Multicenter Technical Validation of 30 Rapid Antigen Tests for the Detection of SARS-CoV-2 (VALIDATE)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Samples
2.3. Validation Protocol
2.4. Technical Validation
2.5. Limits of Detection
2.6. Reference Method
2.7. Performance Acceptance Criteria
2.8. Statistical Analysis
2.9. Ethical Declaration
3. Results
3.1. Sample Characteristics
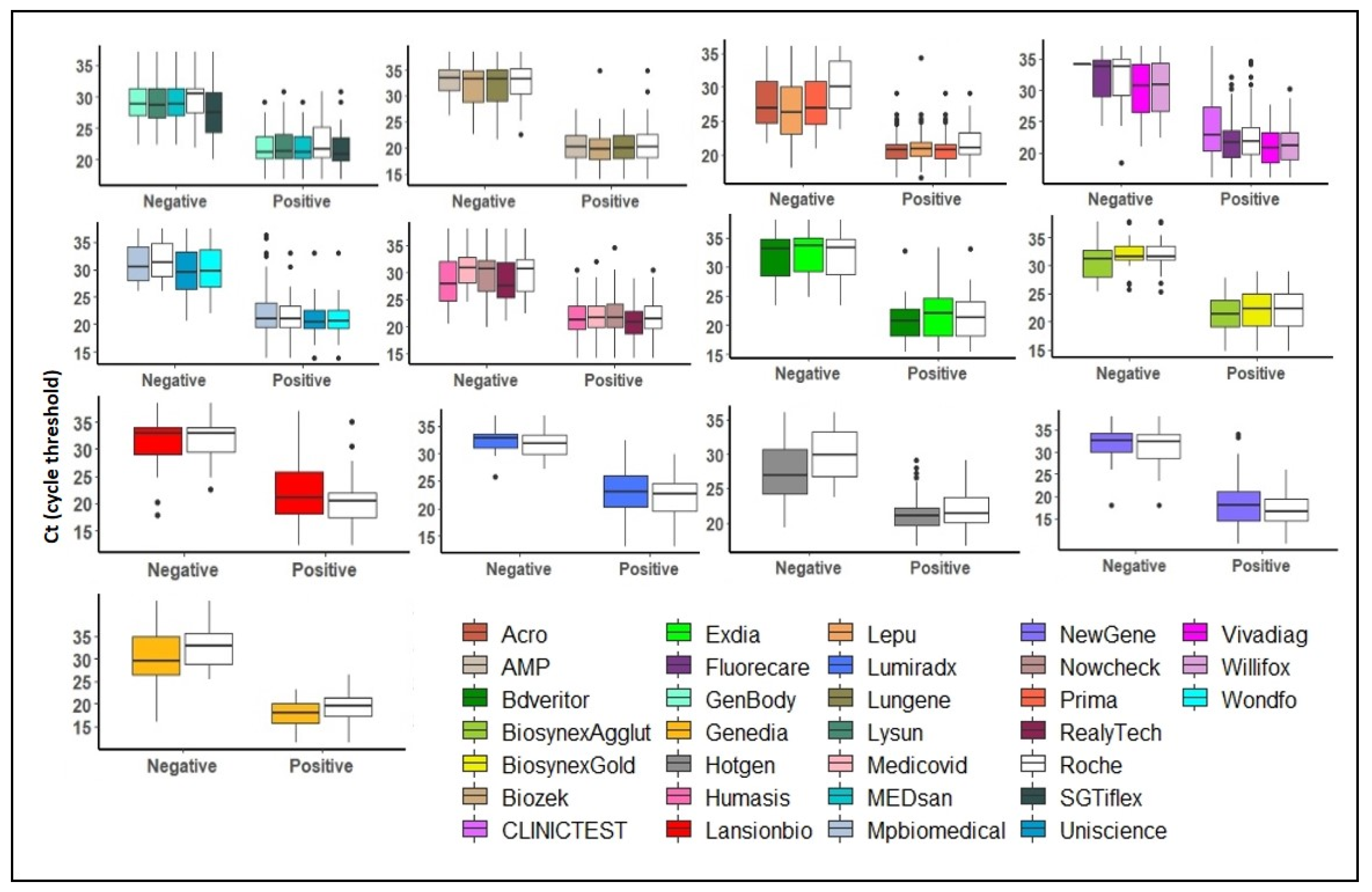
3.2. Sensitivity of RATs
3.3. Specificity of RATs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Rolling Updates on Coronavirus Disease (COVID-19). Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen (accessed on 25 January 2021).
- WHO. Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays. Available online: https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays (accessed on 16 July 2021).
- FIND. SARS-CoV-2 Diagnostic Pipeline. Available online: https://www.finddx.org/covid-19/pipeline/?avance=all&type=Rapid+diagnostic+tests&test_target=Antigen&status=all§ion=show-all&action=default (accessed on 25 January 2021).
- Caruana, G.; Croxatto, A.; Kampouri, E.; Kritikos, A.; Opota, O.; Foerster, M.; Brouillet, R.; Senn, L.; Lienhard, R.; Egli, A.; et al. Implementing SARS-CoV-2 Rapid Antigen Testing in the Emergency Ward of a Swiss University Hospital: The INCREASE Study. Microorganisms 2021, 9, 798. [Google Scholar] [CrossRef] [PubMed]
- Mouliou, D.S.; Gourgoulianis, K.I. False-positive and false-negative COVID-19 cases: Respiratory prevention and management strategies, vaccination, and further perspectives. Expert Rev. Respir. Med. 2021, 15, 1–10. [Google Scholar] [CrossRef]
- Beckmann, C.; Hirsch, H.H. Diagnostic performance of near-patient testing for influenza. J. Clin. Virol. 2015, 67, 43–46. [Google Scholar] [CrossRef]
- Green, D.A.; StGeorge, K. Rapid Antigen Tests for Influenza: Rationale and Significance of the FDA Reclassification. J. Clin. Microbiol. 2018, 56, e00711-18. [Google Scholar] [CrossRef] [Green Version]
- Jacot, D.; Greub, G.; Jaton, K.; Opota, O. Viral load of SARS-CoV-2 across patients and compared to other respiratory viruses. Microbes Infect. 2020, 22, 617–621. [Google Scholar] [CrossRef]
- Albert, E.; Torres, I.; Bueno, F.; Huntley, D.; Molla, E.; Fernandez-Fuentes, M.A.; Martinez, M.; Poujois, S.; Forque, L.; Valdivia, A.; et al. Field evaluation of a rapid antigen test (Panbio COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin. Microbiol. Infect. 2021, 27, 472.e7–472.e10. [Google Scholar] [CrossRef]
- Berger, A.; Ngo Nsoga, M.T.; Perez-Rodriguez, F.J.; Aad, Y.A.; Sattonnet-Roche, P.; Gayet-Ageron, A.; Jaksic, C.; Torriani, G.; Boehm, E.; Kronig, I.; et al. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS ONE 2021, 16, e0248921. [Google Scholar] [CrossRef]
- Cerutti, F.; Burdino, E.; Milia, M.G.; Allice, T.; Gregori, G.; Bruzzone, B.; Ghisetti, V. Urgent need of rapid tests for SARS CoV-2 antigen detection: Evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J. Clin. Virol. 2020, 132, 104654. [Google Scholar] [CrossRef]
- Corman, V.M.; Haage, V.C.; Bleicker, T.; Schmidt, M.L.; Mühlemann, B.; Zuchowski, M.; Jó Lei, W.K.; Tscheak, P.; Möncke-Buchner, E.; Müller, M.A.; et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: A single-centre laboratory evaluation study. Lancet Microbe 2021, 2, e311–e319. [Google Scholar] [CrossRef]
- Diao, B.; Wen, K.; Zhang, J.; Chen, J.; Han, C.; Chen, Y.; Wang, S.; Deng, G.; Zhou, H.; Wu, Y. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin. Microbiol. Infect. 2021, 27, 289.e1–289.e4. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020, 8, Cd013705. [Google Scholar] [CrossRef] [PubMed]
- James, A.E.; Gulley, T.; Kothari, A.; Holder, K.; Garner, K.; Patil, N. Performance of the BinaxNOW coronavirus disease 2019 (COVID-19) Antigen Card test relative to the severe acute respiratory coronavirus virus 2 (SARS-CoV-2) real-time reverse transcriptase polymerase chain reaction (rRT-PCR) assay among symptomatic and asymptomatic healthcare employees. Infect. Control Hosp. Epidemiol. 2021, 20, 1–3. [Google Scholar] [CrossRef]
- Kyosei, Y.; Namba, M.; Yamura, S.; Takeuchi, R.; Aoki, N.; Nakaishi, K.; Watabe, S.; Ito, E. Proposal of De Novo Antigen Test for COVID-19: Ultrasensitive Detection of Spike Proteins of SARS-CoV-2. Diagnostics 2020, 10, 594. [Google Scholar] [CrossRef]
- Mockel, M.; Corman, V.M.; Stegemann, M.S.; Hofmann, J.; Stein, A.; Jones, T.C.; Gastmeier, P.; Seybold, J.; Offermann, R.; Bachmann, U.; et al. SARS-CoV-2 antigen rapid immunoassay for diagnosis of COVID-19 in the emergency department. Biomarkers 2021, 26, 213–220. [Google Scholar] [CrossRef]
- Schwob, J.-M.; Miauton, A.; Petrovic, D.; Perdrix, J.; Senn, N.; Jaton, K.; Opota, O.; Maillard, A.; Minghelli, G.; Cornuz, J.; et al. Antigen rapid tests, nasopharyngeal PCR and saliva PCR to detect SARS-CoV-2: A prospective comparative clinical trial. medRxiv 2020, 1–16. [Google Scholar] [CrossRef]
- Scohy, A.; Anantharajah, A.; Bodeus, M.; Kabamba-Mukadi, B.; Verroken, A.; Rodriguez-Villalobos, H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020, 129, 104455. [Google Scholar] [CrossRef]
- Egli, A.; Lienhard, R.; Jaton, K.; Greub, G. Recommendation of the Swiss Society of Microbiology for usage of SARS-CoV-2 specific antigen tests. Pipette—Swiss Lab. Med. 2020, 6, 18–20. [Google Scholar]
- Opota, O.; Brouillet, R.; Greub, G.; Jaton, K. Comparison of SARS-CoV-2 RT-PCR on a high-throughput molecular diagnostic platform and the cobas SARS-CoV-2 test for the diagnostic of COVID-19 on various clinical samples. Pathog. Dis. 2020, 78, ftaa061. [Google Scholar] [CrossRef] [PubMed]
- Pillonel, T.; Scherz, V.; Jaton, K.; Greub, G.; Bertelli, C. Letter to the editor: SARS-CoV-2 detection by real-time RT-PCR. Eurosurveillance 2020, 25, 2000880. [Google Scholar] [CrossRef]
- Greub, G.; Sahli, R.; Brouillet, R.; Jaton, K. Ten years of R&D and full automation in molecular diagnosis. Future Microbiol. 2016, 11, 403–425. [Google Scholar] [CrossRef] [Green Version]
- Council, T.S.F. Ordinance 3 on Measures to Combat the Coronavirus (COVID-19) (COVID-19 Ordinance 3). In 818.101.24, 2020; p. 46. Available online: https://www.fedlex.admin.ch/eli/cc/2020/438/en (accessed on 1 December 2021).
- Puyskens, A.; Krause, E.; Michel, J.; Nübling, M.; Scheiblauer, H.; Bourquain, D.; Grossegesse, M.; Valusenko, R.; Corman, V.; Drosten, C.; et al. Establishment of an evaluation panel for the decentralized technical evaluation of the sensitivity of 31 rapid detection tests for SARS-CoV-2 diagnostics. medRxiv 2021, 1–29. [Google Scholar] [CrossRef]
- Scheiblauer, H.; Filomena, A.; Nitsche, A.; Puyskens, A.; Corman, V.M.; Drosten, C.; Zwirglmaier, K.; Lange, C.; Emmerich, P.; Müller, M.; et al. Comparative sensitivity evaluation for 122 CE-marked rapid diagnostic tests for SARS-CoV-2 antigen, Germany, September 2020 to April 2021. Eurosurveillance 2021, 26, 2100441. [Google Scholar] [CrossRef] [PubMed]
- Kritikos, A.; Caruana, G.; Brouillet, R.; Miroz, J.P.; Abed-Maillard, S.; Stieger, G.; Opota, O.; Croxatto, A.; Vollenweider, P.; Bart, P.A.; et al. Sensitivity of Rapid Antigen Testing and RT-PCR Performed on Nasopharyngeal Swabs versus Saliva Samples in COVID-19 Hospitalized Patients: Results of a Prospective Comparative Trial (RESTART). Microorganisms 2021, 9, 1910. [Google Scholar] [CrossRef] [PubMed]
- Caruana, G.; Lebrun, L.-L.; Aebischer, O.; Opota, O.; Urbano, L.; de Rham, M.; Marchetti, O.; Greub, G. The dark side of SARS-CoV-2 rapid antigen testing: Screening asymptomatic patients. New Microbes New Infect. 2021, 42, 100899. [Google Scholar] [CrossRef]
- Perchetti, G.A.; Huang, M.-L.; Peddu, V.; Jerome, K.R.; Greninger, A.L.; McAdam, A.J. Stability of SARS-CoV-2 in phosphate-buffered saline for molecular detection. J. Clin. Microbiol. 2020, 58, e01094-20. [Google Scholar] [CrossRef]

| Manufacturer | Antigen Assay | Sensitivity at Ct ≤ 29 | Sensitivity at Ct ≤ 26 | Sensitivity at Ct ≤ 23 | Specificity |
|---|---|---|---|---|---|
| Ref. Value: ≥80% (CI) * | Ref. Value: ≥90% (CI) | Ref. Value: ≥95% (CI) | Ref. Value: ≥99% | ||
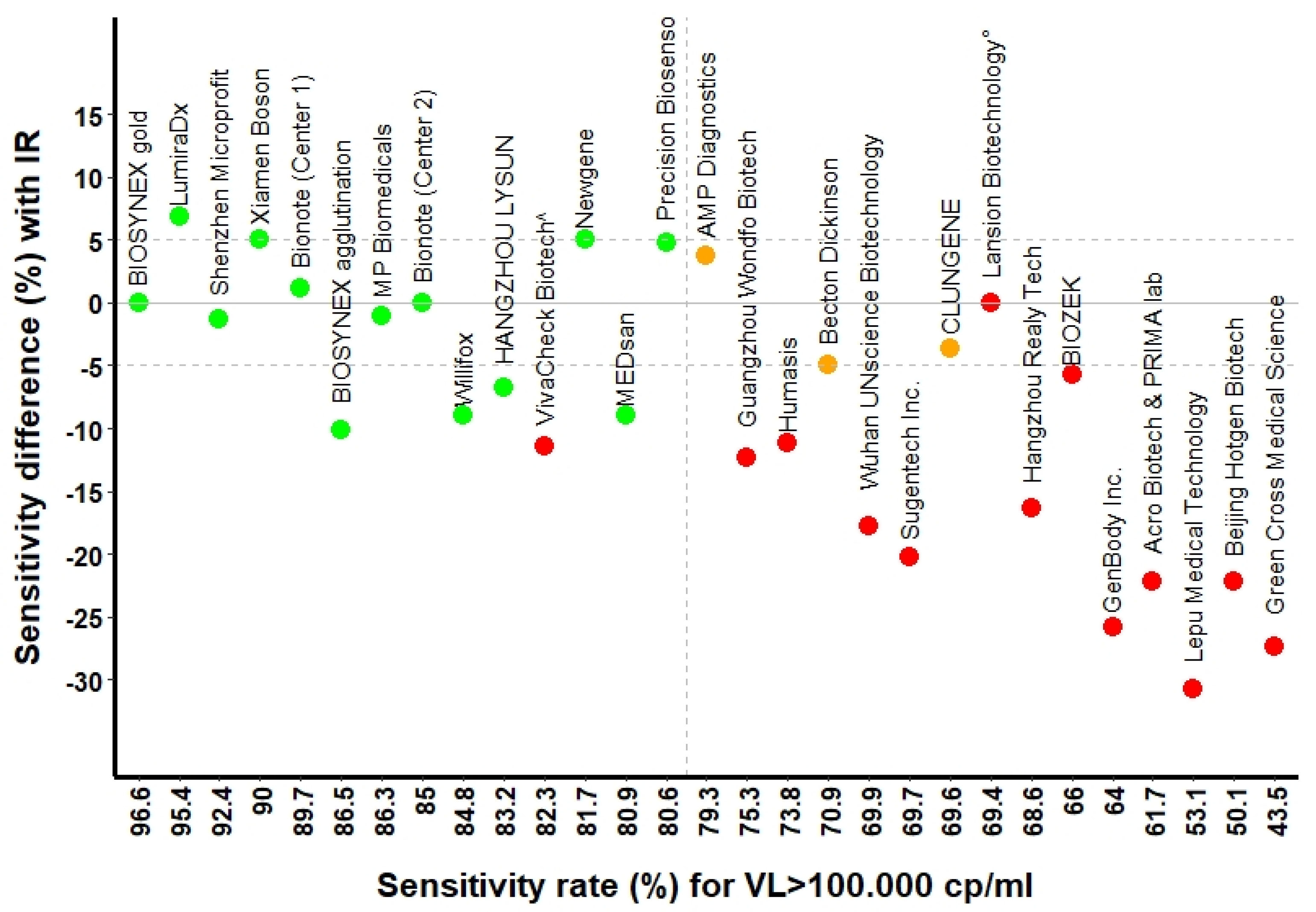
| BIOSYNEX Swiss | Biosnyex COVID-19 Ag + BSS ° | 86.5% (77.6, 92.8) | 98.6% (92.4, 100) | 100% (92.8, 100) | 99.5% |
| BIOSYNEX Swiss | Biosynex COVID-19 Ag BSS ^ | 96.6% (90.5, 99.3) | 98.6% (92.4, 100) | 100% (92.8, 100) | 100% |
| Newgene—Hangzhou-Bioengineering | COVID-19 Antigen Detection Kit | 81.7% (69.6, 90.5) | 94.1% (83.8, 98.8) | 97.8% (88.2, 99.9) | 99% |
| CLUNGENE | COVID-19 Rapid Antigen Test Cassette | 69.6% (55.9, 81.2) | 92.7% (80.1, 98.5) | 94.3% (80.8, 99.3) | 100% |
| Precision Biosensor, Inc. | Exdia COVID-19 Ag | 80.6 (68.6, 89.6) | 97.9% (88.7, 100) | 100% (89.7, 100) | 99.5% |
| Shenzhen Microprofit Biotech Co. | Fluorecare® SARS-CoV-2 Spike Protein Test Kit | 92.4% (84.2, 97.2) | 95.8% (88.0, 99.1) | 100% (93.0, 100) | 100% |
| LumiraDx, Alloa | LumiraDx SARS-CoV-2 Ag | 98.6% (92.6, 100) | 98.3% (91.1, 100) | 100% (91.2, 100) | 99% |
| HANGZHOU LYSUN Biotechnology | LYSUN SARS-CoV-2 Antigen | 83.2% (73.7, 90.3) | 92.1% (83.6, 97.1) | 97.9% (88.9, 100.0) | 100% |
| Xiamen Boson Biotech Co., Ltd. | Medicovid-AG® Test | 90% (81.2, 95.6) | 97.1% (89. 8, 99.6) | 100% (92.8, 100) | 99.3% |
| MEDsan GmbH | MEDsan® SARS-CoV-2 Antigen Rapid Test | 80.9% (71.2, 88.5) | 92.1% (83.6, 97.1) | 97.9% (88.9, 100) | 100% |
| MP Biomedicals GmbH | MP COVID-19 Antigen Rapid Test | 86.3% (76.3, 93.2) | 100% (93.7, 100) | 100% (92.1, 100) | 100% |
| Bionote | NowCheck® COVID-19 Ag Test | 85% (75.3, 92.0) | 92.7% (83. 7, 97.6) | 95.9% (86.0, 99.5) | 99% |
| AMP Diagnostics | Test rapide AMP SARS-CoV-2 Ag | 79.3 (65.9, 89.2) | 100% (90.97, 100) | 100% (90.0, 100) | 100% |
| Becton Dickinson | The BD Veritor™ System | 70.9% (58.1, 81.8) | 93.6% (82.5, 98.7) | 100% (89.7, 100) | 99.7% |
| Willifox | Willi Fox COVID-19 Antigen Test® | 84.8% (75.0, 91.9) | 90% (80.5, 95.9) | 96.1% (86.5, 99.5) | 100% |
| Manufacturer | Antigen Assay | Sensitivity at Ct ≤ 29 | Sensitivity at Ct ≤ 26 | Sensitivity at Ct ≤ 23 | Specificity |
|---|---|---|---|---|---|
| Ref. Value: ≥80% (CI) * | Ref. Value: ≥90% (CI) | Ref. Value: ≥95% (CI) | Ref. Value: ≥99% | ||
| Acro Biotech | Acro COVID-19 Antigen test | 61.7% (50.3, 72.3) | 72.5% (60.4, 82.5) | 88.2% (76.1, 95.6) | 100% |
| Healgen Scientific Limited Liability Company | CLINITEST®, Rapid COVID-19 Antigen Test + | N/A | N/A | N/A | 0% |
| PRIMA Lab SA | COVID-19 Antigen Rapid Test | 61.7% (50.3, 72.3) | 72.5% (60.4, 82.5) | 88.2% (76.1, 95.6) | 100% |
| BIOZEK | COVID-19 Antigen Rapid Test Cassette | 66.0% (51.7, 78.5) | 92.1% (78.6, 98.3) | 96.9% (84.2, 99.9) | 98.7% |
| GenBody Inc. | GenBody COVID-19 Ag | 64.0% (53.2, 74.0) | 72.4% (60.9, 82.0) | 79.2% (65.0, 89.5) | 100% |
| Green Cross Medical Science Corp. | Genedia W COVID-19 Ag § | 43.5% (30.9, 56.7) | 62.8% (46.7, 77.0) | 66.7% (49.8, 80.9) | 100% |
| Humasis | Humasis COVID-19 Ag Test | 73.8% (62.7, 83.0) | 80.9% (69.5, 89.4) | 85.7% (72.8, 94.1) | 99.3% |
| Lansion Biotechnology Co. | Lansionbio® COVID-19 Antigen Test Kit—Dry Fluorescence Immunoassay | 69.4% (54.6, 81.8) | 88.2% (72.6, 96.7) | 93.3% (77.9, 99.2) | 88.8% |
| Beijing Hotgen Biotech Co. | Novel Coronavirus 2019-nCoV Antigen Test—colloidal Gold | 60.5% (49.0, 71.2) | 68.7% (56.2, 79.4) | 85.1% (71.7, 93.8) | 100% |
| Hangzhou Realy Tech Co. | REALY antigen test | 68.8% (57.4, 78.7) | 77.9% (66.2, 87.1) | 89.8% (77.8, 96.6) | 100% |
| Lepu Medical Zechnology Co. | SARS-CoV-2 Antigen Rapid Test—Colloidal Gold | 53.1% (41.7, 64.3) | 62.3% (49.8, 73.7) | 72.5% (58.3, 84.1) | 100% |
| Wuhan UNscience Biotechnology Co. | SARS-CoV-2 Antigen Rapid Test Kit | 69.9% (58.0, 80.1) | 84.2% (72.1, 92.5) | 88.9% (76.0, 96.3) | 100% |
| Sugentech Inc. | SGTi-flex COVID-19 Ag | 69.7% (59.0, 79.0) | 80.3% (69.5, 88.5) | 91.7% (80.0, 97.7) | 100% |
| VivaCheck Biotech (Hangzhou) Co. | VivaDiag™, SARS-CoV-2 Rapid Ag Test | 82.3% (72.1, 90.0) | 88.6% (78.7, 94.9) | 92.2% (81.1, 97.8) | 100% |
| Guangzhou Wondfo Biotech Co. | Wondfo SARS-CoV-2 Antigen Test | 75.3% (63.9, 84.7) | 92.9% (83.0, 98.1) | 97.8% (88.2, 99.9) | 100% |
| Validation Batch | Sensitivity at Ct ≤ 29 | Sensitivity at Ct ≤ 26 | Sensitivity at Ct ≤ 23 | Specificity |
|---|---|---|---|---|
| Serie 1 | 89.9% (93, 98.3) | 93.4% (94.9, 99.3) | 97.9% (97.2, 99.9) | 100% |
| Serie 2 | 83.9% (74.1, 91.2) | 91.3% (82, 96.7) | 100% (93, 100) | 100% |
| Serie 3 | 82.7% (72.7, 90.2) | 89.5% (79.7, 95.7) | 100% (92.5, 100) | 100% |
| Serie 4 | 93.7% (85.8, 97.9) | 97.1% (90.1, 99.7) | 98% (89.6, 100) | 100% |
| Serie 5 | 87.7% (77.9, 94.2) | 100% (93.7, 100) | 100% (92.1, 100) | 100% |
| Serie 6 | 85% (75.3, 92.0) | 94.1% (85.6, 98.4) | 97.9% (89.2, 100) | 99.5% |
| Serie 7 | 93.2% (84.7, 97.7) | 100% (94.1, 100) | 100% (91.2, 100) | 99% |
| Serie 8 | 96.6% (90.5, 99.3) | 98.6% (92.4, 100) | 100% (92.7, 100) | 99.0% |
| Serie 9 | 75.8% (63.3, 85.8) | 95.7% (85.5, 99.5) | 95.7% (85.5, 99.5) | 99.7% |
| Serie 10 | 73.2% (59.7, 84.2) | 95.1% (83.5, 99.4) | 97.1% (85.1, 99.9) | 100% |
| Serie 11 | 69.4% (54.6, 81.8) | 94.1% (80.3, 99.3) | 96.7% (82.8, 99.9) | 100% |
| Serie 12 | 76.7% (64.0, 86.6) | 90.2% (78.6, 96.7) | 97.8% (88.2, 99.9) | 99.5% |
| Serie 13 | 70.9% (58.1. 81.8) | 97.7% (87.7, 99.9) | 100% (90.9, 100) | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greub, G.; Caruana, G.; Schweitzer, M.; Imperiali, M.; Muigg, V.; Risch, M.; Croxatto, A.; Opota, O.; Heller, S.; Albertos Torres, D.; et al. Multicenter Technical Validation of 30 Rapid Antigen Tests for the Detection of SARS-CoV-2 (VALIDATE). Microorganisms 2021, 9, 2589. https://doi.org/10.3390/microorganisms9122589
Greub G, Caruana G, Schweitzer M, Imperiali M, Muigg V, Risch M, Croxatto A, Opota O, Heller S, Albertos Torres D, et al. Multicenter Technical Validation of 30 Rapid Antigen Tests for the Detection of SARS-CoV-2 (VALIDATE). Microorganisms. 2021; 9(12):2589. https://doi.org/10.3390/microorganisms9122589
Chicago/Turabian StyleGreub, Gilbert, Giorgia Caruana, Michael Schweitzer, Mauro Imperiali, Veronika Muigg, Martin Risch, Antony Croxatto, Onya Opota, Stefanie Heller, Diana Albertos Torres, and et al. 2021. "Multicenter Technical Validation of 30 Rapid Antigen Tests for the Detection of SARS-CoV-2 (VALIDATE)" Microorganisms 9, no. 12: 2589. https://doi.org/10.3390/microorganisms9122589
APA StyleGreub, G., Caruana, G., Schweitzer, M., Imperiali, M., Muigg, V., Risch, M., Croxatto, A., Opota, O., Heller, S., Albertos Torres, D., Tritten, M.-L., Leuzinger, K., Hirsch, H. H., Lienhard, R., & Egli, A. (2021). Multicenter Technical Validation of 30 Rapid Antigen Tests for the Detection of SARS-CoV-2 (VALIDATE). Microorganisms, 9(12), 2589. https://doi.org/10.3390/microorganisms9122589







