Exploring Antibiotic Resistance Diversity in Leuconostoc spp. by a Genome-Based Approach: Focus on the lsaA Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Sequence Analysis to Retrieve the Antibiotic Resistance Genes
2.2. Analysis of the lsaA Gene
2.3. Bacterial Strains and Growth Conditions
2.4. Antimicrobial Susceptibility Testing
2.5. RNA Isolation and Real-Time PCR in L. pseudomesenteroides LMG 11482T (=NCDO 768T)
3. Results
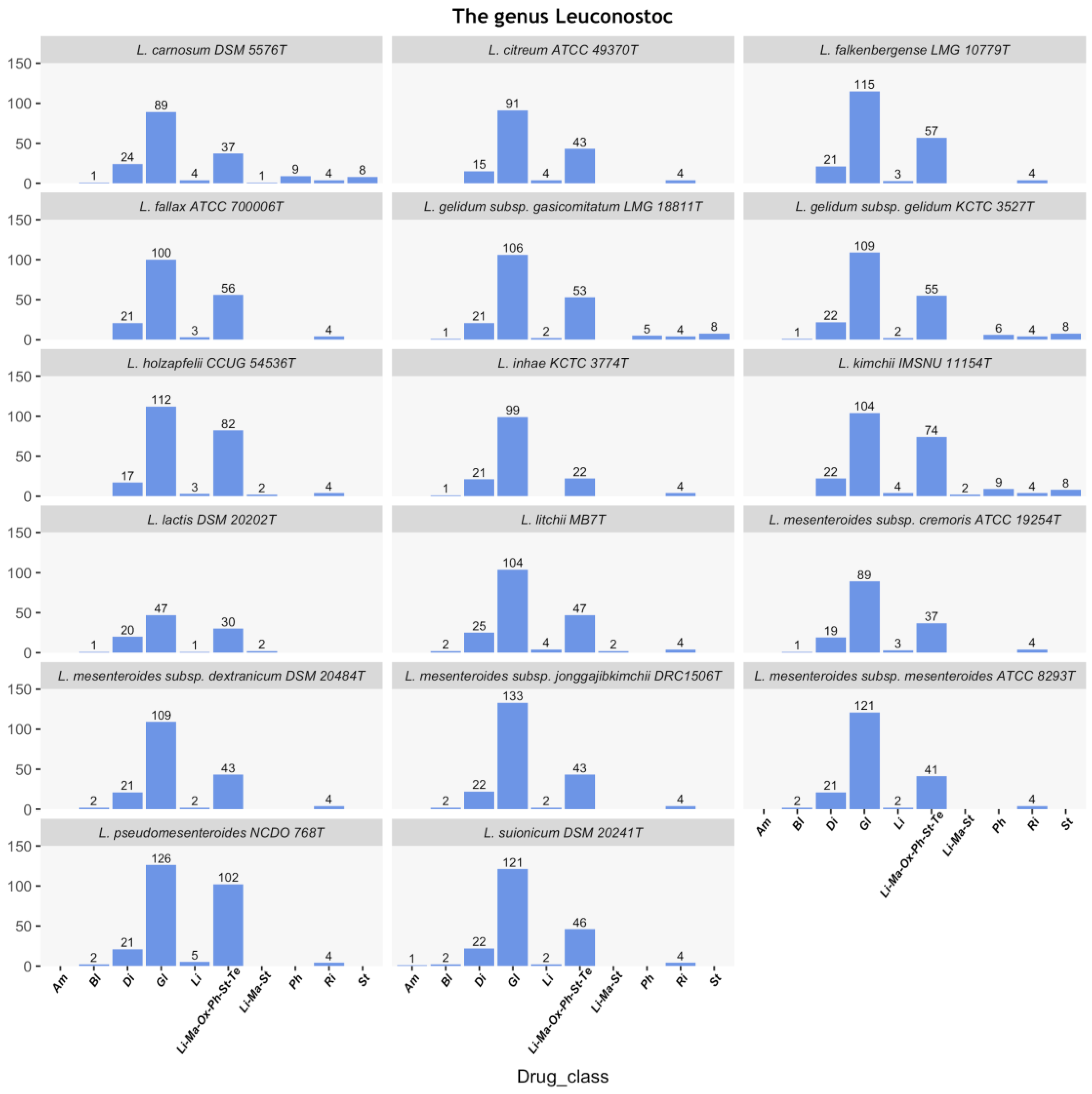
3.1. In Silico Prediction of ARGs in the Leuconostoc Genus
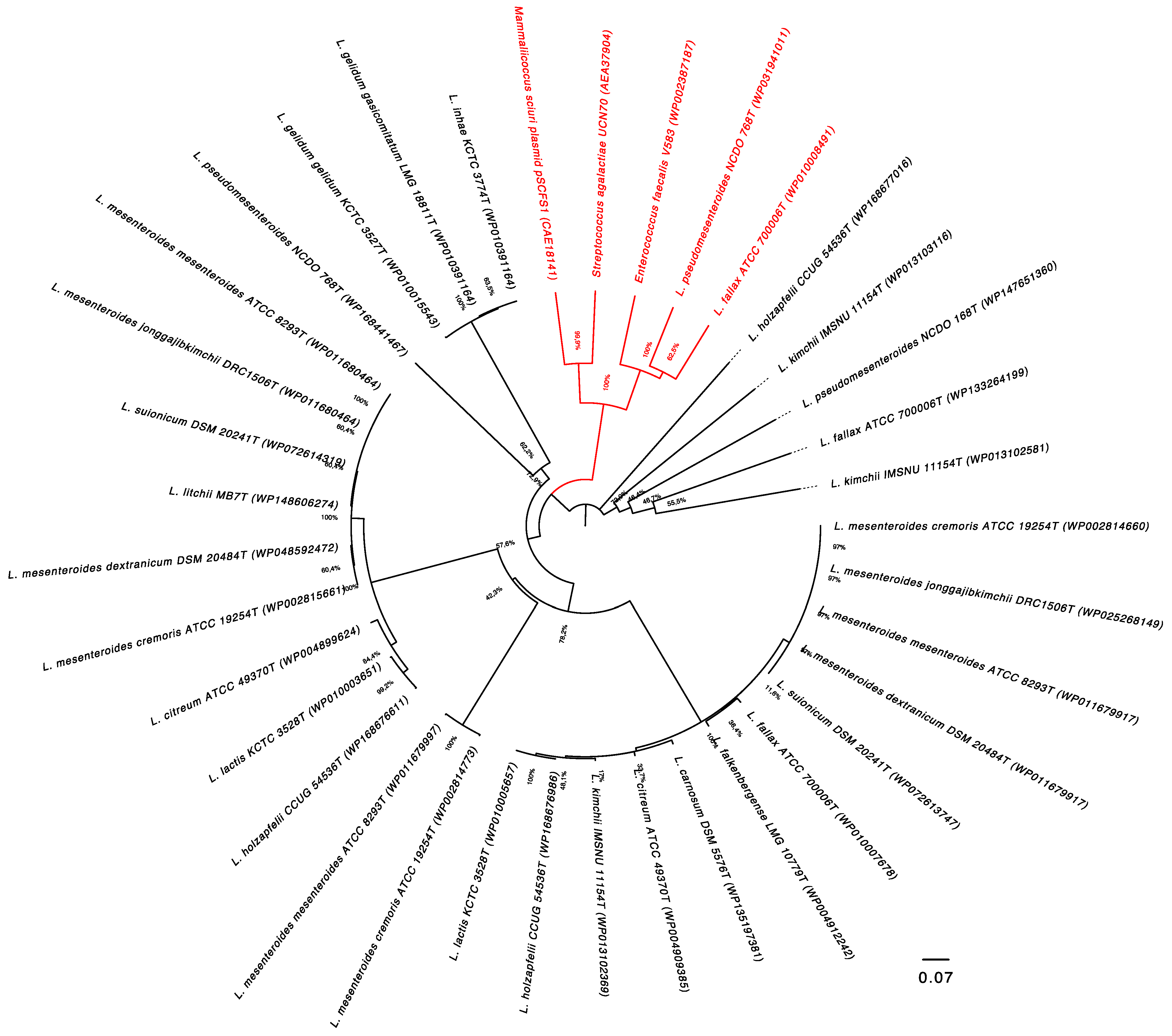
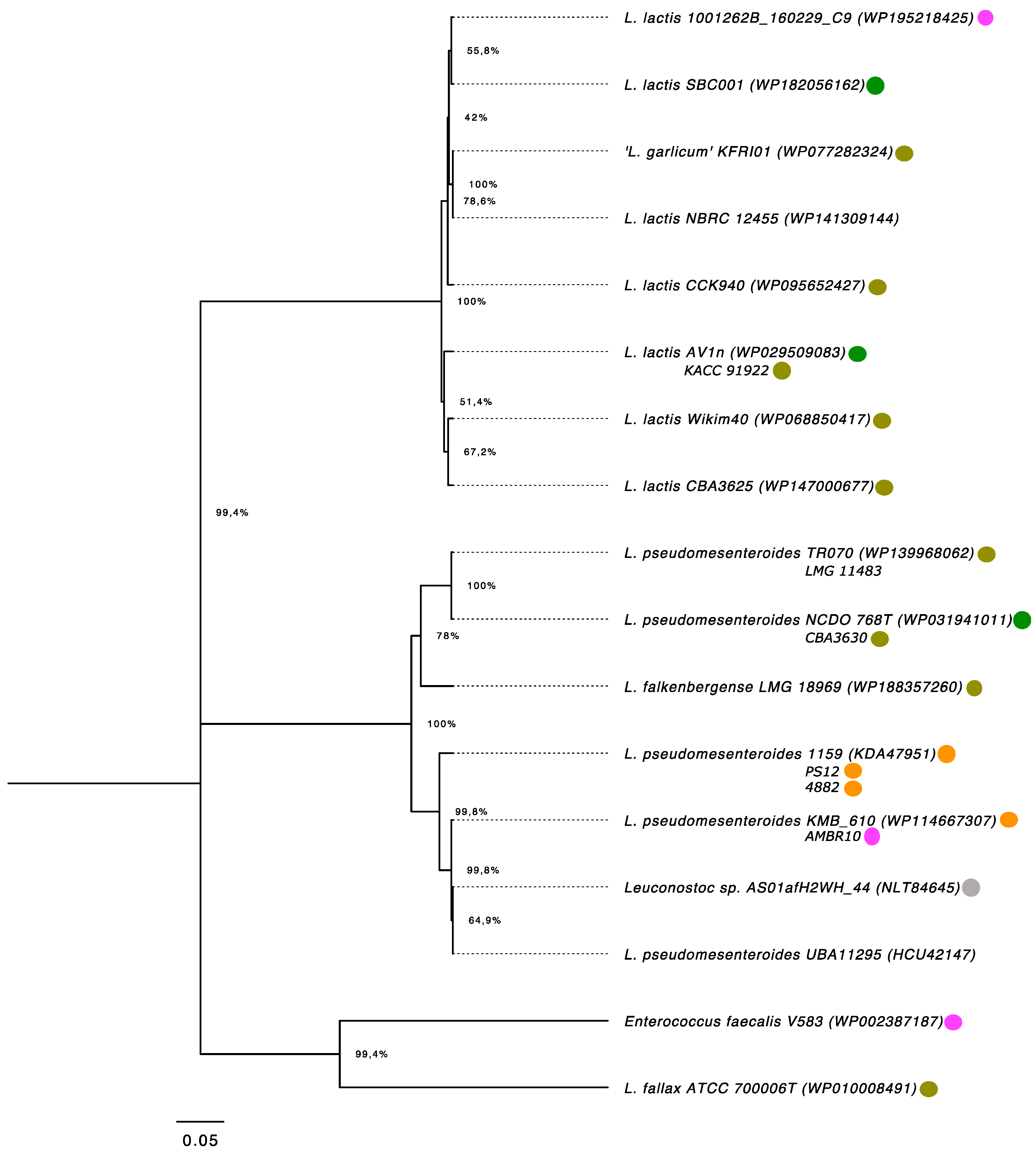
3.2. Distribution Patterns of the lsaA Gene in the Leuconostoc Genus
3.3. Analysis of Flanking Regions of the lsaA Gene
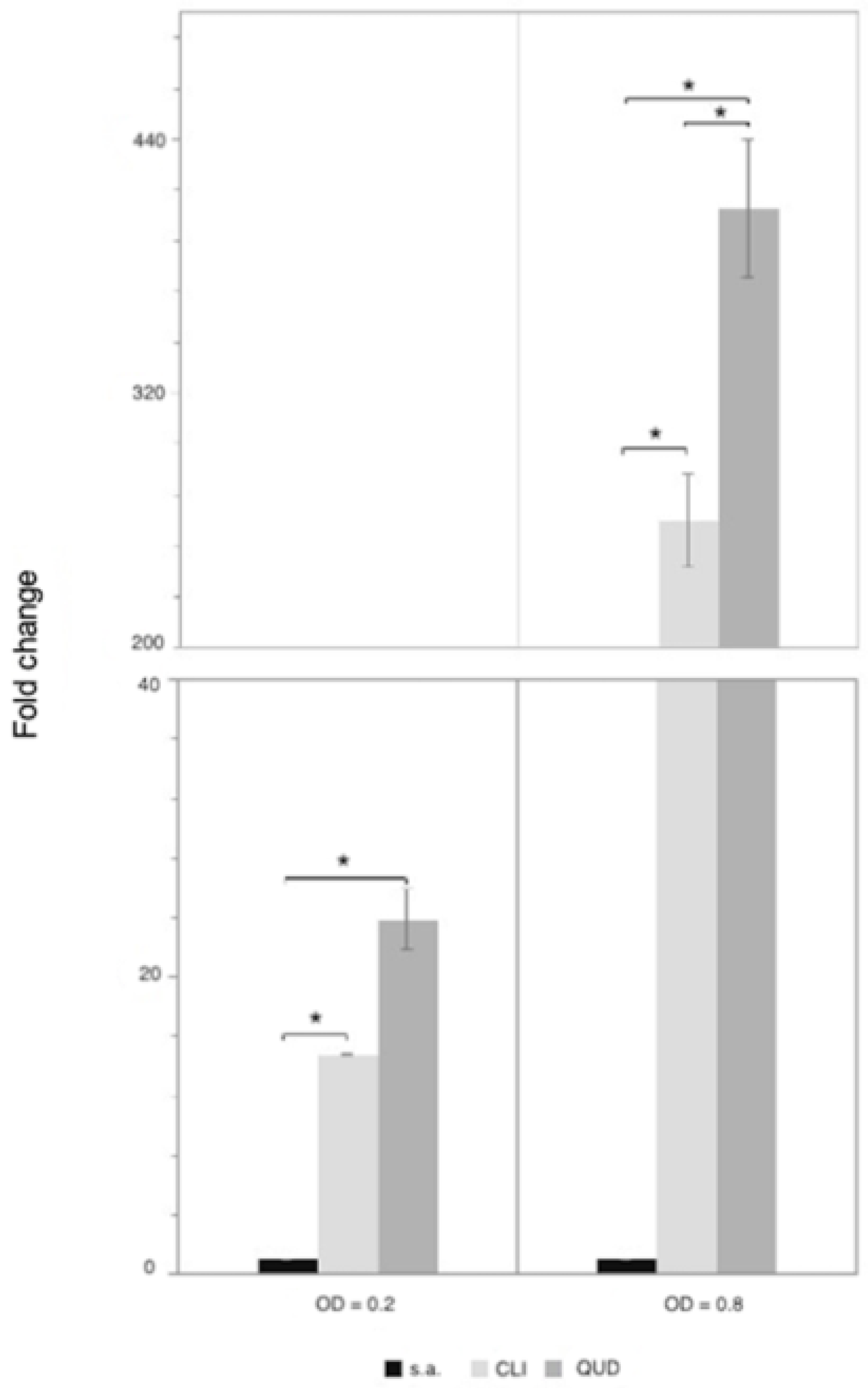
3.4. Determination of Phenotypic Resistance towards CLI and QUD
3.5. Quantification of lsaA Expression in L. pseudomesenteroides LMG 11482T (=NCDO 768T)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.; Wang, L.; Wu, Y.-C.; Mori, K.; Tamura, T.; Chang, C.; Chang, Y.-C.; Wu, H.-C.; Yi, H.-H.; Wang, P.-Y. Leuconostoc litchii sp. nov., a novel lactic acid bacterium isolated from lychee. Int. J. Syst. Evol. Microbiol. 2020, 70. [Google Scholar] [CrossRef] [PubMed]
- Ehrmann, M.A.; Freiding, S.; Vogel, R.F. Leuconostoc palmae sp. nov., a novel lactic acid bacterium isolated from palm wine. Int. J. Syst. Evol. Microbiol. 2009, 59. [Google Scholar] [CrossRef]
- Lyhs, U.; Snauwaert, I.; Pihlajaviita, S.; Vuyst, L.; De Vandamme, P. Leuconostoc rapi sp. nov., isolated from sous-vide-cooked rutabaga. Int. J. Syst. Evol. Microbiol. 2015, 65. [Google Scholar] [CrossRef] [PubMed]
- Beganović, J.; Pavunc, A.L.; Gjuračić, K.; Špoljarec, M.; Šušković, J.; Kos, B. Improved sauerkraut production with probiotic strain Lactobacillus plantarum L4 and Leuconostoc mesenteroides LMG 7954. J. Food Sci. 2011, 76. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Lee, S.; Park, Y.-S.; Sharma, A. RAPD analysis of Leuconostoc mesenteroides strains associated with vegetables and food products from Korea. LWT 2017, 77. [Google Scholar] [CrossRef]
- De Bruyne, K.; Schillinger, U.; Caroline, L.; Boehringer, B.; Cleenwerck, I.; Vancanneyt, M.; De Vuyst, L.; Franz, C.M.A.P.; Vandamme, P. Leuconostoc holzapfelii sp. nov., isolated from Ethiopian coffee fermentation and assessment of sequence analysis of housekeeping genes for delineation of Leuconostoc species. Int. J. Syst. Evol. Microbiol. 2007, 57. [Google Scholar] [CrossRef]
- Vera-Pingitore, E.; Jimenez, M.E.; Dallagnol, A.; Belfiore, C.; Fontana, C.; Fontana, P.; Von Wright, A.; Vignolo, G.; Plumed-Ferrer, C. Screening and characterization of potential probiotic and starter bacteria for plant fermentations. LWT Food Sci. Technol. 2016, 71. [Google Scholar] [CrossRef]
- Martinez-Murcia, A.; Collins, M. A phylogenetic analysis of an atypical leuconostoc: Description of Leuconostoc fallax sp. nov. FEMS Microbiol. Lett. 1991, 82. [Google Scholar] [CrossRef][Green Version]
- Kim, J.; Chun, J.; Han, H.U. Leuconostoc kimchii sp. nov., a new species from kimchi. Int. J. Syst. Evol. Microbiol. 2000, 50. [Google Scholar] [CrossRef]
- Kim, B.; Lee, J.; Jang, J.; Kim, J.; Han, H. Leuconostoc inhae sp. nov., a lactic acid bacterium isolated from kimchi. Int. J. Syst. Evol. Microbiol. 2003, 53, 1123–1126. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.H.; Kim, K.H.; Chun, B.H.; Ryu, B.H.; Han, N.S.; Jeon, C.O. A proposal of Leuconostoc mesenteroides subsp. jonggajibkimchii subsp. nov. and reclassification of Leuconostoc mesenteroides subsp. suionicum (Gu et al., 2012) as Leuconostoc suionicum sp. nov. based on complete genome sequences. Int. J. Syst. Evol. Microbiol. 2017, 67. [Google Scholar] [CrossRef]
- Lee, S.H.; Park, M.S.; Jung, J.Y.; Jeon, C.O. Leuconostoc miyukkimchii sp. nov., isolated from brown algae (Undaria pinnatifida) kimchi. Int. J. Syst. Evol. Microbiol. 2012, 62. [Google Scholar] [CrossRef]
- Inventory of Microbial Food Cultures with Safety Demonstration in Fermented Food Products; 495/2018; Intnational Dairy Federation: Brussels, Belgium, 2018; Available online: https://store.fil-idf.org/product/bulletin-idf-n-495-2018-inventory-microbial-food-cultures-safety-demonstration-fermented-food-products/ (accessed on 26 February 2021).
- Wu, Y.; Gu, C.T. Leuconostoc falkenbergense sp. nov., isolated from a lactic culture, fermentating string beans and traditional yogurt. Int. J. Syst. Evol. Microbiol. 2021, 71. [Google Scholar] [CrossRef] [PubMed]
- Shaw, B.G.; Harding, C.D. Leuconostoc gelidum sp. nov. and Leuconostoc carnosum sp. nov. from chill-stored meats. Int. J. Syst. Bacteriol. 1989, 39. [Google Scholar] [CrossRef]
- Oki, K.; Rai, A.K.; Sato, S.; Watanabe, K.; Tamang, J.P. Lactic acid bacteria isolated from ethnic preserved meat products of the Western Himalayas. Food Microbiol. 2011, 28. [Google Scholar] [CrossRef]
- Rahkila, R.; De Bruyne, K.; Johansson, P.; Vandamme, P.; Björkroth, J. Reclassification of Leuconostoc gasicomitatum as Leuconostoc gelidum subsp. gasicomitatum comb. nov., description of Leuconostoc gelidum subsp. aenigmaticum subsp. nov., designation of Leuconostoc gelidum subsp. gelidum subsp. nov. and emended description of Leuconostoc gelidum. Int. J. Syst. Evol. Microbiol. 2014, 64. [Google Scholar] [CrossRef]
- Ogier, J.; Casalta, E.; Farrok, C.; Saihi, A. Safety assessment of dairy microorganisms: The Leuconostoc genus. Int. J. Food Microbiol. 2008, 126. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic amine production by lactic acid bacteria: A review. Foods 2019, 8, 17. [Google Scholar] [CrossRef]
- Flórez, A.B.; Campedelli, I.; Delgado, S.; Alegría, Á.; Salvetti, E.; Felis, G.E.; Mayo, B.; Torriani, S. Antibiotic susceptibility profiles of dairy Leuconostoc, analysis of the genetic basis of atypical resistances and transfer of genes in vitro and in a food matrix. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Muñoz, M.C.C.; Benomar, N.; Lerma, L.L.; Gálvez, A.; Abriouel, H. Antibiotic resistance of Lactobacillus pentosus and Leuconostoc pseudomesenteroides isolated from naturally-fermented Aloreña table olives throughout fermentation process. Int. J. Food Microbiol. 2014, 172. [Google Scholar] [CrossRef]
- Flórez, A.B.; Delgado, S.; Mayo, B. Antimicrobial susceptibility of lactic acid bacteria isolated from a cheese environment. Can. J. Microbiol. 2005, 51. [Google Scholar] [CrossRef]
- Ammor, M.S.; Gueimonde, M.; Danielsen, M.; Zagorec, M.; van Hoek, A.H.A.M.; de los Reyes-Gavilán, C.G.; Mayo, B.; Margolles, A. Two different tetracycline resistance mechanisms, plasmid-carried tet(L) and chromosomally located transposon-associated tet(M), coexist in Lactobacillus sakei Rits 9. Appl. Environ. Microbiol. 2008, 74. [Google Scholar] [CrossRef]
- Morandi, S.; Cremonesi, P.; Silvetti, T.; Brasca, M. Technological characterisation, antibiotic susceptibility and antimicrobial activity of wild-type Leuconostoc strains isolated from north Italian traditional cheeses. J. Dairy Res. 2013, 80. [Google Scholar] [CrossRef] [PubMed]
- El Issaoui, K.; Khay, E.O.; Abrini, J.; Zinebi, S.; Amajoud, N.; Senhaji, N.S.; Abriouel, H. Molecular identification and antibiotic resistance of bacteriocinogenic lactic acid bacteria isolated from table olives. Arch. Microbiol. 2020. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Salvetti, E.; Orrù, L.; Capozzi, V.; Martina, A.; Lamontanara, A.; Keller, D.; Cash, H.; Felis, G.E.; Cattivelli, L.; Torriani, S.; et al. Integrate genome-based assessment of safety for probiotic strains: Bacillus coagulans GBI-30, 6086 as a case study. Appl. Microbiol. Biotechnol. 2016, 100. [Google Scholar] [CrossRef]
- Cao, L.; Chen, H.; Wang, Q.; Li, B.; Hu, Y.; Zhao, C.; Hu, Y.; Yin, Y. Literature-based phenotype survey and in silico genotype investigation of antibiotic resistance in the genus Bifidobacterium. Curr. Microbiol. 2020, 77, 4104–4113. [Google Scholar] [CrossRef]
- Singh, K.V.; Weinstock, G.M.; Murray, B.E. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 2002, 46, 1845–1850. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3. [Google Scholar] [CrossRef]
- Klare, I.; Konstabel, C.; Müller-Bertling, S.; Reissbrodt, R.; Huys, G.; Vancanneyt, M.; Swings, J.; Goossens, H.; Witte, W. Evaluation of new broth media for microdilution antibiotic susceptibility testing of lactobacilli, pediococci, lactococci, and bifidobacteria. Appl. Environ. Microbiol. 2005, 71. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; de Bastos, M.L.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16. [Google Scholar] [CrossRef]
- Danielsen, M.; Wind, A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 2003, 82. [Google Scholar] [CrossRef]
- Yan, M.; Han, J.; Xu, X.; Liu, L.; Gao, C.; Zheng, H.; Chen, Y.; Tao, Y.; Zhou, H.; Li, Y.; et al. Gsy, a novel glucansucrase from Leuconostoc mesenteroides, mediates the formation of cell aggregates in response to oxidative stress. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3. [Google Scholar] [CrossRef]
- Sharkey, L.K.R.; Edwards, T.A.; O’Neill, A.J. ABC-F proteins mediate antibiotic resistance through ribosomal protection. MBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Sneath, P.H.A.; Sokal, R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; WF Freeman & Co.: San Francisco, CA, USA, 1973. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Elsevier: Amsterdam, The Netherlands, 1965. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37. [Google Scholar] [CrossRef] [PubMed]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Buergmann, H.; Sørum, H.; Norström, M.; Pons, M.-N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic resistance in the food chain: A developing country-perspective. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; De Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial resistance in the food chain: A review. Int. J. Environ. Res. Public Health 2013, 10, 2643. [Google Scholar] [CrossRef]
- Stanhope, M.J.; Lefébure, T.; Walsh, S.L.; Becker, J.A.; Lang, P.; Pavinski Bitar, P.D.; Miller, L.A.; Italia, M.J.; Amrine-Madsen, H. Positive selection in penicillin-binding proteins 1a, 2b, and 2x from Streptococcus pneumoniae and its correlation with amoxicillin resistance development. Infect. Genet. Evol. 2008, 8. [Google Scholar] [CrossRef]
- Rosander, A.; Connolly, E.; Roos, S. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl. Environ. Microbiol. 2008, 74. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 2008, 282. [Google Scholar] [CrossRef]
- Roberts, M.C. Resistance to tetracycline, macrolide-lincosamide-streptogramin, trimethoprim, and sulfonamide drug classes. Mol. Biotechnol. 2002, 20. [Google Scholar] [CrossRef]
- Leech, J.; Cabrera-Rubio, R.; Walsh, A.M.; Macori, G.; Walsh, C.J.; Barton, W.; Finnegan, L.; Crispie, F.; O’Sullivan, O.; Claesson, M.J.; et al. Fermented-food metagenomics reveals substrate-associated differences in taxonomy and health-associated and antibiotic resistance determinants. MSystems 2020, 5. [Google Scholar] [CrossRef]
- Guevarra, R.B.; Magez, S.; Peeters, E.; Chung, M.S.; Kim, K.H.; Radwanska, M. Comprehensive genomic analysis reveals virulence factors and antibiotic resistance genes in Pantoea agglomerans KM1, a potential opportunistic pathogen. PLoS ONE 2021, 16, e0239792. [Google Scholar] [CrossRef]
- Besrour-Aouam, N.; Fhoula, I.; Hernández-Alcántara, A.M.; Mohedano, M.L.; Najjari, A.; Prieto, A.; Ruas-Madiedo, P.; López, P.; Ouzari, H.-I. The role of dextran production in the metabolic context of Leuconostoc and Weissella Tunisian strains. Carbohydr. Polym. 2021, 253. [Google Scholar] [CrossRef]
- Lee, S.; Park, J.; Jang, J.K.; Lee, B.H.; Park, Y.S. Structural analysis of gluco-oligosaccharides produced by Leuconostoc lactis and their prebiotic effect. Molecules 2019, 24, 3998. [Google Scholar] [CrossRef]
- Kim, M.; Jang, J.-K.; Park, Y.-S. Production optimization, structural analysis, and prebiotic- and anti-inflammatory effects of gluco-oligosaccharides produced by Leuconostoc lactis SBC001. Microorganisms 2021, 9, 200. [Google Scholar] [CrossRef]
- Moon, J.S.; Choi, H.S.; Shin, S.Y.; Noh, S.J.; Jeon, C.O.; Han, N.S. Genome sequence analysis of potential probiotic strain Leuconostoc lactis EFEL005 isolated from kimchi. J. Microbiol. 2015, 53, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.P.; Shin, S.C.; Kang, W.-K.; Chin, Y.-W.; Turner, T.L.; Choi, H.W.; Song, K.M.; Kim, H.J. Complete genome sequence of Leuconostoc garlicum KCCM 43211 producing exopolysaccharide. J. Biotechnol. 2017, 246. [Google Scholar] [CrossRef]
- Kumar, A.; Augustine, D.; Mehta, A.; Dinesh, K.R.; Viswam, D.; Philip, R. Leuconostoc garlicum: An unusual pathogen in the era of vancomycin therapy. Indian J. Chest Dis. Allied Sci. 2012, 2, 127–130. [Google Scholar]
- Pedersen, T.B.; Kot, W.P.; Hansen, L.H.; Sørensen, S.J.; Broadbent, J.R.; Vogensen, F.K.; Ardö, Y. Genome sequences of two Leuconostoc pseudomesenteroides strains isolated from Danish dairy starter cultures. Genome Announc. 2014, 2. [Google Scholar] [CrossRef]
- Meslier, V.; Loux, V.; Renault, P. Genome sequence of Leuconostoc pseudomesenteroides strain 4882, isolated from a dairy starter culture. J. Bacteriol. 2012, 194. [Google Scholar] [CrossRef]
- Land, M.; Hauser, L.; Jun, S.R.; Nookaew, I.; Leuze, M.R.; Ahn, T.H.; Karpinets, T.; Lund, O.; Kora, G.; Wassenaar, T.; et al. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genom. 2015, 15, 141–161. [Google Scholar] [CrossRef]
- Mikalsen, T.; Pedersen, T.; Willems, R.; Coque, T.M.; Werner, G.; Sadowy, E.; Van Schaik, W.; Jensen, L.B.; Sundsfjord, A.; Hegstad, K. Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis. BMC Genom. 2015, 16. [Google Scholar] [CrossRef]
- Eraclio, G.; Ricci, G.; Fortina, M.G. Insertion sequence elements in Lactococcus garvieae. Gene 2015, 555. [Google Scholar] [CrossRef]
- Nicoloff, H.; Bringel, F. ISLpl1 is a functional IS30-related insertion element in Lactobacillus plantarum that is also found in other lactic acid bacteria. Appl. Environ. Microbiol. 2003, 69, 6032–6040. [Google Scholar] [CrossRef] [PubMed]
- Ravin, V.; Alatossava, T. Three new insertion sequence elements ISLdl2, ISLdl3, and ISLdl4 in Lactobacillus delbrueckii: Isolation, molecular characterization, and potential use for strain identification. Plasmid 2003, 49. [Google Scholar] [CrossRef]
- Kumar, R.; Grover, S.; Kaushik, J.K.; Batish, V.K. IS30-related transposon mediated insertional inactivation of bile salt hydrolase (bsh1) gene of Lactobacillus plantarum strain Lp20. Microbiol. Res. 2014, 169. [Google Scholar] [CrossRef]
- Shi, Y.-Z.; Yoshida, T.; Fujiwara, A.; Nishiki, I. Characterization of lsa(D), a novel gene responsible for resistance to lincosamides, streptogramins A, and pleuromutilins in fish pathogenic Lactococcus garvieae Serotype II. Microb. Drug Resist. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- McDermott, P.F.; Tyson, G.H.; Kabera, C.; Chen, Y.; Li, C.; Folster, J.P.; Ayers, S.L.; Lam, C.; Tate, H.P.; Zhao, S. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother. 2016, 60. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhu, W.; Cao, Z.; Xu, B.; Wang, G.; Luo, M. High correlation between genotypes and phenotypes of environmental bacteria Comamonas testosteroni strains. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]


| Species | Type Strain | Isolation Source | GenBank Accession Number |
|---|---|---|---|
| Leuconostoc carnosum | DSM 5576T | Chill-stored meat | JACHGL000000000.1 |
| Leuconostoc citreum | ATCC 49370T | Food and clinical sources | PUFH00000000.1 |
| Leuconostoc falkenbergense | LMG 10779T | Lactic culture | BMBS00000000.1 |
| Leuconostoc fallax | ATCC 700006T | Sauerkraut | PUFI00000000.1 |
| Leuconostoc gelidum subsp. gasicomitatum | LMG 18811T | Packaged meat | FN822744.1 |
| Leuconostoc gelidum subsp. gelidum | KCTC 3527T | Kimchi (a traditional Korean food made by fermentation of Chinese cabbage) | AEMI00000000.1 |
| Leuconostoc holzapfelii | CCUG 54536T | Ethiopian coffee fermentation | JAAXPO000000000.1 |
| Leuconostoc inhae | KCTC 3774T | Kimchi | AEMJ00000000.1 |
| Leuconostoc kimchii | IMSNU 11154T | Kimchi | chromosome: NC_014136.1/CP001758.1 plasmid: LkipL4701: NC_014131.1/CP001753.1 LkipL4704: NC_014132.1/CP001754.1 LkipL4719: NC_014133.1/CP001755.1 LkipL4726: NC_014134.1/CP001756.1 LkipL48: NC_014135.1/CP001757.1 |
| Leuconostoc lactis | DSM 20202T | Milk | AEOR00000000.1 |
| Leuconostoc litchii | MB7T | Lychee | SDGY00000000.1 |
| Leuconostoc mesenteroides subsp. cremoris | ATCC 19254T | Hansen’s dried starter powder | ACKV00000000.1 |
| Leuconostoc mesenteroides subsp. cremoris | DSM 20484T | Missing (isolated in 1912) | chromosome: NZ_CP012009.1/CP012009.1 plasmid pDSM20484: NZ_CP012010.1/CP012010.1 |
| Leuconostoc mesenteroides subsp. Jonggajibkimchii | DRC1506T | Kimchi | chromosome: NZ_CP014611.1/CP014611.1 plasmid pDRC1: NZ_CP014612.1/CP014612.1 pDRC2: NZ_CP014613.1/CP014613.1 pDRC3: NZ_CP014614.1/CP014614.1 |
| Leuconostoc mesenteroides subsp. mesenteroides | ATCC 8293T | Fermenting olives | chromosome: NC_008531.1/CP000414.1 plasmid pLEUM1: NC_008496.1/CP000415.1 |
| Leuconostoc pseudomesenteroides | NCDO 768T | Cane juice | JAAXPY000000000.1 |
| Leuconostoc suionicum | DSM 20241T | Missing (isolated in 1972) | chromosome: NZ_CP015247.1/CP015247.1 plasmid unnamed: NZ_CP015248.1/CP015248.1 |
| Species | MIC (µg/mL) | |
|---|---|---|
| CLI | QUD | |
| Leuconostoc carnosum CECT 4024T (=DSM 5576T) | <0.03 | 1 |
| Leuconostoc fallax LMG 13177T (=ATCC 700006T) | 4 | 8 |
| Leuconostoc gelidum subsp. gasicomitatum CECT 5767T (=LMG 18811T) | <0.03 | 1 |
| Leuconostoc gelidum subsp. gelidum CECT 4026T (=KCTC 3527T) | <0.03 | 0.5 |
| Leuconostoc inhae CECT 7026T (=KCTC 3774T) | <0.03 | 1 |
| Leuconostoc lactis LMG 8894T (=DSM 20202T) | <0.03 | 1 |
| Leuconostoc mesenteroides subsp. cremoris LMG 6909T (=ATCC 19254T) | 0.12 | 1 |
| Leuconostoc mesenteroides subsp. dextranicum LMG 6908T (=ATCC 19254T) | 0.12 | 0.5 |
| Leuconostoc mesenteroides subsp. mesenteroides LMG 6893T (=ATCC 8293T) | 0.12 | 1 |
| Leuconostoc pseudomesenteroides LMG 11482T (=NCDO 768T) | 16 | 2 |
| Leuconostoc suionicum CECT 8146T (=DSM 20241T) | 0.12 | 1 |
| ECOFF | 1 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvetti, E.; Campedelli, I.; Larini, I.; Conedera, G.; Torriani, S. Exploring Antibiotic Resistance Diversity in Leuconostoc spp. by a Genome-Based Approach: Focus on the lsaA Gene. Microorganisms 2021, 9, 491. https://doi.org/10.3390/microorganisms9030491
Salvetti E, Campedelli I, Larini I, Conedera G, Torriani S. Exploring Antibiotic Resistance Diversity in Leuconostoc spp. by a Genome-Based Approach: Focus on the lsaA Gene. Microorganisms. 2021; 9(3):491. https://doi.org/10.3390/microorganisms9030491
Chicago/Turabian StyleSalvetti, Elisa, Ilenia Campedelli, Ilaria Larini, Giada Conedera, and Sandra Torriani. 2021. "Exploring Antibiotic Resistance Diversity in Leuconostoc spp. by a Genome-Based Approach: Focus on the lsaA Gene" Microorganisms 9, no. 3: 491. https://doi.org/10.3390/microorganisms9030491
APA StyleSalvetti, E., Campedelli, I., Larini, I., Conedera, G., & Torriani, S. (2021). Exploring Antibiotic Resistance Diversity in Leuconostoc spp. by a Genome-Based Approach: Focus on the lsaA Gene. Microorganisms, 9(3), 491. https://doi.org/10.3390/microorganisms9030491






