S.; Chung, J. Aggregatibacter actinomycetemcomitans-Induced AIM2 Inflammasome Activation Is Suppressed by Xylitol in Differentiated THP-1 Macrophages. J. Periodontol. 2016, 87, e116–e126. [Google Scholar] [CrossRef]
- Belibasakis, G.N.; Johansson, A. Aggregatibacter actinomycetemcomitans targets NLRP3 and NLRP6 inflammasome expression in human mononuclear leukocytes. Cytokine 2012, 59, 124–130. [Google Scholar] [CrossRef]
- Zhao, P.; Liu, J.; Pan, C.; Pan, Y. NLRP3 inflammasome is required for apoptosis of Aggregatibacter actinomycetemcomitans-infected human osteoblastic MG63 cells. Acta Histochem. 2014, 116, 1119–1124. [Google Scholar] [CrossRef]
- Mäkelä, M.; Salo, T.; Uitto, V.J.; Larjava, H. Matrix metalloproteinases (MMP-2 and MMP-9) of the oral cavity: Cellular origin and relationship to periodontal status. J. Dent. Res. 1994, 73, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Kowolik, M.J.; Grant, M. Myeloperoxidase activity in human gingival crevicular neutrophils. Arch. Oral Biol. 1983, 28, 293–295. [Google Scholar] [CrossRef]
- Li, Y.Q.; Yan, J.P.; Xu, W.L.; Wang, H.; Xia, Y.J.; Wang, H.J.; Zhu, Y.Y.; Huang, X.J. ADAM17 mediates MMP9 expression in lung epithelial cells. PLoS ONE 2013, 8, e51701. [Google Scholar] [CrossRef]
- Weiler, J.; Mohr, M.; Zänker, K.S.; Dittmar, T. Matrix metalloproteinase-9 (MMP9) is involved in the TNF-α-induced fusion of human M13SV1-Cre breast epithelial cells and human MDA-MB-435-pFDR1 cancer cells. Cell Commun. Signal. 2018, 16, 14. [Google Scholar] [CrossRef]
- Chang, M.C.; Pan, Y.H.; Wu, H.L.; Lu, Y.J.; Liao, W.C.; Yeh, C.Y.; Lee, J.J.; Jeng, J.H. Stimulation of MMP-9 of oral epithelial cells by areca nut extract is related to TGF-β/Smad2-dependent and -independent pathways and prevented by betel leaf extract, hydroxychavicol and melatonin. Aging 2019, 11, 11624–11639. [Google Scholar] [CrossRef]
- Bodet, C.; Chandad, F.; Grenier, D. Inhibition of host extracellular matrix destructive enzyme production and activity by a high-molecular-weight cranberry fraction. J. Periodontal Res. 2007, 42, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, R.; Usui, M.; Yamamoto, G.; Nishii, K.; Tsukamoto, Y.; Okamatsu, Y.; Sato, T.; Asou, Y.; Nakashima, K.; Yamamoto, M. Tumor necrosis factor-α enhances RANKL expression in gingival epithelial cells via protein kinase A signaling. J. Periodontal Res. 2014, 49, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Usui, M.; Sato, T.; Yamamoto, G.; Okamatsu, Y.; Hanatani, T.; Moritani, Y.; Sano, K.; Yamamoto, M.; Nakashima, K. Gingival epithelial cells support osteoclastogenesis by producing receptor activator of nuclear factor kappa B ligand via protein kinase A signaling. J. Periodontal Res. 2016, 51, 462–470. [Google Scholar] [CrossRef]
- Zhou, L.; Le, Y.; Tian, J.; Yang, X.; Jin, R.; Gai, X.; Sun, Y. Cigarette smoke-induced RANKL expression enhances MMP-9 production by alveolar macrophages. Int. J. Chron. Obstruct. Pulmon. Dis. 2018, 14, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Park, S.R.; Kim, D.J.; Han, S.H.; Kang, M.J.; Lee, J.Y.; Jeong, Y.J.; Lee, S.J.; Kim, T.H.; Ahn, S.G.; Yoon, J.H.; et al. Diverse Toll-like receptors mediate cytokine production by Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans in macrophages. Infect. Immun. 2014, 82, 1914–1920. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Botello, N.R.; García-Hernández, A.L.; Moreno-Fierros, L. Expression of toll-like receptors 2, 4 and 9 is increased in gingival tissue from patients with type 2 diabetes and chronic periodontitis. J. Periodontal Res. 2012, 47, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Lima, H.R.; Gelani, V.; Fernandes, A.P.; Gasparoto, T.H.; Torres, S.A.; Santos, C.F.; Garlet, G.P.; da Silva, J.S.; Campanelli, A.P. The essential role of toll like receptor-4 in the control of Aggregatibacter actinomycetemcomitans infection in mice. J. Clin. Periodontol. 2010, 37, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Gelani, V.; Fernandes, A.P.; Gasparoto, T.H.; Garlet, T.P.; Cestari, T.M.; Lima, H.R.; Ramos, E.S.; de Souza Malaspina, T.S.; Santos, C.F.; Garlet, G.P.; et al. The role of toll-like receptor 2 in the recognition of Aggregatibacter actinomycetemcomitans. J. Periodontol. 2009, 80, 2010–2019. [Google Scholar] [CrossRef] [PubMed]
- Beklen, A.; Hukkanen, M.; Richardson, R.; Konttinen, Y.T. Immunohistochemical localization of Toll-like receptors 1-10 in periodontitis. Oral Microbiol. Immunol. 2008, 23, 425–431. [Google Scholar] [CrossRef]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.S.; Lee, H.; Lee, J.O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, R.; Akashi, S.; Ogata, H.; Nagai, Y.; Fukudome, K.; Miyake, K.; Kimoto, M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 1999, 189, 1777–1782. [Google Scholar] [CrossRef]
- Ando-Suguimoto, E.S.; Benakanakere, M.R.; Mayer, M.P.A.; Kinane, D.F. Distinct Signaling Pathways between Human Macrophages and Primary Gingival Epithelial Cells by Aggregatibacter actinomycetemcomitans. Pathogens 2020, 9, 248. [Google Scholar] [CrossRef]
- Takai, T. TSLP expression: Cellular sources, triggers, and regulatory mechanisms. Allergol. Int. 2012, 61, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Takai, T.; Chen, X.; Xie, Y.; Vu, A.T.; Le, T.A.; Kinoshita, H.; Kawasaki, J.; Kamijo, S.; Hara, M.; Ushio, H.; et al. TSLP expression induced via Toll-like receptor pathways in human keratinocytes. Methods Enzymol. 2014, 535, 371–387. [Google Scholar] [CrossRef]
- Tsutsumi, T.; Nakashima, K.; Isoda, T.; Yokota, M.; Nishihara, T. Involvement of adhesion molecule in in vitro plaque-like formation of macrophages stimulated with Aggregatibacter actinomycetemcomitans lipopolysaccharide. J. Periodontal Res. 2010, 45, 550–556, Erratum in J. Periodontal Res. 2010, 45, 702. [Google Scholar] [CrossRef]
- Shimada, T.; Sugano, N.; Nishihara, R.; Suzuki, K.; Tanaka, H.; Ito, K. Differential effects of five Aggregatibacter actinomycetemcomitans strains on gingival epithelial cells. Oral Microbiol. Immunol. 2008, 23, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Sugano, N.; Ikeda, K.; Shimada, K.; Iizuka, T.; Ito, K. Protease-activated receptor 2 mediates interleukin-8 and intercellular adhesion molecule-1 expression in response to Aggregatibacter actinomycetemcomitans. Oral Microbiol. Immunol. 2009, 24, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Lyck, R.; Enzmann, G. The physiological roles of ICAM-1 and ICAM-2 in neutrophil migration into tissues. Curr. Opin. Hematol. 2015, 22, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Canonica, G.W.; Ciprandi, G.; Pesce, G.P.; Buscaglia, S.; Paolieri, F.; Bagnasco, M. ICAM-1 on epithelial cells in allergic subjects: A hallmark of allergic inflammation. Int. Arch. Allergy Immunol. 1995, 107, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Fine, D.H.; Patil, A.G.; Loos, B.G. Classification and diagnosis of aggressive periodontitis. J. Periodontol. 2018, 89 (Suppl. 1), S103–S119. [Google Scholar] [CrossRef] [PubMed]
- Van Dyke, T.E.; Bartold, P.M.; Reynolds, E.C. The Nexus between Periodontal Inflammation and Dysbiosis. Front. Immunol. 2020, 11, 511. [Google Scholar] [CrossRef]
- Loos, B.G.; Van Dyke, T.E. The role of inflammation and genetics in periodontal disease. Periodontology 2000 2020, 83, 26–39. [Google Scholar] [CrossRef]
- Umeda, J.E.; Longo, P.L.; Simionato, M.R.; Mayer, M.P. Differential transcription of virulence genes in Aggregatibacter actinomycetemcomitans serotypes. J. Oral Microbiol. 2013, 5, 21473. [Google Scholar] [CrossRef]




| Target | Forward | Reverse |
|---|---|---|
| IL-1β | ctgtcctgcgtgttgaaaga | ttgggtaatttttgggatctaca |
| IL-6 | gccagctatgaactccttct | gaaggcagcaggcaacac |
| IL-8 | agatctgaagtgtgatgactc | gaagcttgtgtgctctgctgtctc |
| IL-18 | gatagccagcctagaggtatgg | ccttgatgttatcaggaggattca |
| TNF-α | cagcctcttctccttcctgat | gccagagggctgattagaga |
| MMP-9 | gccactactgtgcctttgagtc | ccctcagagaatcgccagtact |
| TLR-2 | ctctcggtgtcggaatgtc | aggatcagcaggaacagagc |
| TLR-4 | ccctcccctgtacccttct | tccctgccttgaataccttc |
| TLR-6 | actgaccttcctggatgtggca | tgacctcatcttctggcagctc |
| TSLP | gccatgaaaactaaggctgc | cgccacaatccttgtaattg |
| ICAM | agcggctgacgtgtgcagtaat | tctgagacctctggcttcgtca |
| RANKL | tgattcatgtaggagaattaaacagg | gatgtgctgtgatccaacga |
| 18S | ctcaacacgggaaacctcac | cgctccaccaactaagaacg |
| Oral Epithelial Cells (OKF6/TERT2) Mean (S.D.) | Macrophage Cells (THP-1) Mean (S.D.) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 h | 24 h | 2 h | 24 h | |||||||||||||
| a | b | c | n.i. | a | b | c | n.i. | a | b. | c. | n.i. | a | b | c | n.i. | |
| IL-1β | 2.26 (±0.1) | 2.87 (±0.1) | 3.48 (±0.09) | 1 | 5.09 (±0.4) | 3.65 (±0.2) | 3.40 (±0.6) | 1 | 14.32 (±0.09) | 24 (±1.6) | 15.52 (±1.6) | 1 | 159.52 (±7.0) | 333.16 (±2.3) | 262.03 (±12.3) | 1 |
| IL-6 | 11.75 (±1.0) | 15.39 (±0.05) | 22.78 (±0.1) | 1 | 7.91 (±0.1) | 5.48 (±1.3) | 7.73 (±1.0) | 1 | 14.19 (±0.5) | 13.94 (±0.6) | 9.03 (±0.3) | 1 | 124.13 (±10.7) | 149 (±3.0) | 164.62 (±3.4) | 1 |
| IL-8 | 57.69 (±1.4) | 62.75 (±3.0) | 96.01 (±1.6) | 1 | 11.14 (±0.3) | 3.77 (±0.1) | 34.41 (±0.1) | 1 | 15.53 (±0.9) | 8.46 (±1.2) | 4.40 (±0.2) | 1 | 65.93 (±3.6) | 47.60 (±5.1) | 45.63 (±4.0) | 1 |
| IL-18 | 1.20 (±0.1) | 2.11 (±0.4) | 2.44 (±0.1) | 1 | 0.86 (±0.03) | 0.82 (±0.03) | 0.87 (±0.05) | 1 | 5.16 (±0.3) | 3.75 (±0.2) | 3.91 (±0.4) | 1 | 35.98 (±3.2) | 25.91 (±1.4) | 26.85 (±1.0) | 1 |
| TNF-α | 18.90 (±0.6) | 20.97 (±0.5) | 50.64 (±6.6) | 1 | 2.61 (±0.2) | 2.37 (±0.09) | 2.11 (±0.009) | 1 | 18.51 (±0.4) | 20.99 (±0.8) | 20.29 (±0.07) | 1 | 29.54 (±0.3) | 26.74 (±1.0) | 26.15 (±0.7) | 1 |
| MMP-9 | 2.59 (±0.2) | 3.31 (±0.3) | 4.66 (±0.2) | 1 | 3.64 (±0.1) | 11.23 (±0.07) | 35.21 (±2.3) | 1 | 3.49 (±0.1) | 6.86 (±0.4) | 3.20 (±0.01) | 1 | 4.40 (±0.2) | 4.62 (±0.1) | 4.3 (±0.09) | 1 |
| RANKL | 2.53 (±0.2) | 4.77 (±0.1) | 2.99 (±0.4) | 1 | 3.10 (±0.01) | 4.17 (±0.05) | 3.12 (±0.04) | 1 | 12.10 (±0.6) | 12.63 (±0.7) | 12.29 (±1.5) | 1 | 37.40 (±1.3) | 32.99 (±1.3) | 38.34 (±0.9) | 1 |
| TLR-2 | 1.66 (±0.1) | 2.01 (±0.2) | 1.32 (±0.1) | 1 | 1.55 (±0.2) | 1.58 (±0.01) | 1.47 (±0.02) | 1 | 3.31 (±0.1) | 3.36 (±0.3) | 3.41 (±0.4) | 1 | 4.37 (±0.3) | 4.55 (±0.1) | 4.40 (±0.3) | 1 |
| TLR-4 | 2.65 (±0.1) | 2.62 (±0.2) | 2.85 (±0.1) | 1 | 4.80 (±0.8) | 4.75 (±1.0) | 4.46 (±0.3) | 1 | 2.28 (±0.1) | 2.26 (±0.02) | 2.23 (±0.2) | 1 | 3.27 (±0.1) | 3.27 (±0.2) | 3.31 (±0.1) | 1 |
| TLR-6 | 0.97 (±0.1) | 1.50 (±0.2) | 1.61 (±0.09) | 1 | 1.71 (±0.03) | 1.36 (±0.01) | 1.71 (±0.03) | 1 | 2.40 (±0.3) | 2.22 (±0.04) | 2.14 (±0.02) | 1 | 2.29 (±0.06) | 2.77 (±0.1) | 2.15 (±0.07) | 1 |
| TSLP | 11.49 (±0.1) | 7.38 (±0.9) | 10.14 (±1.8) | 1 | 5.57 (±0.7) | 3.04 (±0.4) | 8.15 (±0.4) | 1 | 64.22 (±2.2) | 58.37 (±0.5) | 52.62 (±1.1) | 1 | 66.54 (±0.8) | 67.38 (±0.8) | 65.44 (±1.3) | 1 |
| ICAM-1 | 14.29 (±0.01) | 12.59 (±0.1) | 16.87 (±0.7) | 1 | 1.95 (±0.1) | 2.33 (±0.2) | 2.15 (±0.08) | 1 | 12.72 (±0.8) | 15.42 (±0.5) | 12.42 (±0.6) | 1 | 46.69 (±1.1) | 45.48 (±1.2) | 47.50 (±1.5) | 1 |
| 18S | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betancur, D.; Muñoz Grez, C.; Oñate, A. Comparative Analysis of Cytokine Expression in Oral Keratinocytes and THP-1 Macrophages in Response to the Most Prevalent Serotypes of Aggregatibacter actinomycetemcomitans. Microorganisms 2021, 9, 622. https://doi.org/10.3390/microorganisms9030622
Betancur D, Muñoz Grez C, Oñate A. Comparative Analysis of Cytokine Expression in Oral Keratinocytes and THP-1 Macrophages in Response to the Most Prevalent Serotypes of Aggregatibacter actinomycetemcomitans. Microorganisms. 2021; 9(3):622. https://doi.org/10.3390/microorganisms9030622
Chicago/Turabian StyleBetancur, Daniel, Camila Muñoz Grez, and Angel Oñate. 2021. "Comparative Analysis of Cytokine Expression in Oral Keratinocytes and THP-1 Macrophages in Response to the Most Prevalent Serotypes of Aggregatibacter actinomycetemcomitans" Microorganisms 9, no. 3: 622. https://doi.org/10.3390/microorganisms9030622
APA StyleBetancur, D., Muñoz Grez, C., & Oñate, A. (2021). Comparative Analysis of Cytokine Expression in Oral Keratinocytes and THP-1 Macrophages in Response to the Most Prevalent Serotypes of Aggregatibacter actinomycetemcomitans. Microorganisms, 9(3), 622. https://doi.org/10.3390/microorganisms9030622






