Assessment of Marine Gill Disease in Farmed Atlantic Salmon (Salmo salar) in Chile Using a Novel Total Gross Gill Scoring System: A Case Study
Abstract
:1. Introduction
2. Materials and Methods
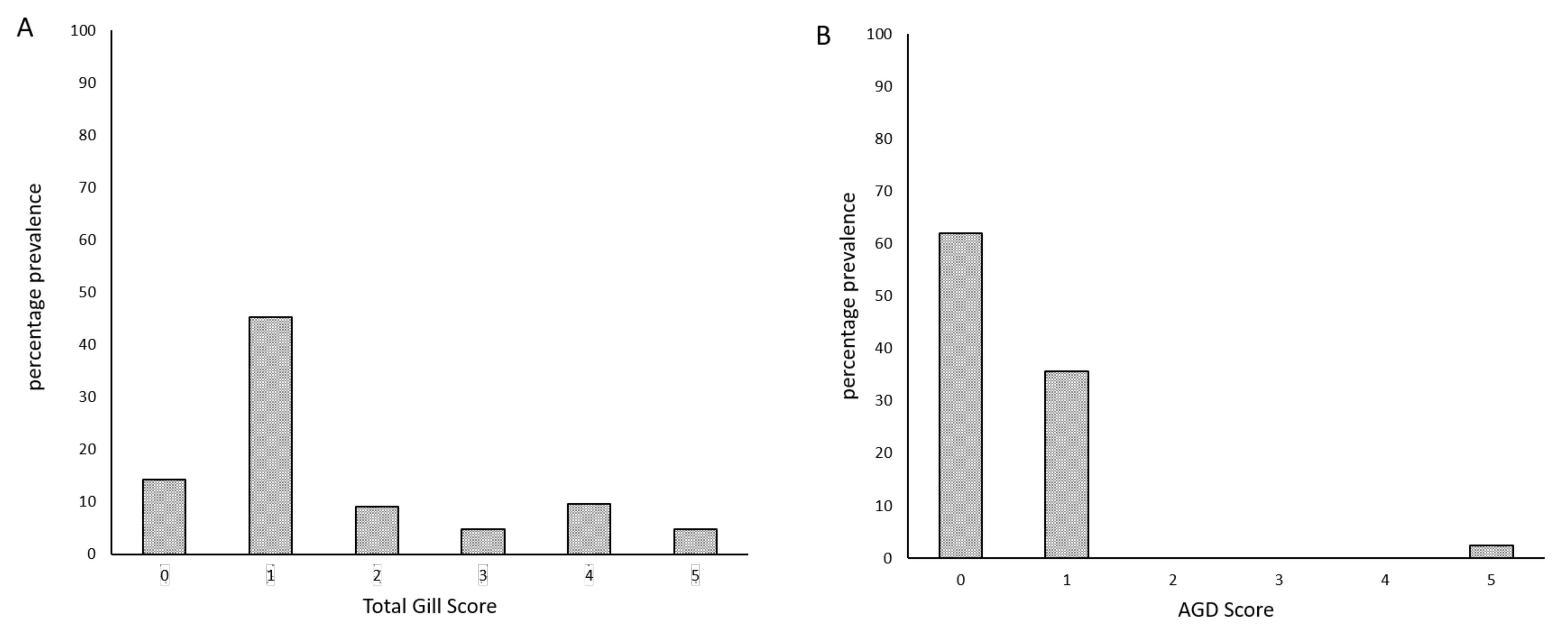
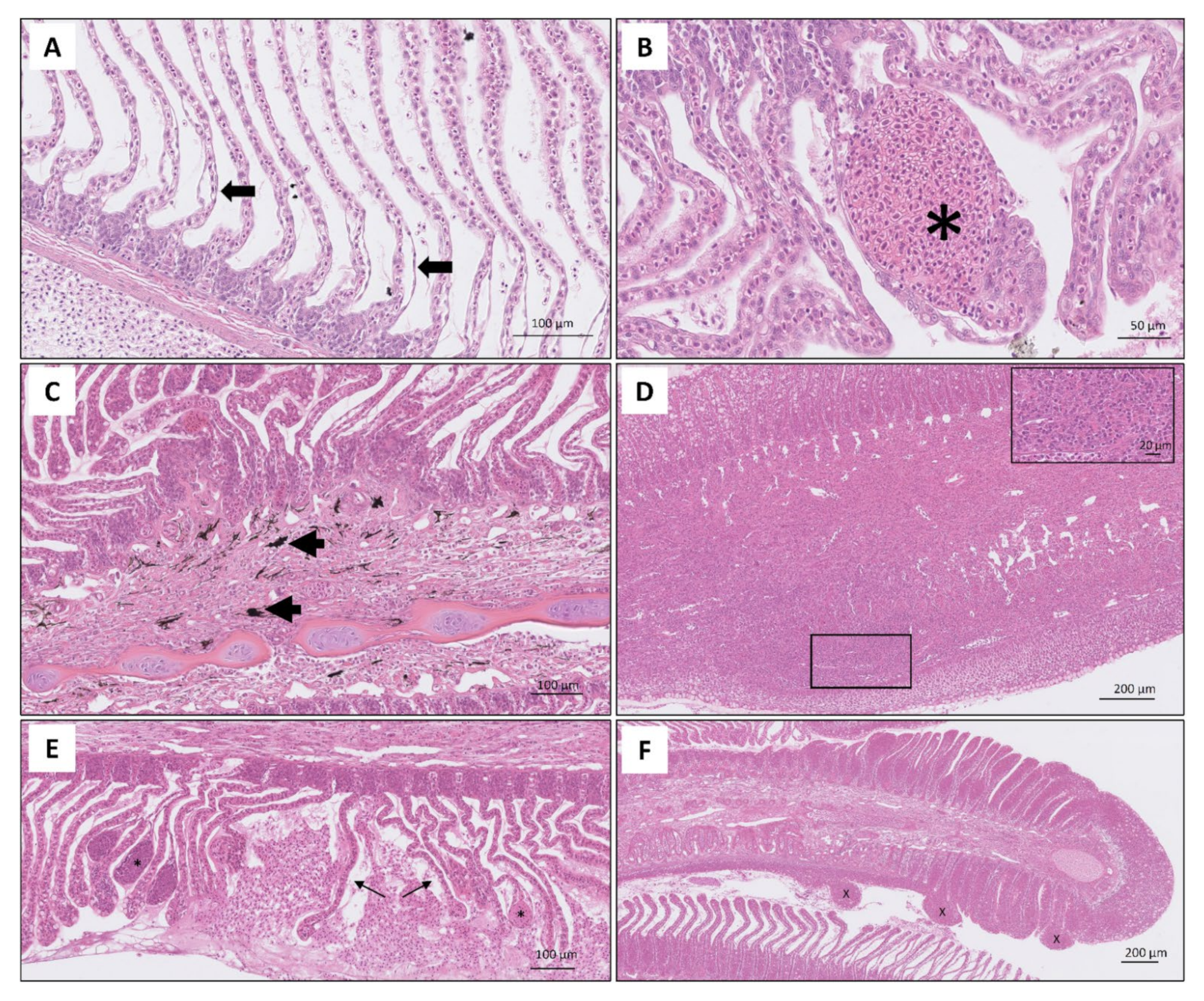
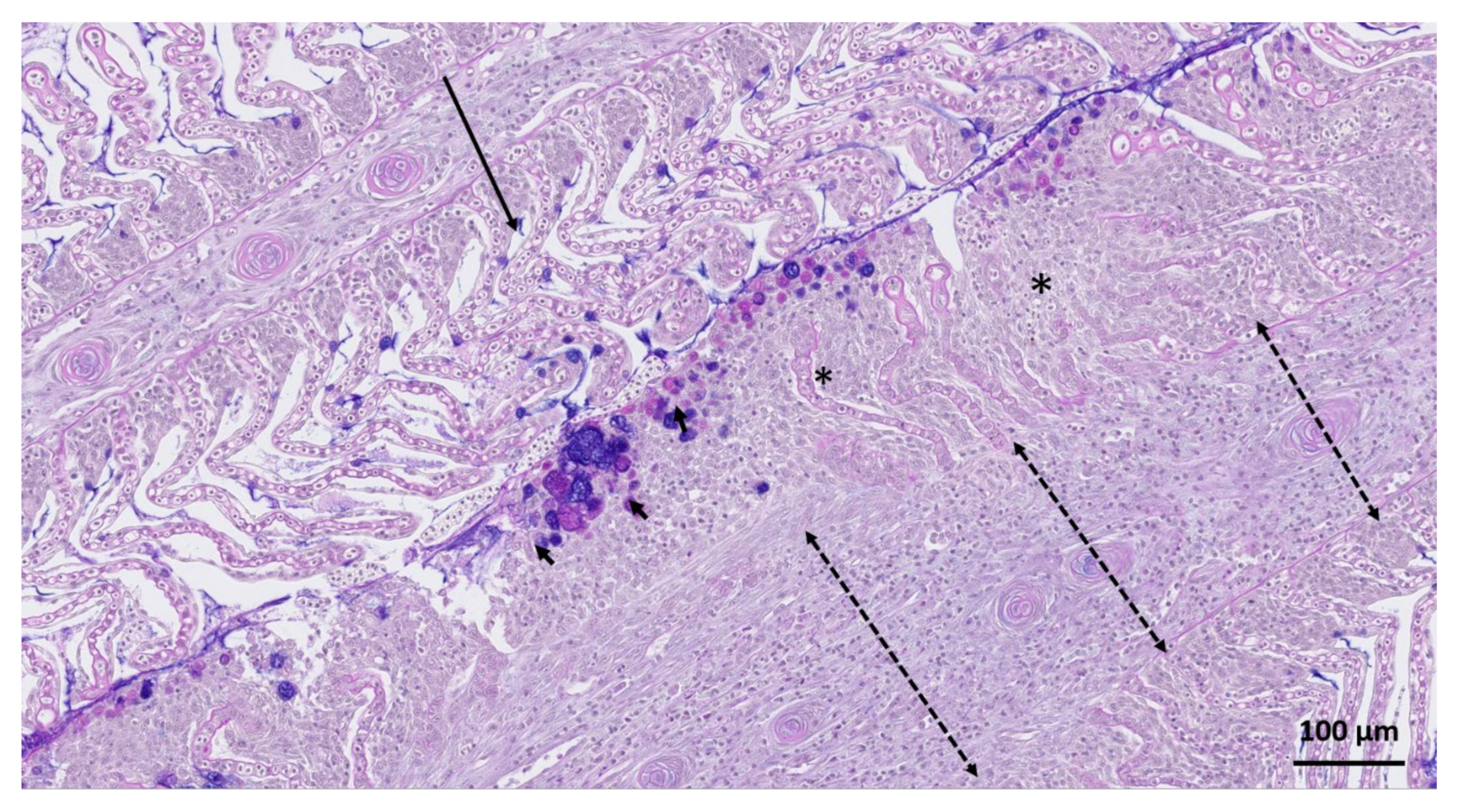
3. Results
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitchell, S.O.; Rodger, H.D. A review of infectious gill disease in marine salmonid fish. J. Fish Dis. 2011, 34, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.; Thompson, K.D.; Ashby, A.; Rodger, H.D.; Dagleish, M.P. Complex gill disease: An emerging syndrome in farmed atlantic salmon (Salmo salar L.). J. Comp. Pathol. 2018, 163, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Rozas-Serri, M.; Bohle, H.; Ildefonso, R.; Bustos, P. Development of pcr assay for detection of neoparamoeba perurans and comparison of histological diagnosis. Bull. Eur. Assoc. Fish Pathol. 2011, 31, 211–218. [Google Scholar]
- Munday, B.L.; Lange, K.; Foster, C.; Lester, R.; Handlinger, J. Amoebic gill disease of sea-caged salmonids in tasmanian waters. Tasman. Fish Res. 1993, 28, 14–19. [Google Scholar]
- Clark, A.; Nowak, B.F. Field investigations of amoebic gill disease in atlantic salmon, Salmo salar L., in tasmania. J. Fish Dis. 1999, 22, 433–443. [Google Scholar] [CrossRef]
- Taylor, R.S.; Muller, W.J.; Cook, M.T.; Kube, P.D.; Elliott, N.G. Gill observations in atlantic salmon (Salmo salar L.) during repeated amoebic gill disease (agd) field exposure and survival challenge. Aquaculture 2009, 290, 1–8. [Google Scholar] [CrossRef]
- Persson, D.; Kolstø, S.; Pedersen, T.M. The use of a double gill scoring system for field evaluation of gill health in atlantic salmon (Salmo salar L.). In Proceedings of the EAFP Conference, Las Palmas, Spain, 7–11 September 2015. [Google Scholar]
- Haugland, G.T.; Olsen, A.-B.; Rønneseth, A.; Andersen, L. Lumpfish (Cyclopterus lumpus L.) develop amoebic gill disease (agd) after experimental challenge with paramoeba perurans and can transfer amoebae to atlantic salmon (Salmo salar L.). Aquaculture 2017, 478, 48–55. [Google Scholar] [CrossRef]
- Król, E.; Noguera, P.; Shaw, S.; Costelloe, E.; Gajardo, K.; Valdenegro, V.; Bickerdike, R.; Douglas, A.; Martin, S.A.M. Integration of transcriptome, gross morphology and histopathology in the gill of sea farmed atlantic salmon (Salmo salar): Lessons from multi-site sampling. Front. Genet. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.B.; Ellard, K.; Nowak, B.F. Gross pathology and its relationship with histopathology of amoebic gill disease (agd) in farmed atlantic salmon, Salmo salar L. J. Fish Dis. 2004, 27, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.; Huynh, C.; Cameron, D.; Evans, B.; Cook, M.; Ritchie, G. Gill Score Guide: Amoebic Gill Disease (Agd) Management; Training Document; Tassal Ltd, Marine Harvest, CSIRO and Contributors: Hobart, TS, Australia, 2016. [Google Scholar]
- Yáñez, J.M.; Houston, R.D.; Newman, S. Genetics and genomics of disease resistance in salmonid species. Front. Genet. 2014, 5, 415. [Google Scholar]
- Chalmers, L.; Taylor, J.F.; Roy, W.; Preston, A.C.; Migaud, H.; Adams, A. A comparison of disease susceptibility and innate immune response between diploid and triploid atlantic salmon (Salmo salar) siblings following experimental infection with neoparamoeba perurans, causative agent of amoebic gill disease. Parasitology 2017, 144, 1229–1242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fringuelli, E.; Gordon, A.W.; Rodger, H.; Welsh, M.D.; Graham, D.A. Detection of neoparamoeba perurans by duplex quantitative taqman real-time pcr in formalin-fixed, paraffin-embedded atlantic salmonid gill tissues. J. Fish Dis. 2012, 35, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Steinum, T.; Kvellestad, A.; Colquhoun, D.J.; Heum, M.; Mohammad, S.; Grøntvedt, R.N.; Falk, K. Microbial and pathological findings in farmed atlantic salmon Salmo salar with proliferative gill inflammation. Dis. Aquat. Organ. 2010, 91, 201–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, S.O.; Steinum, T.M.; Toenshoff, E.R.; Kvellestad, A.; Falk, K.; Horn, M.; Colquhoun, D.J. ‘Candidatus Branchiomonas cysticola’ is a common agent of epitheliocysts in seawater-farmed atlantic salmon Salmo salar in norway and ireland. Dis. Aquat. Organ. 2013, 103, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Gjessing, M.C.; Yutin, N.; Tengs, T.; Senkevich, T.; Koonin, E.; Rønning, H.P.; Alarcon, M.; Ylving, S.; Lie, K.I.; Saure, B.; et al. Salmon gill poxvirus, the deepest representative of the chordopoxvirinae. J. Virol. 2015, 89, 9348–9367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nylund, S.; Nylund, A.; Watanabe, K.; Arnesen, C.E.; Karlsbakk, E. Paranucleospora theridion n. Gen., n. Sp. (microsporidia, enterocytozoonidae) with a life cycle in the salmon louse (Lepeophtheirus salmonis, copepoda) and atlantic salmon (Salmo salar). J. Eukaryot. Microbiol. 2010, 57, 95–114. [Google Scholar] [CrossRef] [PubMed]
- Isaksen, T.E.; Karlsbakk, E.; Repstad, O.; Nylund, A. Molecular tools for the detection and identification of ichthyobodo spp. (kinetoplastida), important fish parasites. Parasitol. Int. 2012, 61, 675–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Álvarez, C.; González, S.F.; Santos, Y. Quantitative pcr coupled with melting curve analysis for rapid detection and quantification of tenacibaculum maritimum in fish and environmental samples. Aquaculture 2019, 498, 289–296. [Google Scholar] [CrossRef]
- Gutierrez Rabadan, C.; Spreadbury, C.; Consuegra, S.; Garcia de Leaniz, C. Development, validation and testing of an operational welfare score index for farmed lumpfish Cyclopterus lumpus L. Aquaculture 2021, 531, 735777. [Google Scholar] [CrossRef]
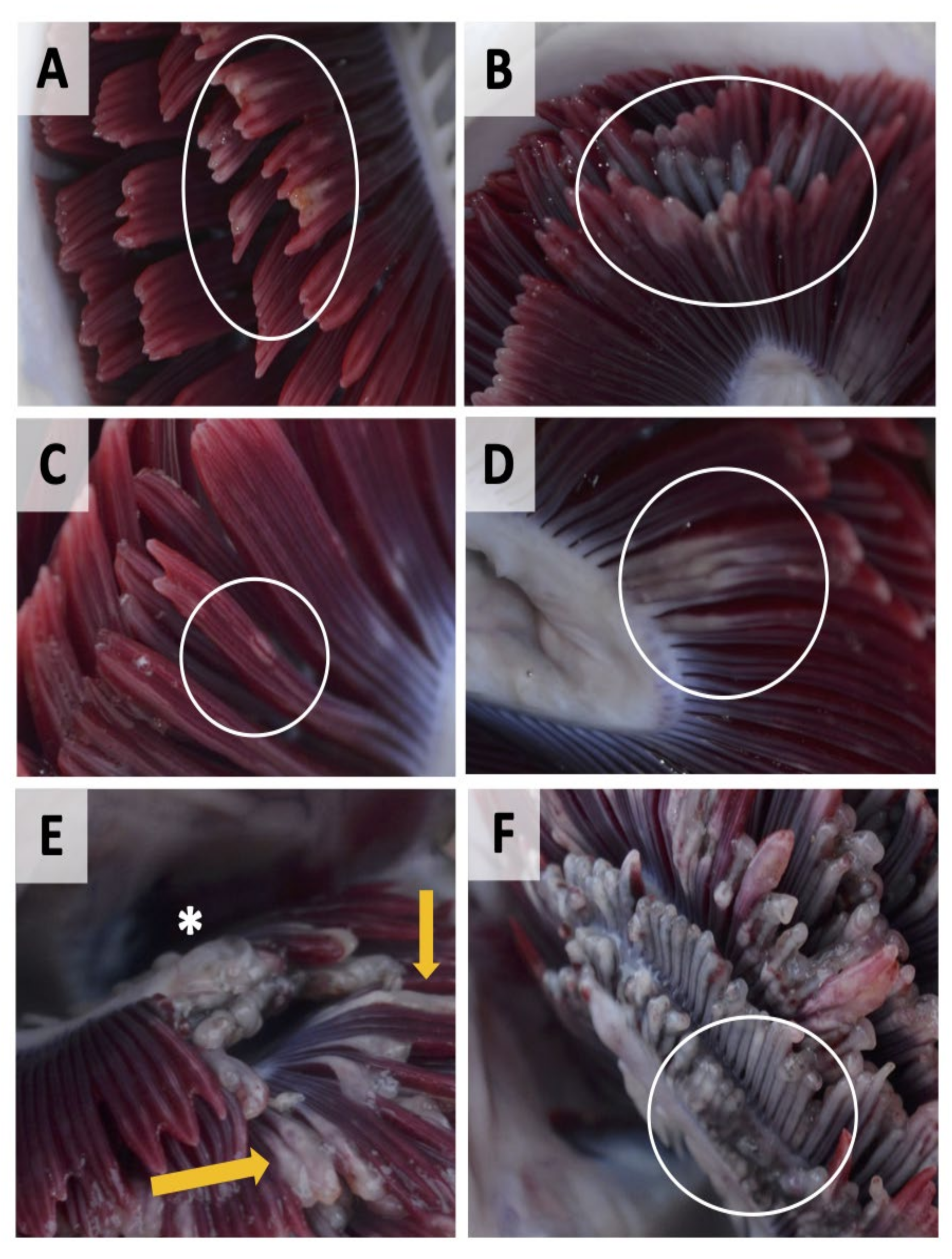
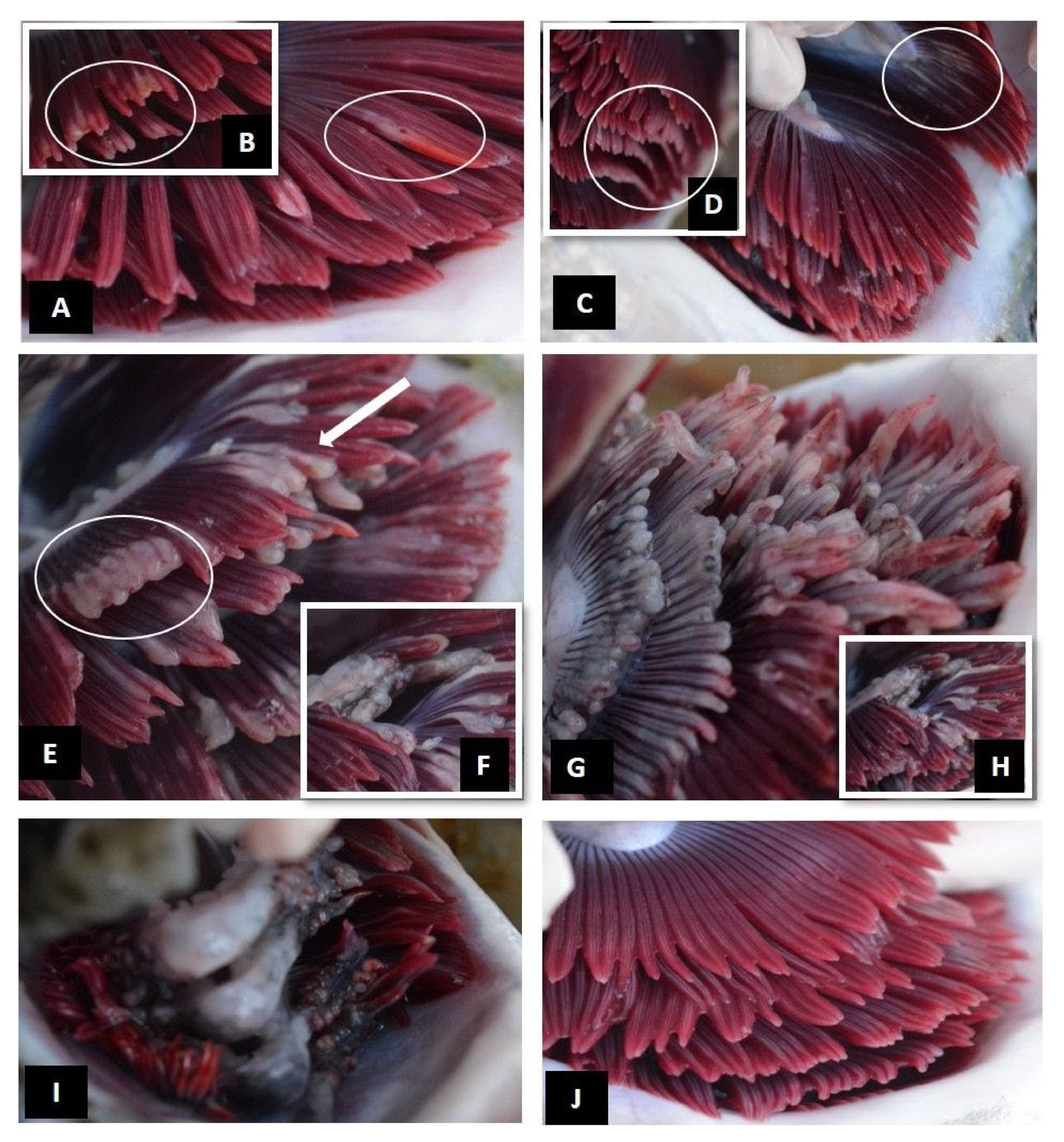
| Level of Infection | Total Gill Score | Description | Mean % of Gill Surface Covered |
|---|---|---|---|
| Clear | 0 | No visible pathology, healthy red coloured gills | 0 |
| Very light | 1 | Discrete focal white streaks or patches on individual filaments and slight erosion/damage to distal ends of filaments | 1–5% |
| Light | 2 | More extensive coalescing white streaks or white focal patches on filaments, more extended erosion/damage to distal ends of filaments | 5–20% |
| Moderate | 3 | Extensive multifilamental peripheral erosion, grossly swollen or thickened filaments with localised areas of necrotic epithelium | 20–50% |
| Advanced | 4 | Extensive grossly swollen or thickened filaments, shortened filaments (>50% of filament length affected), pallor and areas of melanisation | 50–75% |
| Severe | 5 | Widespread necrotic patches, extensive melanisation, almost total destruction of gill architecture due to severe loss of epithelium | >75% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fridman, S.; Tsairidou, S.; Jayasuriya, N.; Sobolewska, H.; Hamilton, A.; Lobos, C.; Houston, R.D.; Rodger, H.; Bron, J.; Herath, T. Assessment of Marine Gill Disease in Farmed Atlantic Salmon (Salmo salar) in Chile Using a Novel Total Gross Gill Scoring System: A Case Study. Microorganisms 2021, 9, 2605. https://doi.org/10.3390/microorganisms9122605
Fridman S, Tsairidou S, Jayasuriya N, Sobolewska H, Hamilton A, Lobos C, Houston RD, Rodger H, Bron J, Herath T. Assessment of Marine Gill Disease in Farmed Atlantic Salmon (Salmo salar) in Chile Using a Novel Total Gross Gill Scoring System: A Case Study. Microorganisms. 2021; 9(12):2605. https://doi.org/10.3390/microorganisms9122605
Chicago/Turabian StyleFridman, Sophie, Smaragda Tsairidou, Nilantha Jayasuriya, Halina Sobolewska, Alastair Hamilton, Carlos Lobos, Ross D. Houston, Hamish Rodger, James Bron, and Tharangani Herath. 2021. "Assessment of Marine Gill Disease in Farmed Atlantic Salmon (Salmo salar) in Chile Using a Novel Total Gross Gill Scoring System: A Case Study" Microorganisms 9, no. 12: 2605. https://doi.org/10.3390/microorganisms9122605
APA StyleFridman, S., Tsairidou, S., Jayasuriya, N., Sobolewska, H., Hamilton, A., Lobos, C., Houston, R. D., Rodger, H., Bron, J., & Herath, T. (2021). Assessment of Marine Gill Disease in Farmed Atlantic Salmon (Salmo salar) in Chile Using a Novel Total Gross Gill Scoring System: A Case Study. Microorganisms, 9(12), 2605. https://doi.org/10.3390/microorganisms9122605







