New Insights into the Co-Occurrences of Glycoside Hydrolase Genes among Prokaryotic Genomes through Network Analysis
Abstract
1. Introduction
2. Methods
2.1. Information Acquisition and Matrix Construction
2.2. Calculations on the Distribution of GHs
2.3. Network Construction and Functional Group Classification
2.4. Heatmap Construction
2.5. Co-Evolution Analysis
3. Results
3.1. GH Information Acquisition and Matrix Construction
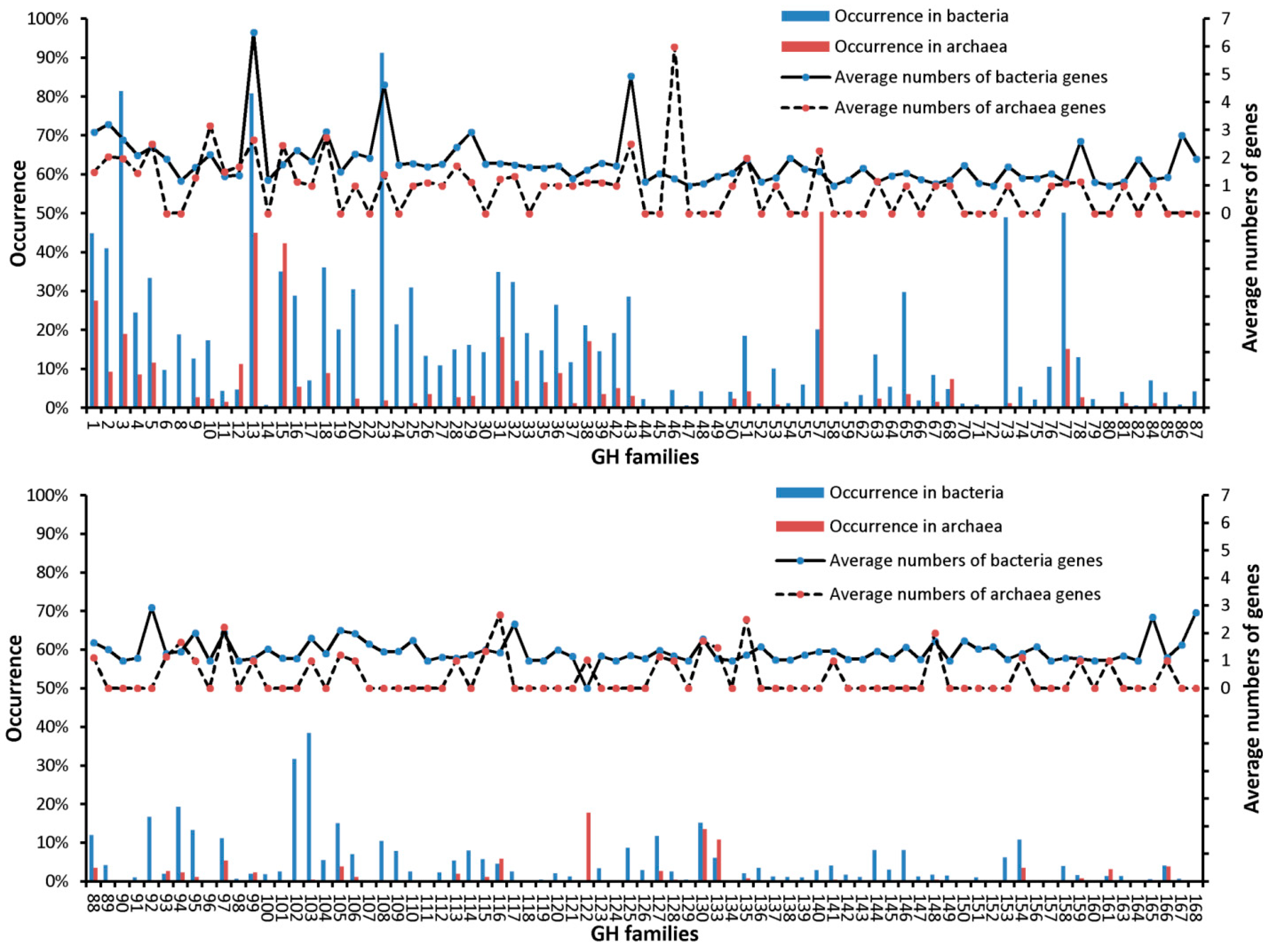
3.2. Occurrences of Genes from Various GHFs
3.3. Co-Occurrence and Network Analysis of GHs
3.4. Network Analysis of Microbes
3.5. Classification of Functional Categories
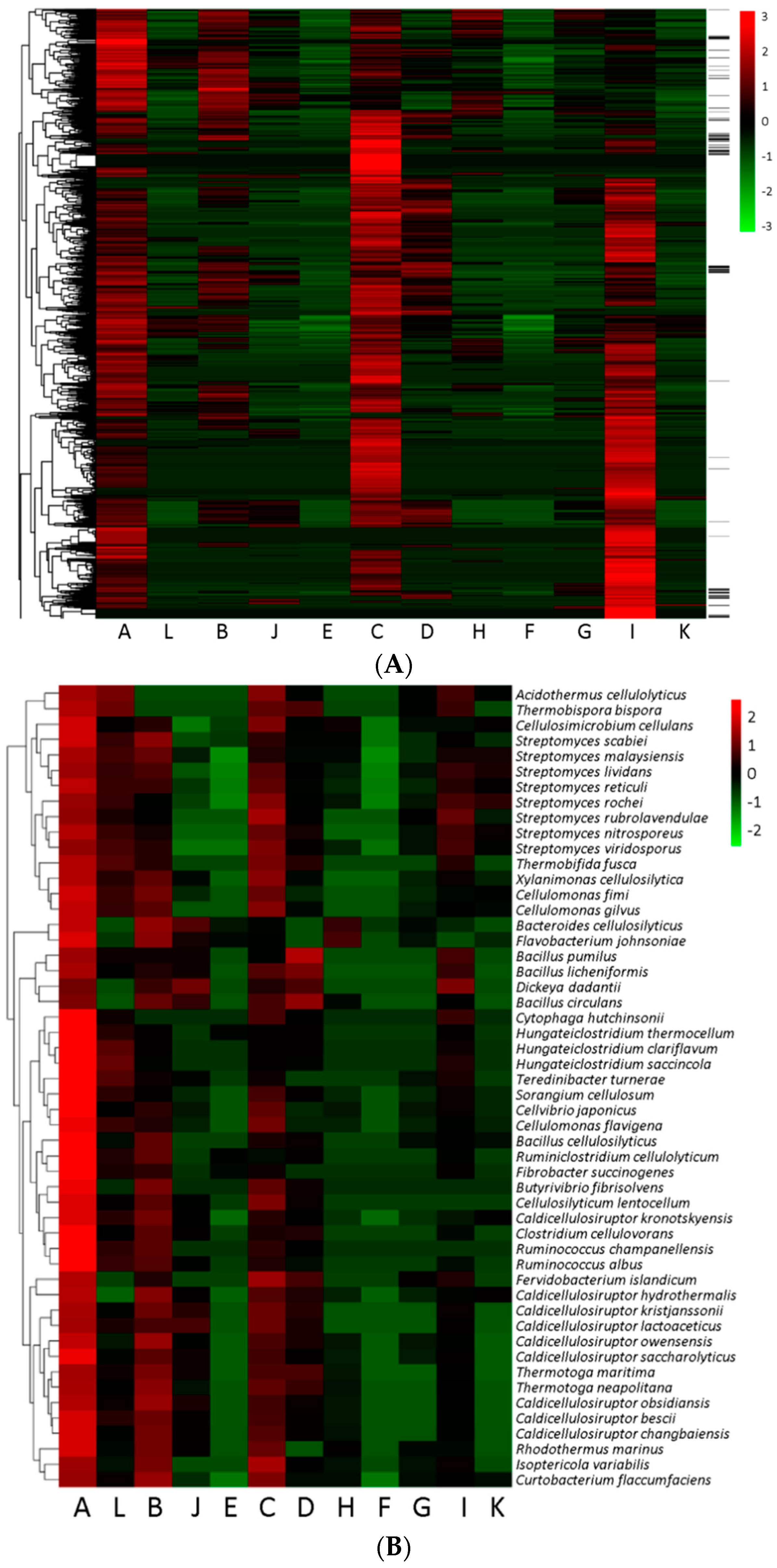
3.6. Heatmap Illustration of Gene Doses in Various Functional Categories
3.7. Co-Evolution Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gougoulias, C.; Clark, J.M.; Shaw, L.J. The role of soil microbes in the global carbon cycle: Tracking the below-ground microbial processing of plant-derived carbon for manipulating carbon dynamics in agricultural systems. J. Sci. Food Agric. 2014, 94, 2362–2371. [Google Scholar] [CrossRef]
- Park, B.H.; Karpinets, T.V.; Syed, M.H.; Leuze, M.R.; Uberbacher, E.C. CAZymes Analysis Toolkit (CAT): Web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 2010, 20, 1574–1584. [Google Scholar] [CrossRef]
- Kirk, O.; Borchert, T.V.; Fuglsang, C.C. Industrial enzyme applications. Curr. Opin. Biotechnol. 2002, 13, 345–351. [Google Scholar] [CrossRef]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef]
- Shendure, J.; Ji, H.L. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- Garron, M.L.; Henrissat, B. The continuing expansion of CAZymes and their families. Curr. Opin. Chem. Biol. 2019, 53, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Aspeborg, H.; Coutinho, P.M.; Wang, Y.; Brumer, H.; Henrissat, B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 2012, 12, 186. [Google Scholar] [CrossRef] [PubMed]
- Mewis, K.; Lenfant, N.; Lombard, V.; Henrissat, B. Dividing the large glycoside hydrolase family 43 into subfamilies: A motivation for detailed enzyme characterization. Appl. Environ. Microbiol. 2016, 82, 1686–1692. [Google Scholar] [CrossRef] [PubMed]
- Cordero, O.X.; Snel, B.; Hogeweg, P. Coevolution of gene families in prokaryotes. Genome Res. 2008, 18, 462–468. [Google Scholar] [CrossRef][Green Version]
- de Juan, D.; Pazos, F.; Valencia, A. Emerging methods in protein co-evolution. Nat. Rev. Genet. 2013, 14, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Fraser, H.B.; Hirsh, A.E.; Wall, D.P.; Eisen, M.B. Coevolution of gene expression among interacting proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 9033–9038. [Google Scholar] [CrossRef]
- Cope, A.L.; O’Meara, B.C.; Gilchrist, M.A. Gene expression of functionally-related genes coevolves across fungal species: Detecting coevolution of gene expression using phylogenetic comparative methods. BMC Genom. 2020, 21, 370. [Google Scholar] [CrossRef] [PubMed]
- Berlemont, R.; Martiny, A.C. Glycoside hydrolases across environmental microbial communities. PLoS Comput. Biol. 2016, 12, e1005300. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.T.C.; Freund, H.L.; Kasanjian, J.; Berlemont, R. Function, distribution, and annotation of characterized cellulases, xylanases, and chitinases from CAZy. Appl. Microbiol. Biotechnol. 2018, 102, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Berlemont, R.; Martiny, A.C. Genomic potential for polysaccharide deconstruction in bacteria. Appl. Environ. Microbiol. 2015, 81, 1513–1519. [Google Scholar] [CrossRef]
- Jacomy, M.; Venturini, T.; Heymann, S.; Bastian, M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE 2014, 9, e98679. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Abe, K.; Nakajima, M.; Yamashita, T.; Matsunaga, H.; Kamisuki, S.; Nihira, T.; Takahashi, Y.; Sugimoto, N.; Miyanaga, A.; Nakai, H.; et al. Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family. J. Biol. Chem. 2017, 292, 7487–7506. [Google Scholar] [CrossRef]
- Koeck, D.E.; Pechtl, A.; Zverlov, V.V.; Schwarz, W.H. Genomics of cellulolytic bacteria. Curr. Opin. Biotechnol. 2014, 29, 171–183. [Google Scholar] [CrossRef]
- Ochoa, D.; Pazos, F. Studying the co-evolution of protein families with the Mirrortree web server. Bioinformatics 2010, 26, 1370–1371. [Google Scholar] [CrossRef]
- Naumoff, D.G. Hierarchical classification of glycoside hydrolases. Biochemistry 2011, 76, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Blohs, M.; Moissl-Eichinger, C.; Mahnert, A.; Spang, A.; Dombrowski, N.; Krupovic, M.; Klingl, A. Encyclopedia of Microbiology; Schmidt, T.M., Ed.; Academic Press: New York, NY, USA, 2019; p. 243. [Google Scholar]
- Oslowski, D.M.; Jung, J.H.; Seo, D.H.; Park, C.S.; Holden, J.F. Production of hydrogen from alpha-1,4-and beta-1,4-linked saccharides by marine hyperthermophilic archaea. Appl. Environ. Microbiol. 2011, 77, 3169–3173. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Toshchakov, S.V.; Kolganova, T.V.; Kublanov, I.V. Halo(natrono)archaea isolated from hypersaline lakes utilize cellulose and chitin as growth substrates. Front Microbiol. 2015, 6, 942. [Google Scholar] [CrossRef]
- Naumoff, D.G.; Dedysh, S.N. Bacteria from poorly studied phyla as a potential source of new enzymes: Beta-galactosidases from planctomycetes and verrucomicrobia. Microbiology. 2018, 87, 796–805. [Google Scholar] [CrossRef]
- Popa, O.; Dagan, T. Trends and barriers to lateral gene transfer in prokaryotes. Curr. Opin. Microbiol. 2011, 14, 615–623. [Google Scholar] [CrossRef]
- Ndeh, D.; Rogowski, A.; Cartmell, A.; Luis, A.S.; Basle, A.; Gray, J.; Venditto, I.; Briggs, J.; Zhang, X.Y.; Labourel, A.; et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 2017, 544, 65–70. [Google Scholar] [CrossRef]
- Maehara, T.; Yagi, H.; Sato, T.; Ohnishi-Kameyama, M.; Fujimoto, Z.; Kamino, K.; Kitamura, Y.; St John, F.; Yaoi, K.; Kaneko, S. GH30 glucuronoxylan-specific xylanase from Streptomyces turgidiscabies C56. Appl. Environ. Microbiol. 2018, 84, e01850-17. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.J.; Price, N.D. Genetic co-occurrence network across sequenced microbes. PLoS Comput. Biol. 2011, 7, e1002340. [Google Scholar] [CrossRef]
- Maamar, H.; Abdou, L.; Boileau, C.; Valette, O.; Tardif, C. Transcriptional analysis of the cip-cel gene cluster from Clostridium cellulolyticum. J. Bacteriol. 2006, 188, 2614–2624. [Google Scholar] [CrossRef][Green Version]
- Svartström, O.; Alneberg, J.; Terrapon, N.; Lombard, V.; de Bruijn, I.; Malmsten, J.; Dalin, A.-M.; Muller, E.E.; Shah, P.; Wilmes, P. Ninety-nine de novo assembled genomes from the moose (Alces alces) rumen microbiome provide new insights into microbial plant biomass degradation. ISME J. 2017, 11, 2538–2551. [Google Scholar] [CrossRef]
- Marynowska, M.; Goux, X.; Sillam-Dussès, D.; Rouland-Lefèvre, C.; Halder, R.; Wilmes, P.; Gawron, P.; Roisin, Y.; Delfosse, P.; Calusinska, M. Compositional and functional characterisation of biomass-degrading microbial communities in guts of plant fibre-and soil-feeding higher termites. Microbiome 2020, 8, 1–18. [Google Scholar] [CrossRef]
- Andrade, A.C.; Fróes, A.; Lopes, F.Á.C.; Thompson, F.L.; Krüger, R.H.; Dinsdale, E.; Bruce, T. Diversity of microbial carbohydrate-active enZYmes (CAZYmes) associated with freshwater and soil samples from Caatinga biome. Microb. Ecol. 2017, 74, 89–105. [Google Scholar] [CrossRef]
- Faysal, M.A.M.; Arifuzzaman, S. A comparative analysis of large-scale network visualization tools. In Proceedings of the 2018 IEEE International Conference on Big Data (Big Data), Seattle, WA, USA, 10–13 December 2018; IEEE: Seattle, WA, USA, 2018; pp. 4837–4843. [Google Scholar]
- Barberan, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef]
- Williams, R.J.; Howe, A.; Hofmockel, K.S. Demonstrating microbial co-occurrence pattern analyses within and between ecosystems. Front Microbiol. 2014, 5, 358. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Y.; Wang, W.B. Benzyldimethyldodecyl ammonium chloride shifts the proliferation of functional genes and microbial community in natural water from eutrophic lake. Environ. Pollut. 2018, 236, 355–365. [Google Scholar] [CrossRef]
- Blennow, A.; Engelsen, S.B.; Nielsen, T.H.; Baunsgaard, L.; Mikkelsen, R. Starch phosphorylation: A new front line in starch research. Trends Plant Sci. 2002, 7, 445–450. [Google Scholar] [CrossRef]
- Scheurwater, E.; Reid, C.W.; Clarke, A.J. Lytic transglycosylases: Bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 2008, 40, 586–591. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Li, N.; Yuan, X.F.; Hua, B.B.; Wang, J.G.; Ishii, M.; Igarashi, Y.; Cui, Z.J. Enhancing the cellulose-degrading activity of cellulolytic bacteria CTL-6 (Clostridium thermocellum) by co-culture with non-cellulolytic bacteria W2-10 (Geobacillus sp.). Appl. Biochem. Biotechnol. 2013, 171, 1578–1588. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Cao, G.L.; Zheng, J.; Fu, D.F.; Song, J.Z.; Zhang, J.Z.; Zhao, L.; Yang, Q. Developing a mesophilic co-culture for direct conversion of cellulose to butanol in consolidated bioprocess. Biotechnol. Biofuels 2015, 8, 84. [Google Scholar] [CrossRef]
- Cui, J.M.; Mai, G.Q.; Wang, Z.W.; Liu, Q.; Zhou, V.; Ma, Y.F.; Liu, C.L. Metagenomic insights into a cellulose-rich niche reveal microbial cooperation in cellulose degradation. Front Microbiol. 2019, 10, 618. [Google Scholar] [CrossRef]
- Akinosho, H.; Yee, K.; Close, D.; Ragauskas, A. The emergence of Clostridium thermocellum as a high utility candidate for consolidated bioprocessing applications. Front Chem. 2014, 2, 66. [Google Scholar] [CrossRef]
- Chung, D.; Cha, M.; Guss, A.M.; Westpheling, J. Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc. Natl. Acad. Sci. USA 2014, 111, 8931–8936. [Google Scholar] [CrossRef]
- McBride, M.J.; Xie, G.; Martens, E.C.; Lapidus, A.; Henrissat, B.; Rhodes, R.G.; Goltsman, E.; Wang, W.; Xu, J.; Hunnicutt, D.W.; et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl. Environ. Microbiol. 2009, 75, 6864–6875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.P.; Ding, S.Y.; Mielenz, J.R.; Cui, J.B.; Elander, R.T.; Laser, M.; Himmel, M.E.; McMillan, J.R.; Lynd, L.R. Fractionating recalcitrant lignocellulose at modest reaction conditions. Biotechnol. Bioeng. 2007, 97, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Rettenmaier, R.; Schneider, M.; Munk, B.; Lebuhn, M.; Jünemann, S.; Sczyrba, A.; Maus, I.; Zverlov, V.; Liebl, W. Importance of Defluviitalea raffinosedens for hydrolytic biomass degradation in co-culture with Hungateiclostridium thermocellum. Microorganisms 2020, 8, 915. [Google Scholar] [CrossRef] [PubMed]
- Gullert, S.; Fischer, M.A.; Turaev, D.; Noebauer, B.; Ilmberger, N.; Wemheuer, B.; Alawi, M.; Rattei, T.; Daniel, R.; Schmitz, R.A.; et al. Deep metagenome and metatranscriptome analyses of microbial communities affiliated with an industrial biogas fermenter, a cow rumen, and elephant feces reveal major differences in carbohydrate hydrolysis strategies. Biotechnol. Biofuels 2016, 9, 121. [Google Scholar] [CrossRef]



| Groups | Ghfs | Profiles of the Enzymatic Functions | No. of Species (Phyla) Largely Fitted a | No. of Species (Phyla) Partially Fitted b | Sources c | Predominant Phyla (No. of Species, Frequency) d |
|---|---|---|---|---|---|---|
| A | 3, 5, 9, 10, 16, 30, 31, 43, 51, 67, 115 | Widely distributed GHs for the decomposition of bulk lignocellulose components, such as cellulose, β-glucan, and glucuronoarabinoxylan, xyloglucan | 283 (12) | 2281 (28) | m | Actinobacteria (516, 78%), Bacteroidetes (262, 88%), Firmicutes (337, 48%), Proteobacteria (895, 50%) |
| B | 2, 27, 35, 36, 42, 53, 78, 95, 106, 127, 146 | Widely distributed GHs for the debranching of pectic polysaccharides, specifically Rhamnogalacturonan II | 154 (10) | 1475 (24) | m | Actinobacteria (341, 53%), Bacteroidetes (202, 72%), Firmicutes (335, 48%), Proteobacteria (413, 23%) |
| C | 13, 77 | GHs for the decomposition or modification of starch | 2007 (30) e | / | m | Actinobacteria (457, 69%), Cyanobacteria (89, 98%), Deinococcus-Thermus (27, 100%), Proteobacteria (896, 50%) |
| D | 1, 4 | Glycosidases | 844 (16) e | / | m | Actinobacteria (222, 34%), Firmicutes (269, 38%), Proteobacteria (262, 15%) |
| E | 137, 138, 139, 141, 142, 143 | GHs for the debranching of pectic polysaccharides, specifically Rhamnogalacturonan II | 37(1) | 73 (4) | s | Acidobacteria (5, 38%), Bacteroidetes (52, 18%) |
| F | 82, 86, 117, 150, 167 | GHs for the decomposition of cell wall polysaccharides from red algae and seaweeds | 13 (4) | 43 (6) | s | Bacteroidetes (19, 6%), Planctomycetes (6, 17%) |
| G | 20, 109 | GHs for the decomposition of hexosamine | 294 (10) e | / | s | Actinobacteria (70, 10%), Bacteroidetes (162, 55%) |
| H | 29, 92, 97, 125 | α-Glycosidases | 314 (9) | 579 (14) | m | Actinobacteria (132, 20%), Bacteroidetes (221, 75%) |
| I | 23, 102, 103 | GHs for the decomposition of peptidoglycan | 1080 (4) e | 1701 (9) | m | Cyanobacteria (74, 81%), Proteobacteria (1608, 90%) |
| J | 28, 88, 105, 154 | GHs for the decomposition of the main chain of pectic polysaccharides, including Homogalacturonan, Rhamnogalacturonan I, and Rhamnogalacturonan II | 312 (11) | 623 (15) | m | Acidobacteria (10, 77%), Bacteroidetes (144, 49%), Firmicutes (114, 16%), Proteobacteria (231, 13%) |
| K | 46, 55, 64, 75, 87, 114 | GHs for the decomposition of fungal cell wall polysaccharides, including alpha-1,3- glucan, beta-1,3-glucan, chitosan, and polygalactosamine | 78 (2) | 231 (9) | s | Acidobacteria (6, 46%), Actinobacteria (128, 26%) |
| L | 6, 11, 12, 48, 55, 62, 64, 74 | Supplemental GHs of group A for more efficient decomposition of bulk lignocellulose components, such as cellulose, beta-glucan, and glucuronoarabinoxylan, xyloglucan | 93 (2) | 338 (12) | m | Actinobacteria (201, 30%), Firmicutes (49, 7%), Proteobacteria (59, 3%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, A.; Jin, M.; Li, N.; Zhu, D.; Xie, R.; Wang, Q.; Lin, H.; Sun, J. New Insights into the Co-Occurrences of Glycoside Hydrolase Genes among Prokaryotic Genomes through Network Analysis. Microorganisms 2021, 9, 427. https://doi.org/10.3390/microorganisms9020427
Geng A, Jin M, Li N, Zhu D, Xie R, Wang Q, Lin H, Sun J. New Insights into the Co-Occurrences of Glycoside Hydrolase Genes among Prokaryotic Genomes through Network Analysis. Microorganisms. 2021; 9(2):427. https://doi.org/10.3390/microorganisms9020427
Chicago/Turabian StyleGeng, Alei, Meng Jin, Nana Li, Daochen Zhu, Rongrong Xie, Qianqian Wang, Huaxing Lin, and Jianzhong Sun. 2021. "New Insights into the Co-Occurrences of Glycoside Hydrolase Genes among Prokaryotic Genomes through Network Analysis" Microorganisms 9, no. 2: 427. https://doi.org/10.3390/microorganisms9020427
APA StyleGeng, A., Jin, M., Li, N., Zhu, D., Xie, R., Wang, Q., Lin, H., & Sun, J. (2021). New Insights into the Co-Occurrences of Glycoside Hydrolase Genes among Prokaryotic Genomes through Network Analysis. Microorganisms, 9(2), 427. https://doi.org/10.3390/microorganisms9020427







