Bacterial Communities and Enzymatic Activities in Sediments of Long-Term Fish and Crab Aquaculture Ponds
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sampling Produce
2.2. Sediment Characterization
2.3. Sediment Extracellular Enzymatic Activities
2.4. Sediment DNA Extraction, 16S rRNA Gene Amplification
2.5. Sequencing Data Analysis
2.6. Statistical Analysis
3. Results
3.1. General Characteristics and Nutrients in Pond Sediments
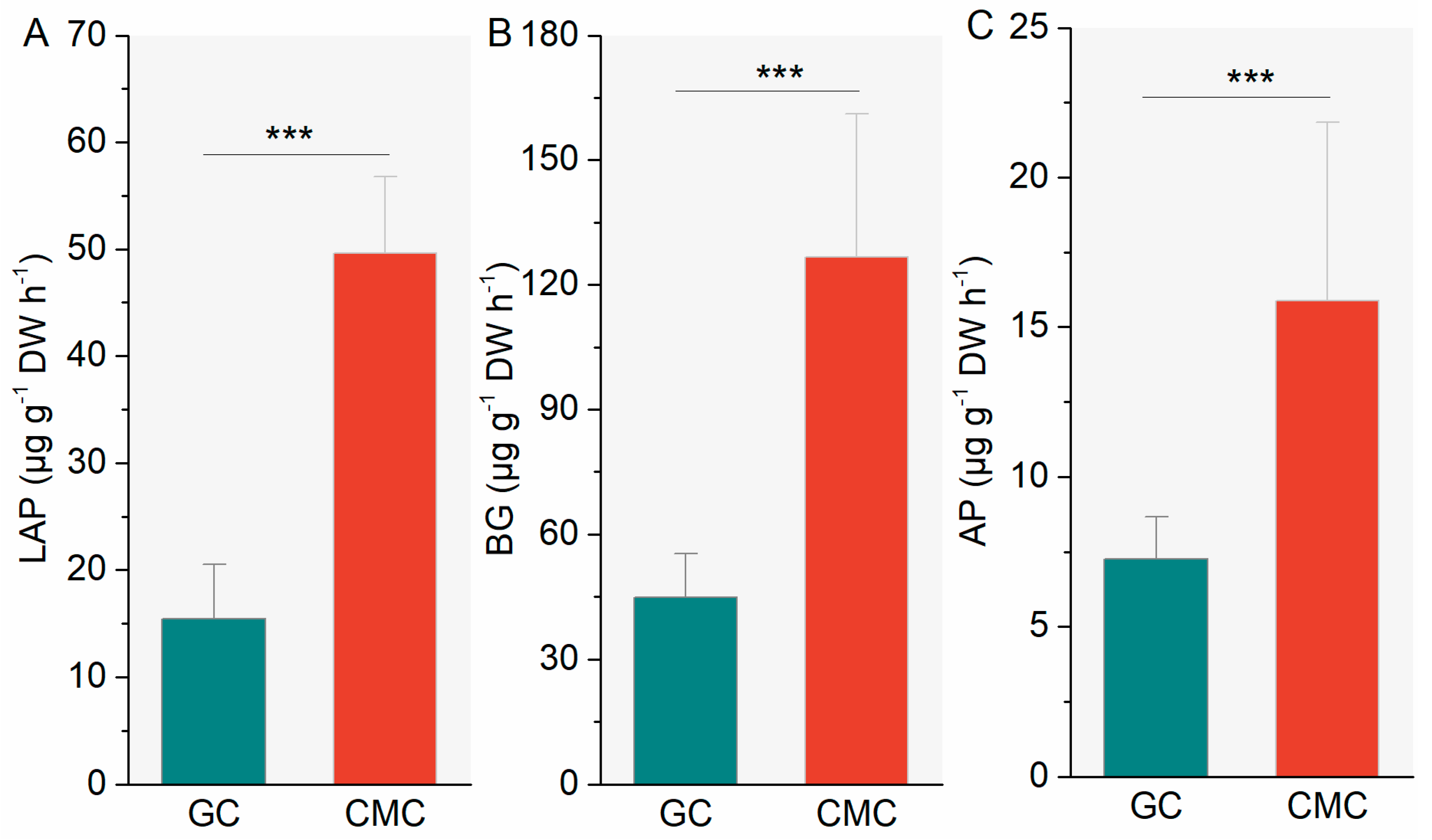
3.2. Sediment Enzymatic Activities
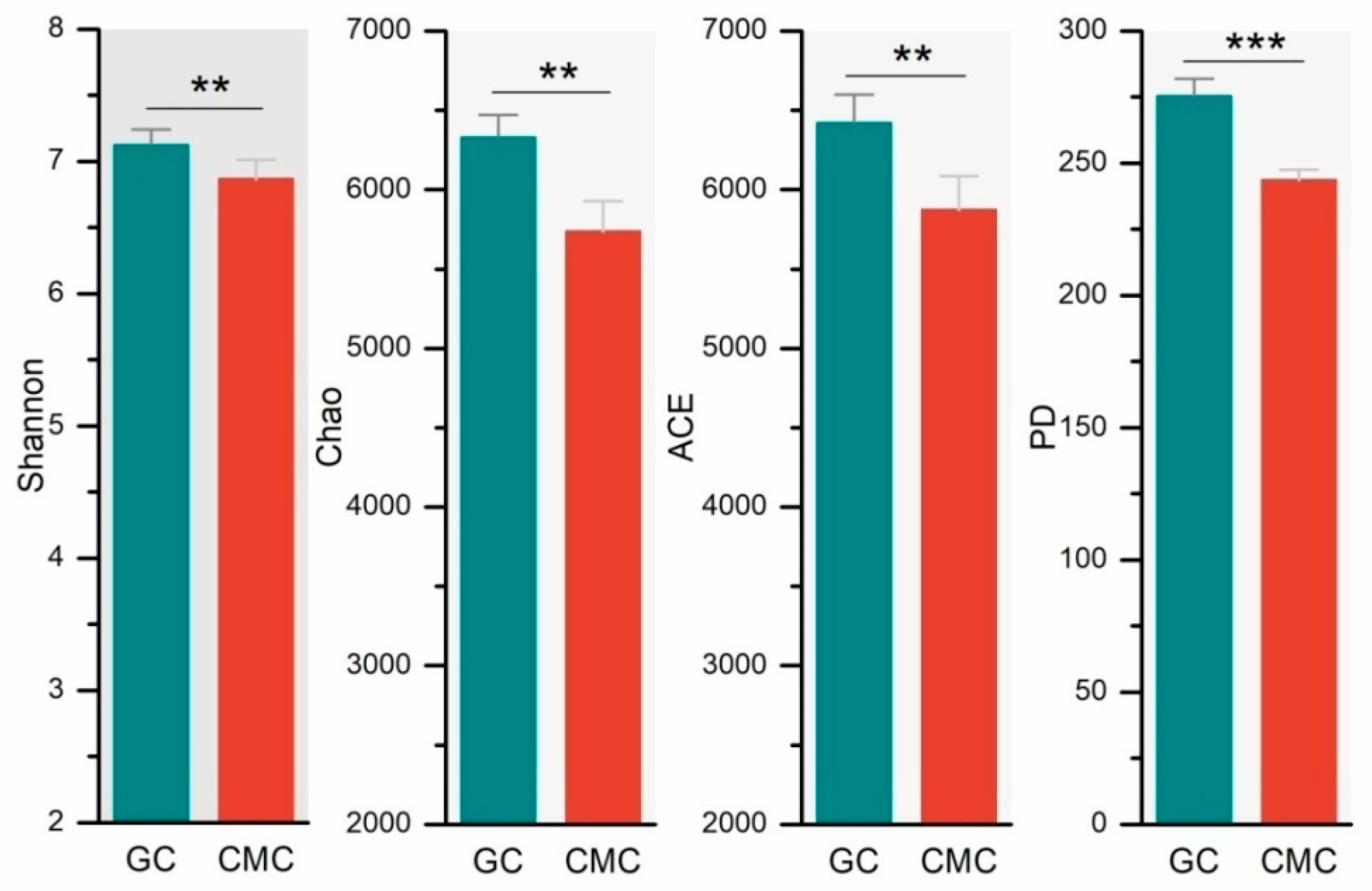
3.3. Sediment Bacterial Richness and Diversity
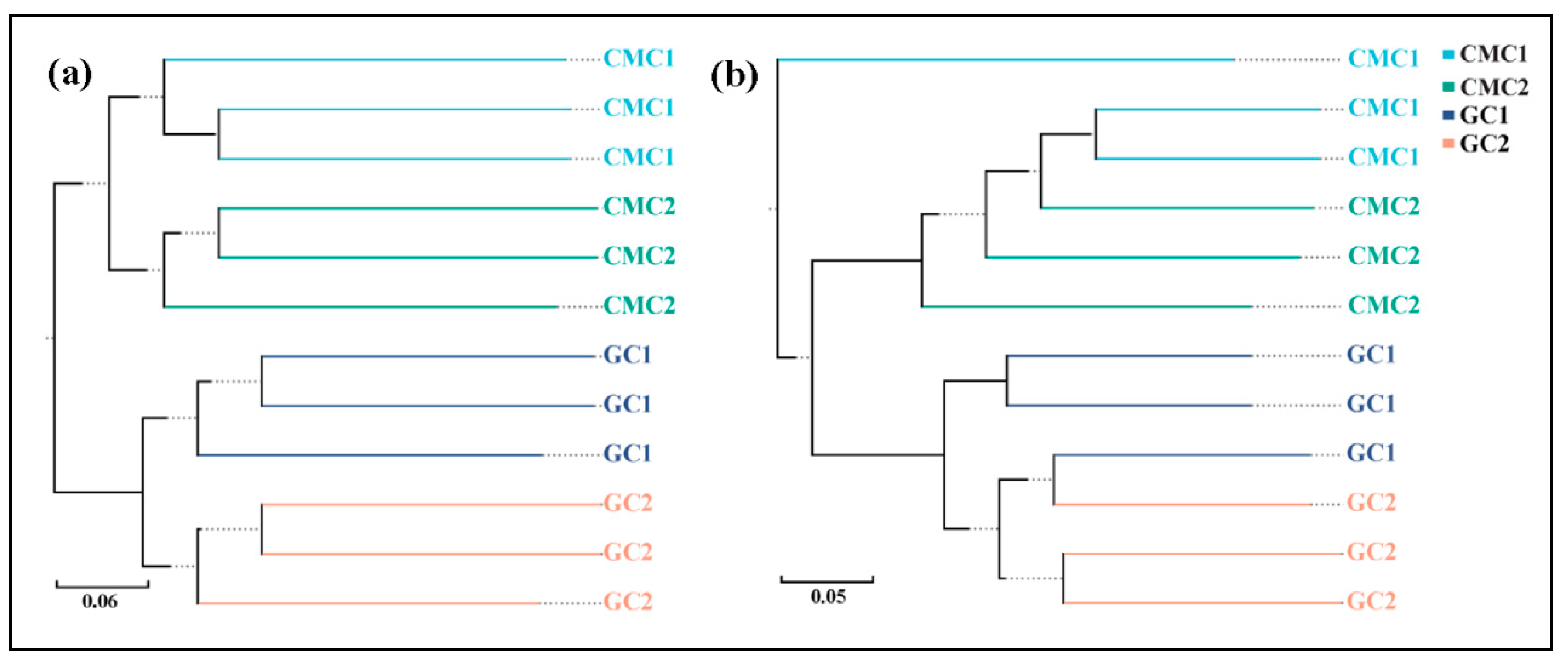
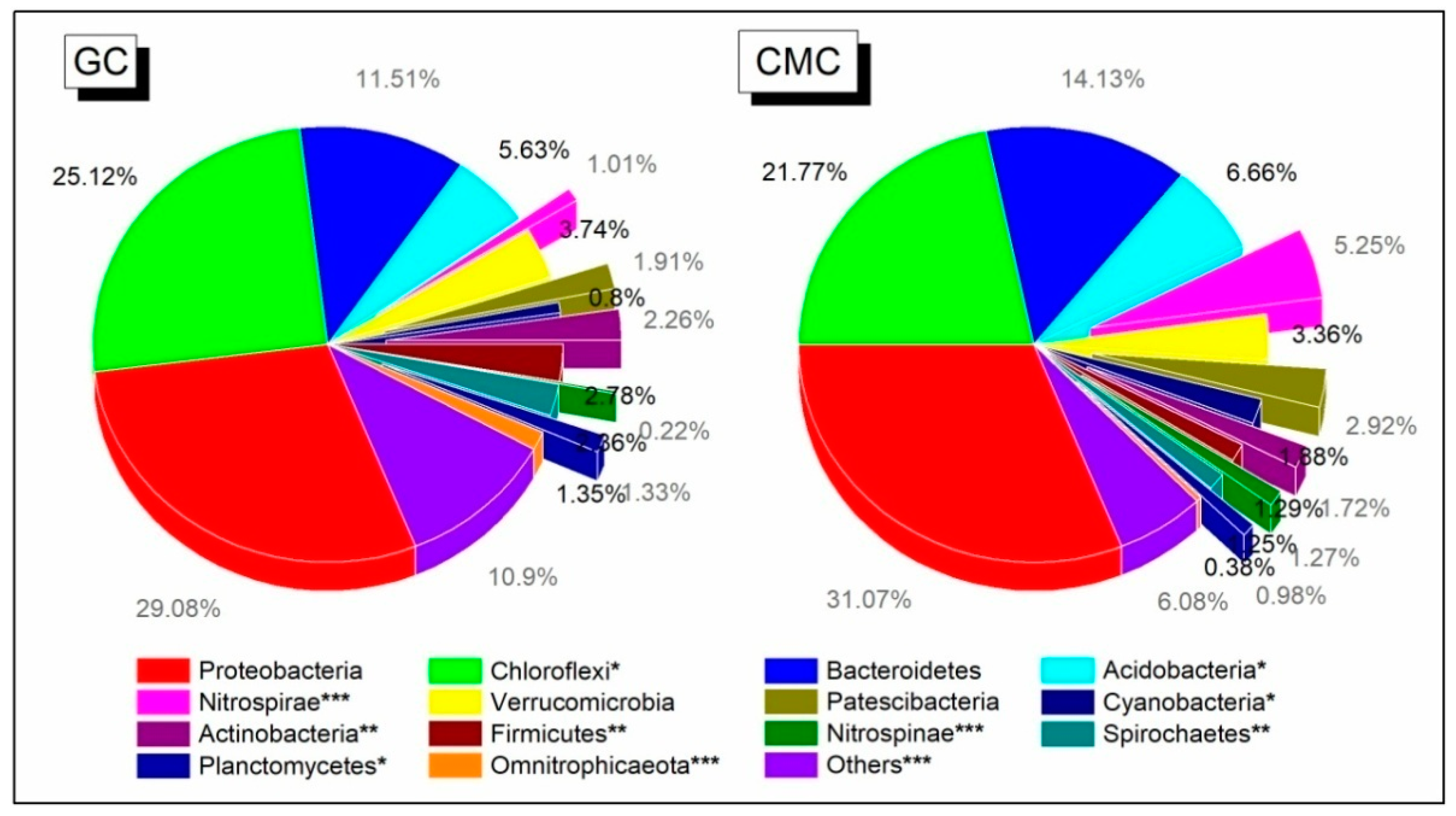
3.4. Sediment Bacterial Community Structure and Composition
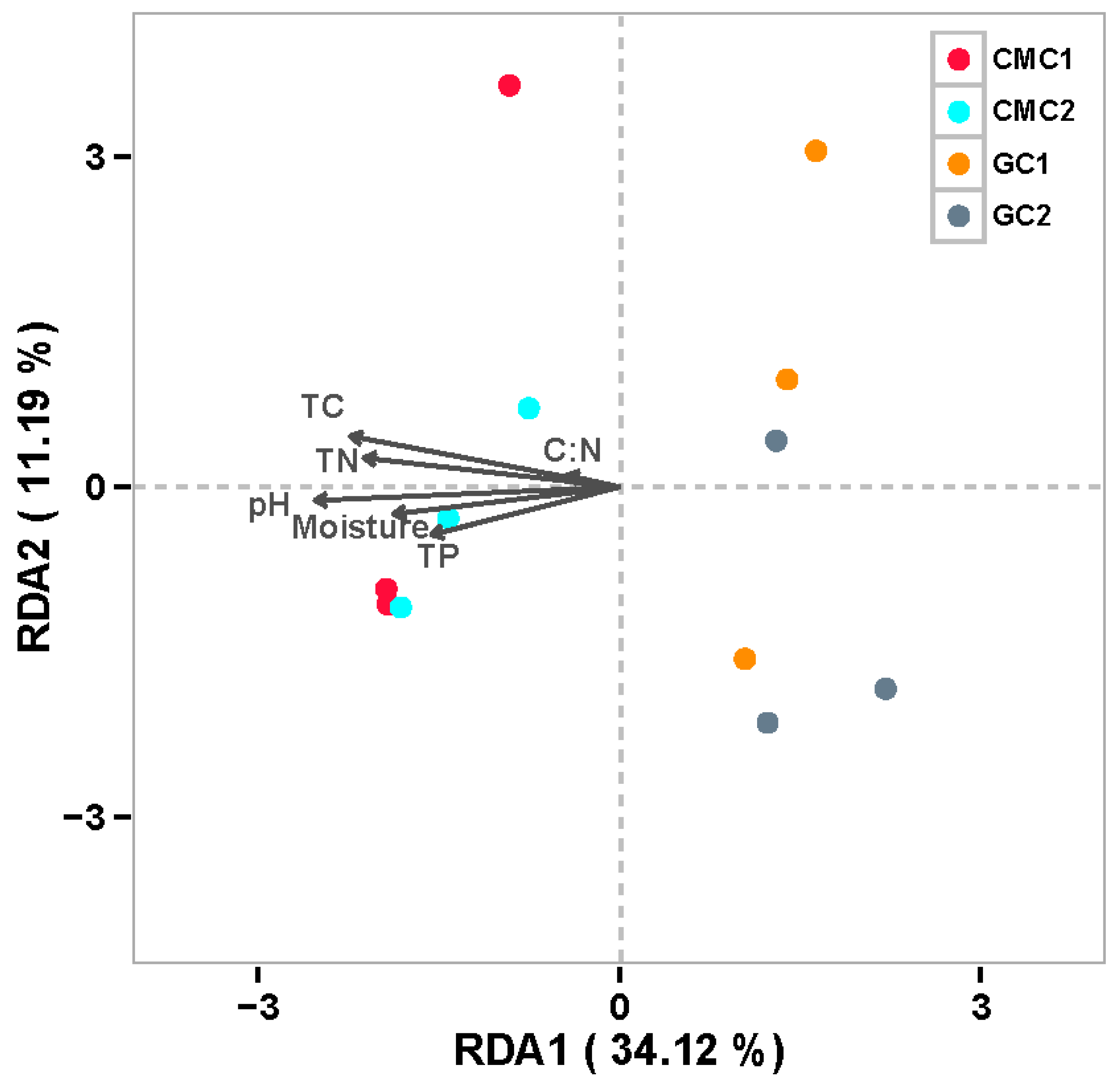
3.5. Key Factors Affecting Sediment Enzyme Activities and Bacterial Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2016, Contributing to Food Security and Nutrition for All. Available online: http//www.fao.org/3/a-i5555e.pdf (accessed on 6 May 2020).
- Boyd, C.E.; Wood, C.W.; Chaney, P.L.; Queiroz, J.F. Role of aquaculture pond sediments in sequestration of annual global carbon emissions. Environ. Pollut. 2010, 158, 2537–2540. [Google Scholar] [CrossRef]
- Chatvijitkul, S.; Boyd, C.E.; Davis, D.A.; McNevin, A.A. Pollution potential indicators for feed-based fish and shrimp culture. Aquaculture 2017, 477, 43–49. [Google Scholar] [CrossRef]
- Hargreaves, J.A. Nitrogen biogeochemistry of aquaculture ponds. Aquaculture 1998, 166, 181–212. [Google Scholar] [CrossRef]
- Pouil, S.; Samsudin, R.; Slembrouck, J.; Sihabuddin, A.; Sundari, G.; Khazaidan, K.; Kristanto, A.H.; Pantjara, B.; Caruso, D. Nutrient budgets in a small-scale freshwater fish pond system in Indonesia. Aquaculture 2019, 504, 267–274. [Google Scholar] [CrossRef]
- Miranda, A.; Voltolina, D.; Frías-Espericueta, M.G.; Izaguirre-Fierro, G.; Rivas-Vega, M.E. Budget and discharges of nutrients to the Gulf of California of a semi-intensive shrimp farm (NW Mexico). Hidrobiológica 2009, 19, 43–48. [Google Scholar]
- Jackson, C.; Preston, N.; Thompson, P.J.; Burford, M. Nitrogen budget and effluent nitrogen components at an intensive shrimp farm. Aquaculture 2003, 218, 397–411. [Google Scholar] [CrossRef]
- Avnimelech, Y.; Mozes, N.; Diab, S.; Kochba, M. Rates of organic carbon and nitrogen degradation in intensive fish ponds. Aquaculture 1995, 134, 211–216. [Google Scholar] [CrossRef]
- Yang, H.; Hu, B. Introduction of Chinese integrated fish farming and major models. In Integrated Fish Farming in China; NACA Technical Manual 7. A World Food Day Publication of the NACA; Network of Aquaculture Centers in Asia-Pacific: Bangkok, Thailand, 1989; pp. 153–196. [Google Scholar]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Knight, R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef] [PubMed]
- Bissett, A.; Burke, C.; Cook, P.L.M.; Bowman, J.P. Bacterial community shifts in organically perturbed sediments. Environ Microbiol. 2007, 9, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Vezzulli, L.; Chelossi, E.; Riccardi, G.; Fabiano, M. Bacterial community structure and activity in fish farm sediments of the Ligurian sea (Western Mediterranean). Aquac. Int. 2002, 10, 123–141. [Google Scholar] [CrossRef]
- Duarte, L.N.; Coelho, F.J.R.C.; Cleary, D.F.R.; Bonifácio, D.; Martins, P.; Gomes, N.C.M. Bacterial and microeukaryotic plankton communities in a semi-intensive aquaculture system of sea bass (Dicentrarchus labrax): A seasonal survey. Aquaculture 2019, 503, 59–69. [Google Scholar] [CrossRef]
- Fan, L.; Qiu, L.; Song, C.; Meng, S.; Zheng, Y.; Li, D.; Hu, G.; Chen, J. Effects of feed input and planting of submerged aquatic vegetation on methanotrophic communities in the surface sediments of aquaculture ponds. Appl. Soil Ecol. 2019, 143, 10–16. [Google Scholar] [CrossRef]
- Zhang, M.; Pan, L.; Huang, F.; Gao, S.; Su, C.; Zhang, M.; He, Z. Metagenomic analysis of composition, function and cycling processes of microbial community in water, sediment and effluent of Litopenaeus vannamei farming environments under different culture modes. Aquaculture 2019, 506, 280–293. [Google Scholar] [CrossRef]
- Rubio-Portillo, E.; Villamor, A.; Fernandez-Gonzalez, V.; Antón, J.; Sanchez-Jerez, P. Exploring changes in bacterial communities to assess the influence of fish farming on marine sediments. Aquaculture 2019, 506, 459–464. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Lauber, C.L.; Weintraub, M.N.; Ahmed, B.; Zeglin, L.H. Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 2008, 11, 1252–1264. [Google Scholar] [CrossRef]
- AOAC. Official Method 2001.11 Protein (Crude) in animal feed, forage (plant tissue), grain, and oilseeds. In Official Methods of Analysis of AOAC Internationl (OMA); AOAC International: Gaithersburg, MD, USA, 2005. [Google Scholar]
- Hoppe, H.G. Significance of exoenzymatic activities in the ecology of brackish water, measurements by means of methylumbelliferyl-substrates. Mar. Ecol. Prog. Ser. 1983, 11, 299–308. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH, fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.T.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Karakassis, I.; Tsapakis, M.; Hatziyanni, E. Seasonal variability in sediment profiles beneath fish farm cages in the Mediterranean. Mar. Ecol. Prog. Ser. 1998, 162, 243–252. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Zhang, S.; Lian, Y.; Ding, H.; Du, X.; Li, Z.; De Silva, S.S. Sustainable farming practices of the Chinese mitten crab (Eriocheir sinensis) around Hongze Lake, lower Yangtze River Basin, China. Ambio 2016, 45, 361–373. [Google Scholar] [CrossRef]
- Tacon, A.G.J.; Metian, M. Global overview on the use of fish meal and fish oil in industrially compounded aquafeeds: Trends and future prospects. Aquaculture 2008, 285, 146–158. [Google Scholar] [CrossRef]
- Herath, S.S.; Satoh, S. Environmental impact of phosphorus and nitrogen from aquaculture. In Woodhead Publishing Series in Food Science, Technology and Nutrition, Feed and Feeding Practices in Aquaculture; Davis, D.A., Ed.; Woodhead Publishing: Cambridge, UK, 2015; pp. 369–386. [Google Scholar]
- Tamminen, M.; Karkman, A.; Corander, J.; Paulin, L.; Virta, M. Differences in bacterial community composition in Baltic sea sediment in response to fish farming. Aquaculture 2011, 313, 15–23. [Google Scholar] [CrossRef]
- Hou, D.; Huang, Z.; Zeng, S.; Liu, J.; He, J. Comparative analysis of the bacterial community compositions of the shrimp intestine, surrounding water and sediment. J. Appl. Microbiol. 2018, 125, 792–799. [Google Scholar] [CrossRef]
- Fan, L.; Wang, Z.; Chen, M.; Qu, Y.; Li, J.; Zhou, A.; Xie, S.; Zeng, F.; Zou, J. Microbiota comparison of pacific white shrimp intestine and sediment at freshwater and marine cultured environment. Sci. Total Environ. 2019, 657, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.J.; Sheik, C.S.; Taylor, C.A.; Jain, S.; Dick, G.J. Community transcriptomic assembly reveals microbes that contribute to deep-sea carbon and nitrogen cycling. ISME J. 2013, 7, 1962–1973. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Chen, D.; Guo, H.; Wei, J.; Bai, Y.; Shen, Q.; Hu, S. Differential responses of soil bacterial communities to long-term N and P inputs in a semi-arid steppe. Geoderma 2017, 292, 25–33. [Google Scholar] [CrossRef]
- Birgander, J.; Reischke, S.; Jones, D.L.; Rousk, J. Temperature adaptation of bacterial growth and 14C-glucose mineralisation in a laboratory study. Soil Biol. Biochem. 2013, 65, 294–303. [Google Scholar] [CrossRef]
- Meisner, A.; Leizeaga, A.; Rousk, J.; Bååth, E. Partial drying accelerates bacterial growth recovery to rewetting. Soil Biol. Biochem. 2017, 112, 269–276. [Google Scholar] [CrossRef]
- Alvarez, G.; Shahzad, T.; Andanson, L.; Bahn, M.; Fontaine, S. Catalytic power of enzymes decreases with temperature: New insights for understanding soil C cycling and microbial ecology under warming. Glob. Chang. Biol. 2018, 24, 4238–4250. [Google Scholar] [CrossRef]
- Rousk, J.; Brookes, P.C.; Bååth, E. Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl. Environ. Microbiol. 2009, 75, 1589–1596. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Liu, J.; Sui, Y.; Yu, Z.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol. Biochem. 2014, 70, 113–122. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Hill, B.H.; Follstad Shah, J.J. Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 2010, 468, 122. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Follstad Shah, J.J.; Elonen, H.C.M. Ecoenzymatic stoichiometry of stream sediments with comparison to terrestrial soils. Biogeochemistry 2012, 111, 455–467. [Google Scholar] [CrossRef]
- Demoling, F.; Figueroa, D.; Bååth, E. Comparison of factors limiting bacterial growth in different soils. Soil Biol. Biochem. 2007, 39, 2485–2495. [Google Scholar] [CrossRef]
- Xu, X.; Thornton, P.E.; Post, W.M. A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems. Glob. Ecol. Biogeogr. 2013, 22, 737–749. [Google Scholar] [CrossRef]

| Ponds | pH | Moisture (%) | TC (g kg−1) | TN (g kg−1) | TP (g kg−1) | C:N |
|---|---|---|---|---|---|---|
| GC | 7.19 ± 0.11 | 52.56 ± 5.45 | 21.35 ± 1.53 | 2.49 ± 0.31 | 1.14 ± 0.35 | 8.97 ± 0.89 |
| CMC | 7.55 ± 0.13 | 59.87 ± 2.61 | 27.46 ± 1.90 | 2.98 ± 0.20 | 1.62 ± 0.37 | 9.24 ± 0.71 |
| p | *** | * | *** | ** | * | ns |
| Enzymatic Indices | pH | Moisture | TC | TN | C:N | TP |
|---|---|---|---|---|---|---|
| LAP | 0.874 ** | 0.769 ** | 0.803 ** | 0.788 ** | 0.141 | 0.629 * |
| BG | 0.696 * | 0.616 * | 0.774 ** | 0.683 * | 0.223 | 0.429 |
| AP | 0.623 ** | 0.691 * | 0.826 ** | 0.650 * | 0.338 | 0.620 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Deng, Q.; Wan, L.; Cao, X.; Zhou, Y.; Song, C. Bacterial Communities and Enzymatic Activities in Sediments of Long-Term Fish and Crab Aquaculture Ponds. Microorganisms 2021, 9, 501. https://doi.org/10.3390/microorganisms9030501
Zhang Z, Deng Q, Wan L, Cao X, Zhou Y, Song C. Bacterial Communities and Enzymatic Activities in Sediments of Long-Term Fish and Crab Aquaculture Ponds. Microorganisms. 2021; 9(3):501. https://doi.org/10.3390/microorganisms9030501
Chicago/Turabian StyleZhang, Zhimin, Qinghui Deng, Lingling Wan, Xiuyun Cao, Yiyong Zhou, and Chunlei Song. 2021. "Bacterial Communities and Enzymatic Activities in Sediments of Long-Term Fish and Crab Aquaculture Ponds" Microorganisms 9, no. 3: 501. https://doi.org/10.3390/microorganisms9030501
APA StyleZhang, Z., Deng, Q., Wan, L., Cao, X., Zhou, Y., & Song, C. (2021). Bacterial Communities and Enzymatic Activities in Sediments of Long-Term Fish and Crab Aquaculture Ponds. Microorganisms, 9(3), 501. https://doi.org/10.3390/microorganisms9030501





