Crossing the Wall: Characterization of the Multiheme Cytochromes Involved in the Extracellular Electron Transfer Pathway of Thermincola ferriacetica
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.1.1. ImdcA
2.1.2. PdcA
2.1.3. CwcA
2.2. Protein Purification
2.2.1. ImdcA
2.2.2. PdcA
2.3. Mass Spectrometry
2.4. Spectroscopic Techniques
2.4.1. UV Visible Spectroscopy
2.4.2. EPR Spectroscopy
2.4.3. NMR Spectroscopy
2.5. Electrochemical Measurements of PdcA and ImdcA
2.6. Homology-Based Modelling
2.6.1. CwcA
2.6.2. OcwA
3. Results
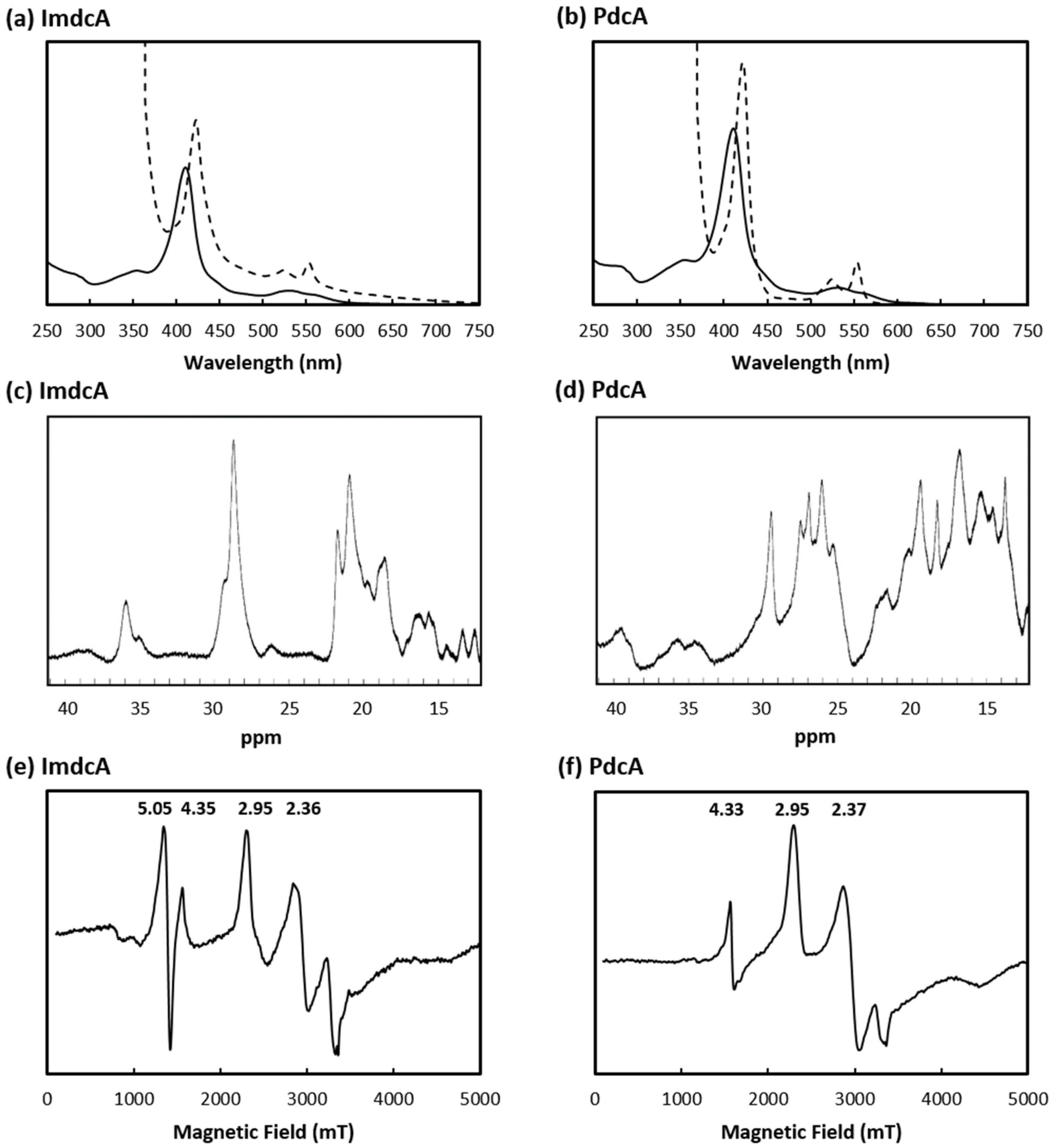
3.1. Production of MHC Involved in EET of T. ferriacetica
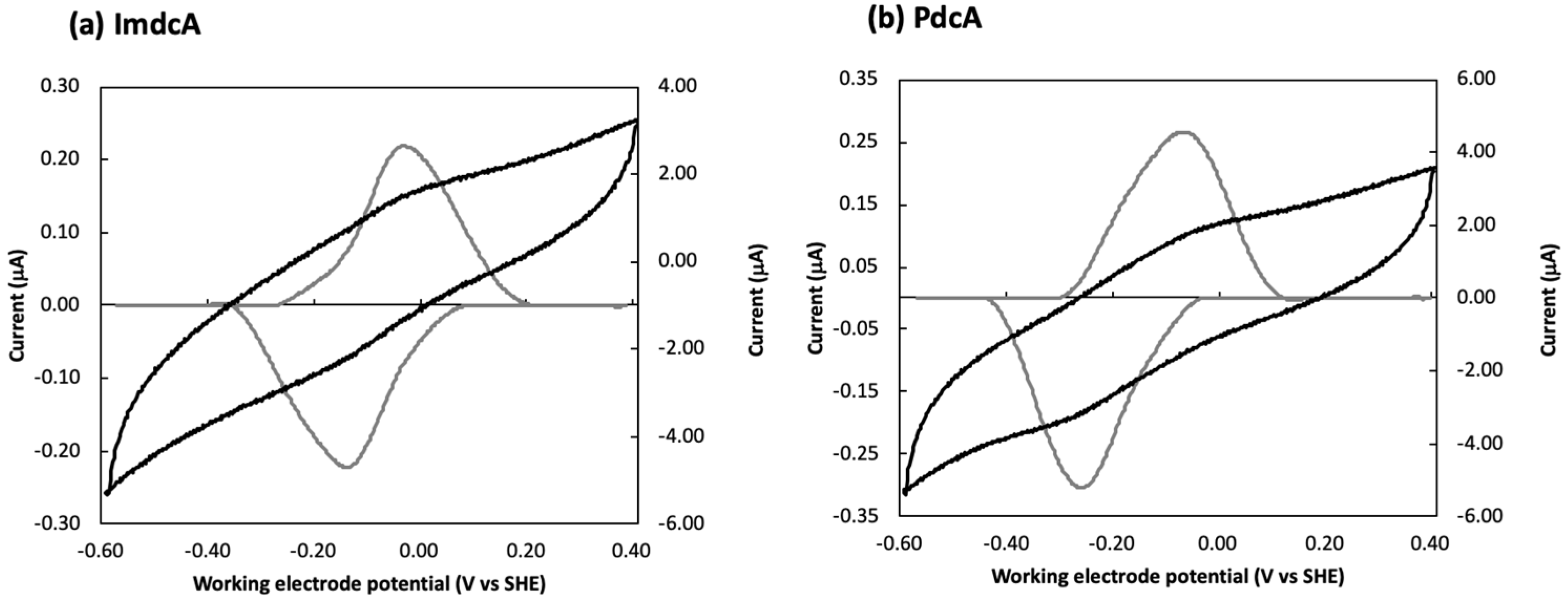
3.2. Electrochemical Behavior of ImdcA and PdcA
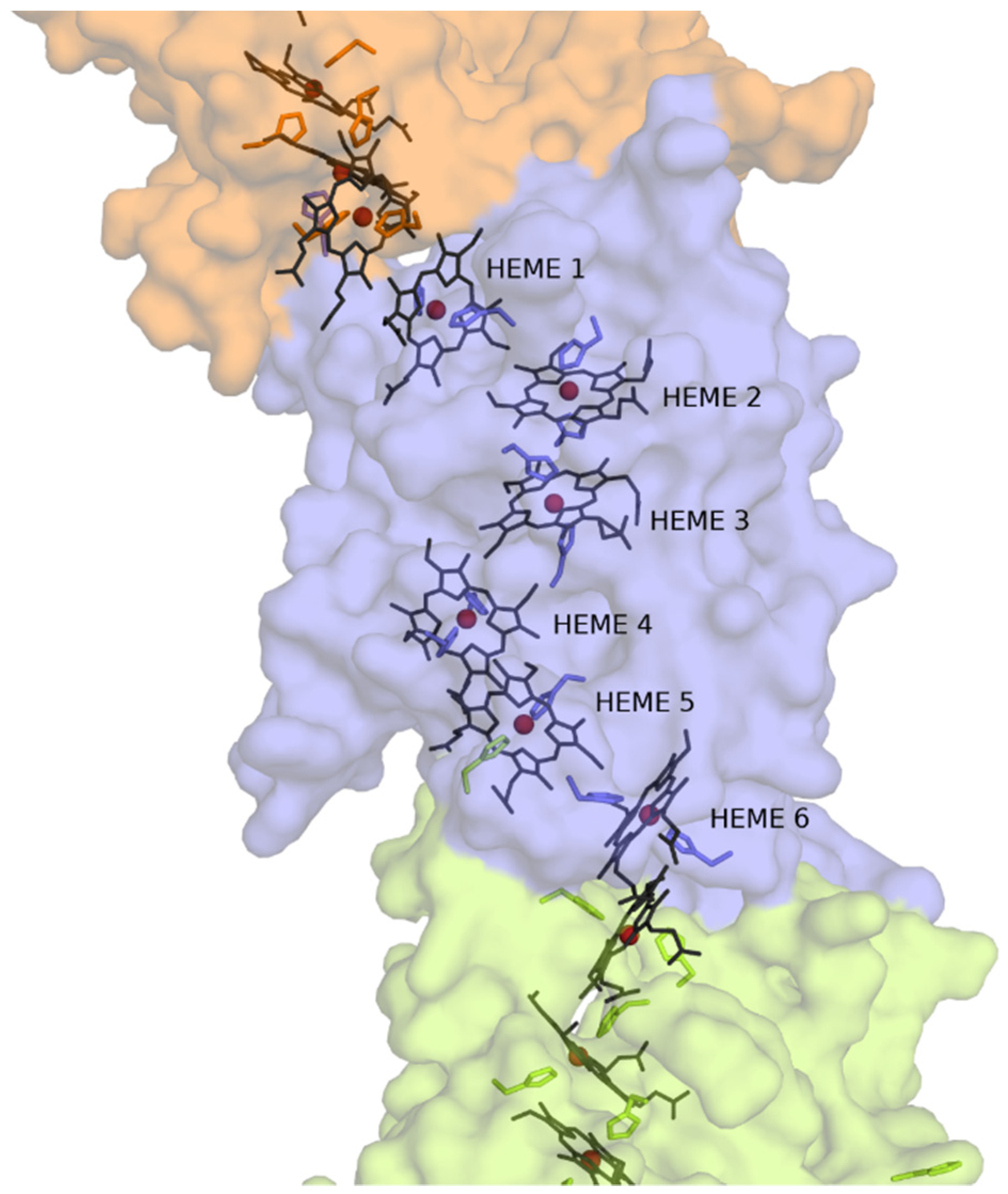
3.3. Structural Model of CwcA
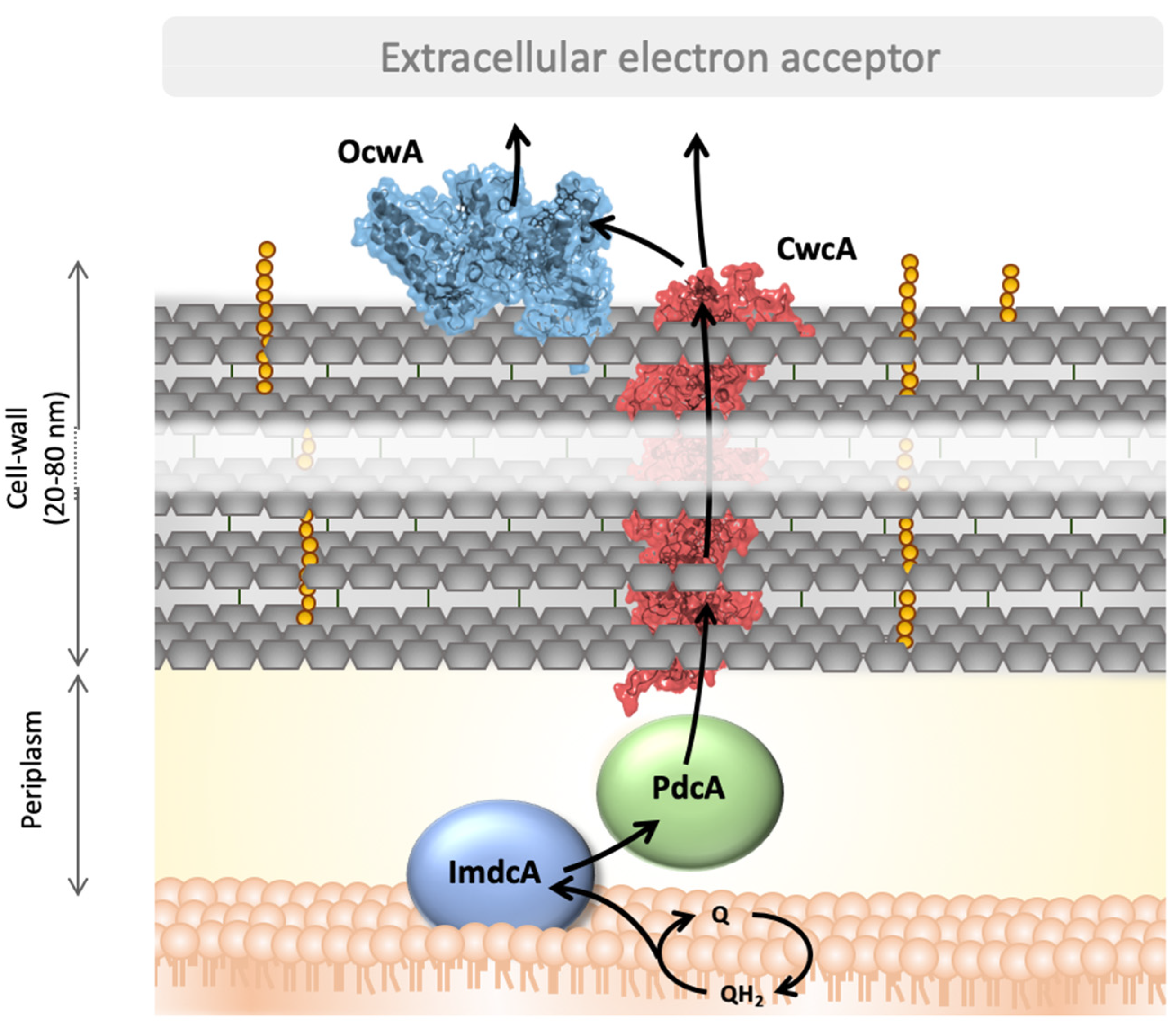
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, A.; Hsu, L.H.H.; Kavanagh, P.; Barrière, F.; Lens, P.N.L.; Lapinsonnière, L.; Lienhard, J.H.; Schröder, U.; Jiang, X.; Leech, D. The ins and outs of microorganism-electrode electron transfer reactions. Nat. Rev. Chem. 2017, 1, 1–13. [Google Scholar] [CrossRef]
- Logan, B.E.; Rossi, R.; Ragab, A.; Saikaly, P.E. Electroactive microorganisms in bioelectrochemical systems. Nat. Rev. Microbiol. 2019, 17, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Potter, M.C. Electrical Effects Accompanying the Decomposition of Organic Compounds. Proc. R. Soc. Lond. B Biol. Sci. 1911, 84, 571. [Google Scholar] [CrossRef]
- Paquete, C.M. Electroactivity across the cell wall of Gram-positive bacteria. Comput. Struct. Biotechnol. J. 2020, 18, 3796–3802. [Google Scholar] [CrossRef] [PubMed]
- White, G.F.; Edwards, M.J.; Gomez-Perez, L.; Richardson, D.J.; Butt, J.N.; Clarke, T.A. Mechanisms of Bacterial Extracellular Electron Exchange. Adv. Microb. Physiol. 2016, 87–138. [Google Scholar] [CrossRef]
- Beblawy, S.; Bursac, T.; Paquete, C.; Louro, R.; Clarke, T.A.; Gescher, J. Extracellular reduction of solid electron acceptors by Shewanella oneidensis. Mol. Microbiol. 2018, 109, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Louro, R.O.; Costa, N.L.; Fernandes, A.P.; Silva, A.V.; Trindade, I.B.; Fonseca, B.M.; Paquete, C.M. Exploring the Molecular Mechanisms of Extracellular Electron Transfer for Harnessing Reducing Power in METs. In Microbial Electrochemical Technology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 261–293. [Google Scholar]
- Nealson, K.H.; Rowe, A.R. Electromicrobiology: Realities, grand challenges, goals and predictions. Microb. Biotechnol. 2016, 9, 595–600. [Google Scholar] [CrossRef]
- Schievano, A.; Pepé Sciarria, T.; Vanbroekhoven, K.; De Wever, H.; Puig, S.; Andersen, S.J.; Rabaey, K.; Pant, D. Electro-Fermentation—Merging Electrochemistry with Fermentation in Industrial Applications. Trends Biotechnol. 2016, 34, 866–878. [Google Scholar] [CrossRef]
- Mathuriya, A.S.; Yakhmi, J.V. Microbial fuel cells—Applications for generation of electrical power and beyond. Crit. Rev. Microbiol. 2016, 42, 127–143. [Google Scholar] [CrossRef]
- Santoro, C.; Arbizzani, C.; Erable, B.; Ieropoulos, I. Microbial fuel cells: From fundamentals to applications. A review. J. Power Sources 2017, 356, 225–244. [Google Scholar] [CrossRef]
- Goswami, R.; Mishra, V.K. A review of design, operational conditions and applications of microbial fuel cells. Biofuels 2018, 9, 203–220. [Google Scholar] [CrossRef]
- Koch, C.; Harnisch, F. Is there a Specific Ecological Niche for Electroactive Microorganisms? ChemElectroChem 2016, 3, 1282–1295. [Google Scholar] [CrossRef]
- Wrighton, K.C.; Agbo, P.; Warnecke, F.; Weber, K.A.; Brodie, E.L.; DeSantis, T.Z.; Hugenholtz, P.; Andersen, G.L.; Coates, J.D. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2008, 2, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.I.; Marcus, A.K.; Parameswaran, P.; Rittmann, B.E. Kinetic experiments for evaluating the nernst-monod model for anode-respiring bacteria (ARB) in a biofilm anode. Environ. Sci. Technol. 2008, 42, 6593–6597. [Google Scholar] [CrossRef] [PubMed]
- Lusk, B.G. Thermophiles; or, the Modern Prometheus: The Importance of Extreme Microorganisms for Understanding and Applying Extracellular Electron Transfer. Front. Microbiol. 2019, 10, 818. [Google Scholar] [CrossRef] [PubMed]
- Lusk, B.G.; Peraza, I.; Albal, G.; Marcus, A.K.; Popat, S.C.; Torres, C.I. pH Dependency in Anode Biofilms of Thermincola ferriacetica Suggests a Proton-Dependent Electrochemical Response. J. Am. Chem. Soc. 2018, 140, 5527–5534. [Google Scholar] [CrossRef]
- Costa, N.L.; Hermann, B.; Fourmond, V.; Faustino, M.M.; Teixeira, M.; Einsle, O.; Paquete, C.M.; Louro, R.O. How Thermophilic Gram-Positive Organisms Perform Extracellular Electron Transfer: Characterization of the Cell Surface Terminal Reductase OcwA. MBio 2019, 10, e01210. [Google Scholar] [CrossRef]
- Lusk, B.G.; Badalamenti, J.P.; Parameswaran, P.; Bond, D.R.; Torres, C.I. Draft Genome Sequence of the Gram-Positive Thermophilic Iron Reducer Thermincola ferriacetica Strain Z-0001T. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Pankratova, G.; Hederstedt, L.; Gorton, L. Extracellular electron transfer features of Gram-positive bacteria. Anal. Chim. Acta 2019, 1076, 32–47. [Google Scholar] [CrossRef]
- Marshall, C.W.; May, H.D. Electrochemical evidence of direct electrode reduction by a thermophilic Gram-positive bacterium, Thermincola ferriacetica. Energy Environ. Sci. 2009, 2, 699–705. [Google Scholar] [CrossRef]
- Byrne-Bailey, K.G.; Wrighton, K.C.; Melnyk, R.A.; Agbo, P.; Hazen, T.C.; Coates, J.D. Complete Genome Sequence of the Electricity-Producing “Thermincola potens” Strain JR. J. Bacteriol. 2010, 192, 4078–4079. [Google Scholar] [CrossRef] [PubMed]
- Lusk, B.G.; Parameswaran, P.; Popat, S.C.; Rittmann, B.E.; Torres, C.I. The effect of pH and buffer concentration on anode biofilms of Thermincola ferriacetica. Bioelectrochemistry 2016, 112, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Wrighton, K.C.; Thrash, J.C.; Melnyk, R.A.; Bigi, J.P.; Byrne-Bailey, K.G.; Remis, J.P.; Schichnes, D.; Auer, M.; Chang, C.J.; Coates, J.D.; et al. Evidence for direct electron transfer by a gram-positive bacterium isolated from a microbial fuel cell. Appl. Environ. Microbiol. 2011, 77, 7633–7639. [Google Scholar] [CrossRef] [PubMed]
- Parameswaran, P.; Bry, T.; Popat, S.C.; Lusk, B.G.; Rittmann, B.E.; Torres, C.I. Kinetic, electrochemical, and microscopic characterization of the thermophilic, anode-respiring bacterium Thermincola ferriacetica. Environ. Sci. Technol. 2013, 47, 4934–4940. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Qian, X.; Morgado, L.; Kim, B.C.; Mester, T.; Izallalen, M.; Salgueiro, C.A.; Lovley, D.R. Purification and characterization of OmcZ, an outer-surface, octaheme c-type cytochrome essential for optimal current production by Geobacter sulfurreducens. Appl. Environ. Microbiol. 2010, 76, 3999–4007. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Mester, T.; Morgado, L.; Arakawa, T.; Sharma, M.L.; Inoue, K.; Joseph, C.; Salgueiro, C.A.; Maroney, M.J.; Lovley, D.R. Biochemical characterization of purified OmcS, a c-type cytochrome required for insoluble Fe(III) reduction in Geobacter sulfurreducens. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 404–412. [Google Scholar] [CrossRef]
- Edwards, M.J.; Richardson, D.J.; Paquete, C.M.; Clarke, T.A. Role of multiheme cytochromes involved in extracellular anaerobic respiration in bacteria. Protein Sci. 2020, 29, 830–842. [Google Scholar] [CrossRef]
- Costa, N.L.; Clarke, T.A.; Philipp, L.-A.; Gescher, J.; Louro, R.O.; Paquete, C.M. Electron transfer process in microbial electrochemical technologies: The role of cell-surface exposed conductive proteins. Bioresour. Technol. 2018, 255, 308–317. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, E.; Zhang, J.; Dai, Y.; Yang, Z.; Christensen, H.E.M.M.; Ulstrup, J.; Zhao, F. Extracellular polymeric substances are transient media for microbial extracellular electron transfer. Sci. Adv. 2017, 3, 1–9. [Google Scholar] [CrossRef]
- Pirbadian, S.; Barchinger, S.E.; Leung, K.M.; Byun, H.S.; Jangir, Y.; Bouhenni, R.A.; Reed, S.B.; Romine, M.F.; Saffarini, D.A.; Shi, L.; et al. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc. Natl. Acad. Sci. USA 2014, 111, 12883–12888. [Google Scholar] [CrossRef]
- Feliciano, G.T.; Steidl, R.J.; Reguera, G. Structural and functional insights into the conductive pili of Geobacter sulfurreducens revealed in molecular dynamics simulations. Phys. Chem. Chem. Phys. 2015, 17, 22217–22226. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gu, Y.; O’Brien, J.P.; Yi, S.M.; Yalcin, S.E.; Srikanth, V.; Shen, C.; Vu, D.; Ing, N.L.; Hochbaum, A.I.; et al. Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers. Cell 2019, 177, 361–369.e10. [Google Scholar] [CrossRef] [PubMed]
- Carlson, H.K.; Iavarone, A.T.; Gorur, A.; Yeo, B.S.; Tran, R.; Melnyk, R.A.; Mathies, R.A.; Auer, M.; Coates, J.D. Surface multiheme c-type cytochromes from Thermincola potens and implications for respiratory metal reduction by Gram-positive bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 1702–1707. [Google Scholar] [CrossRef] [PubMed]
- Marsili, E.; Baron, D.B.; Shikhare, I.D.; Coursolle, D.; Gralnick, J.A.; Bond, D.R. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. USA 2008, 105, 3968–3973. [Google Scholar] [CrossRef] [PubMed]
- von Canstein, H.; Ogawa, J.; Shimizu, S.; Lloyd, J.R. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 2008, 74, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.K.; Kolter, R. A role for excreted quinones in extracellular electron transfer. Nature 2000, 405, 94–97. [Google Scholar] [CrossRef]
- Pham, T.H.; Boon, N.; Aelterman, P.; Clauwaert, P.; De Schamphelaire, L.; Vanhaecke, L.; De Maeyer, K.; Höfte, M.; Verstraete, W.; Rabaey, K. Metabolites produced by Pseudomonas sp. enable a Gram-positive bacterium to achieve extracellular electron transfer. Appl. Microbiol. Biotechnol. 2008, 77, 1119–1129. [Google Scholar] [CrossRef]
- Milliken, C.E.; May, H.D. Sustained generation of electricity by the spore-forming, Gram-positive, Desulfitobacterium hafniense strain DCB2. Appl. Microbiol. Biotechnol. 2007, 73, 1180–1189. [Google Scholar] [CrossRef]
- Edwards, M.J.; White, G.F.; Butt, J.N.; Richardson, D.J.; Clarke, T.A. The Crystal Structure of a Biological Insulated Transmembrane Molecular Wire. Cell 2020, 181, 665–673.e10. [Google Scholar] [CrossRef]
- Costa, N.L.; Carlson, H.K.; Coates, J.D.; Louro, R.O.; Paquete, C.M. Heterologous expression and purification of a multiheme cytochrome from a Gram-positive bacterium capable of performing extracellular respiration. Protein Expr. Purif. 2015, 111, 48–52. [Google Scholar] [CrossRef]
- Fourmond, V. QSoas: A Versatile Software for Data Analysis. Anal. Chem. 2016, 88, 5050–5052. [Google Scholar] [CrossRef] [PubMed]
- Friis, E.P.; Andersen, J.E.T.; Madsen, L.L.; Bonander, N.; Møller, P.; Ulstrup, J. Dynamics of Pseudomonas aeruginosa azurin and its Cys3Ser mutant at single-crystal gold surfaces investigated by cyclic voltammetry and atomic force microscopy. Electrochim. Acta 1998, 43, 1114–1122. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Palmer, G. The electron paramagnetic resonance of metalloproteins. Biochem. Soc. Trans. 1985, 13, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Louro, R.O. Proton thrusters: Overview of the structural and functional features of soluble tetrahaem cytochromes c3. J. Biol. Inorg. Chem. 2007, 12, 1–10. [Google Scholar] [CrossRef]
- Fonseca, B.M.; Saraiva, I.H.; Paquete, C.M.; Soares, C.M.; Pacheco, I.; Salgueiro, C.A.; Louro, R.O. The tetraheme cytochrome from Shewanella oneidensis MR-1 shows thermodynamic bias for functional specificity of the hemes. J. Biol. Inorg. Chem. 2009, 14, 375–385. [Google Scholar] [CrossRef]
- Firer-sherwood, M.; Pulcu, G.S.; Elliott, S.J. Electrochemical interrogations of the Mtr cytochromes from Shewanella: Opening a potential window. J. Biol. Inorg. Chem. 2008, 13, 849–854. [Google Scholar] [CrossRef]
- Paquete, C.M.; Louro, R.O. Unveiling the details of electron transfer in multicenter redox proteins. Acc. Chem. Res. 2014, 47. [Google Scholar] [CrossRef]
- Reis, C.; Louro, R.O.; Pacheco, I.; Catarino, T.; Turner, D.L.; Xavier, A.V. Redox-Bohr effect in the nine haem cytochrome from Desulfovibrio desulfuricans 27774. Inorg. Chim. Acta 2002, 339, 248–252. [Google Scholar] [CrossRef]
- Fonseca, B.M.; Paquete, C.M.; Salgueiro, C.A.; Louro, R.O. The role of intramolecular interactions in the functional control of multiheme cytochromes c. FEBS Lett. 2012, 586. [Google Scholar] [CrossRef]
- Mowat, C.G.; Chapman, S.K. Multi-heme cytochromes—New structures, new chemistry. Dalton Trans. 2005, 21, 3381–3389. [Google Scholar] [CrossRef] [PubMed]
- Paquete, C.M.; Rusconi, G.; Silva, A.V.; Soares, R.; Louro, R.O. A brief survey of the “cytochromome”. Adv. Microb. Physiol. 2019, 75, 69–135. [Google Scholar] [PubMed]
- Moser, C.C.; Chobot, S.E.; Page, C.C.; Dutton, P.L. Distance metrics for heme protein electron tunneling. Biochim. Biophys. Acta Bioenerg. 2008, 1777, 1032–1037. [Google Scholar] [CrossRef] [PubMed]
- Morgado, L.; Brulx, M.; Pessanha, M.; Londer, Y.Y.; Salgueiro, C.A.; Bruix, M.; Pessanha, M.; Londer, Y.Y.; Salgueiro, C.A.; Brulx, M.; et al. Thermodynamic characterization of a triheme cytochrome family from Geobacter sulfurreducens reveals mechanistic And functional diversity. Biophys. J. 2010, 99, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.L.; Oliveira, T.F.; Pereira, I.A.C.; Archer, M. X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J. 2006, 25, 5951–5960. [Google Scholar] [CrossRef]
- McMillan, D.G.G.; Marritt, S.J.; Firer-Sherwood, M.A.; Shi, L.; Richardson, D.J.; Evans, S.D.; Elliott, S.J.; Butt, J.N.; Jeuken, L.J.C. Protein–Protein Interaction Regulates the Direction of Catalysis and Electron Transfer in a Redox Enzyme Complex. J. Am. Chem. Soc. 2013, 135, 10550–10556. [Google Scholar] [CrossRef] [PubMed]
- Saffarini, D.A.; Blumerman, S.L.; Mansoorabadi, K.J. Role of Menaquinones in Fe (III) Reduction by Membrane Fractions of Shewanella putrefaciens. J. Bacteriol. 2002, 184, 846–848. [Google Scholar] [CrossRef]
- Lovley, D.R.; Giovannoni, S.J.; White, D.C.; Champine, J.E.; Phillips, E.J.; Gorby, Y.A.; Goodwin, S. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 1993, 159, 336–344. [Google Scholar] [CrossRef]
- Ehrlich, H.L. Are gram-positive bacteria capable of electron transfer across their cell wall without an externally available electron shuttle? Geobiology 2008, 6, 220–224. [Google Scholar] [CrossRef]
- Light, S.H.; Méheust, R.; Ferrell, J.L.; Cho, J.; Deng, D.; Agostoni, M.; Iavarone, A.T.; Banfield, J.F.; D’Orazio, S.E.F.; Portnoy, D.A. Extracellular electron transfer powers flavinylated extracellular reductases in Gram-positive bacteria. Proc. Natl. Acad. Sci. USA 2019, 116, 26892–26899. [Google Scholar] [CrossRef]
- Méheust, R.; Huang, S.; Rivera-Lugo, R.; Banfield, J.F.; Light, S.H. Widespread bacterial protein flavinylation in functionally distinct extracytosolic redox biochemistries. bioRxiv 2021. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faustino, M.M.; Fonseca, B.M.; Costa, N.L.; Lousa, D.; Louro, R.O.; Paquete, C.M. Crossing the Wall: Characterization of the Multiheme Cytochromes Involved in the Extracellular Electron Transfer Pathway of Thermincola ferriacetica. Microorganisms 2021, 9, 293. https://doi.org/10.3390/microorganisms9020293
Faustino MM, Fonseca BM, Costa NL, Lousa D, Louro RO, Paquete CM. Crossing the Wall: Characterization of the Multiheme Cytochromes Involved in the Extracellular Electron Transfer Pathway of Thermincola ferriacetica. Microorganisms. 2021; 9(2):293. https://doi.org/10.3390/microorganisms9020293
Chicago/Turabian StyleFaustino, Marisa M., Bruno M. Fonseca, Nazua L. Costa, Diana Lousa, Ricardo O. Louro, and Catarina M. Paquete. 2021. "Crossing the Wall: Characterization of the Multiheme Cytochromes Involved in the Extracellular Electron Transfer Pathway of Thermincola ferriacetica" Microorganisms 9, no. 2: 293. https://doi.org/10.3390/microorganisms9020293
APA StyleFaustino, M. M., Fonseca, B. M., Costa, N. L., Lousa, D., Louro, R. O., & Paquete, C. M. (2021). Crossing the Wall: Characterization of the Multiheme Cytochromes Involved in the Extracellular Electron Transfer Pathway of Thermincola ferriacetica. Microorganisms, 9(2), 293. https://doi.org/10.3390/microorganisms9020293






