Isolation and Identification of Wild Yeast from Malaysian Grapevine and Evaluation of Their Potential Antimicrobial Activity against Grapevine Fungal Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Epiphytic Yeast Strain Isolation and Taxonomic Classification
2.2. Pathogen Mould Strains and Growth Conditions
2.3. Antifungal In-Vitro Assays
2.4. Yeast Volatile Organic Compound (VOC) Profiles
2.5. Detached Berry Antifungal Assay
2.6. Statistical Analysis
3. Results
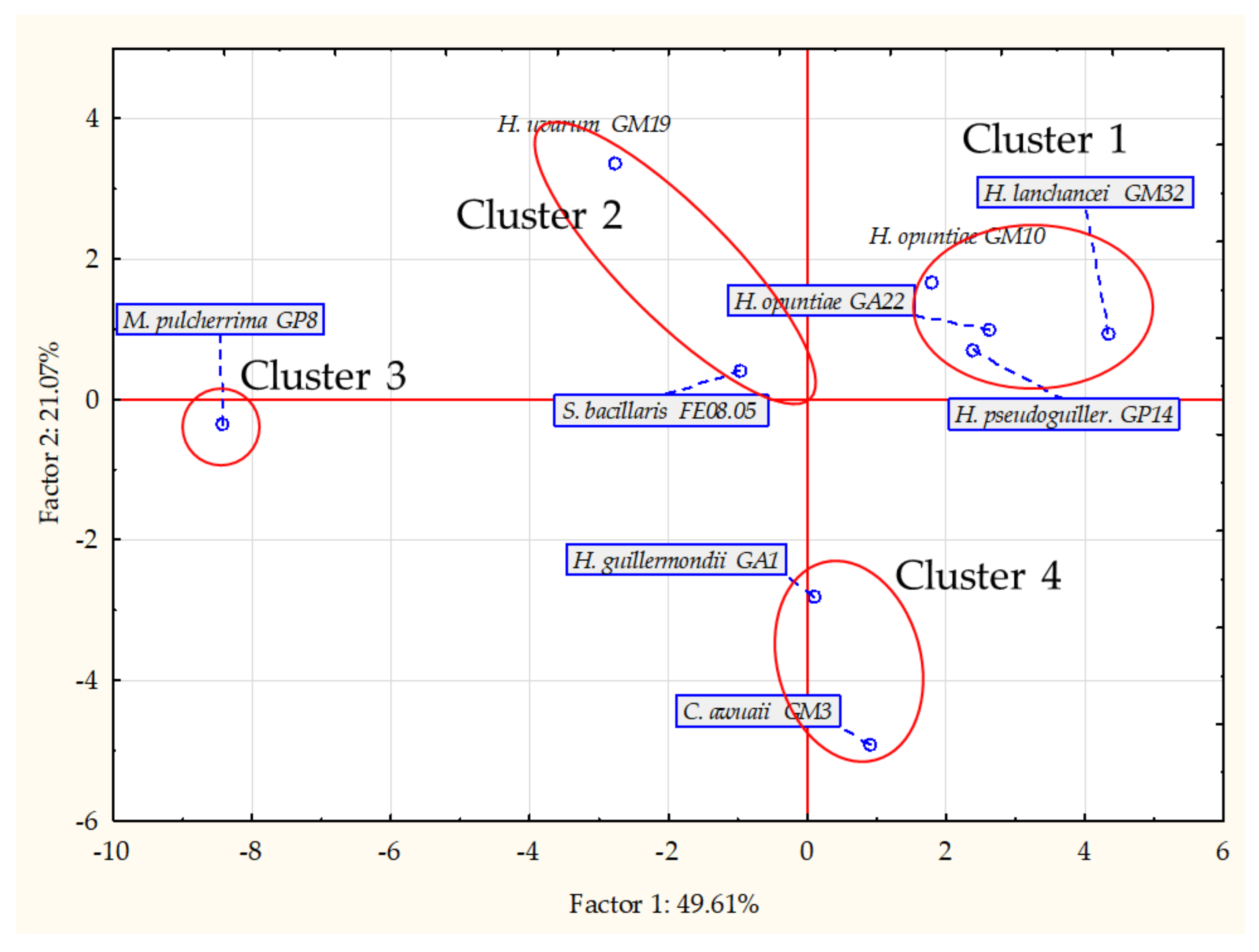
3.1. Molecular Analysis of Yeast Isolates
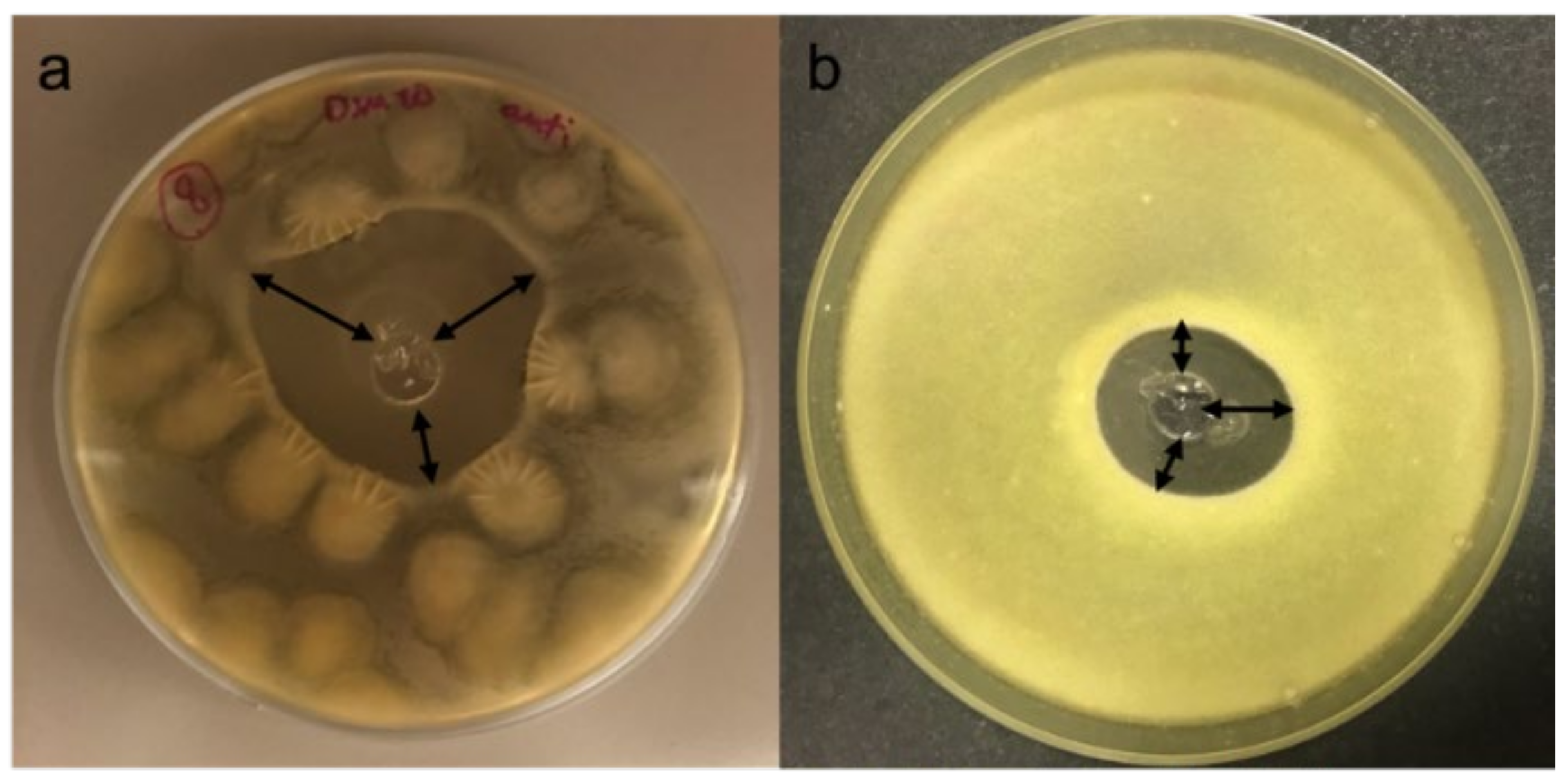
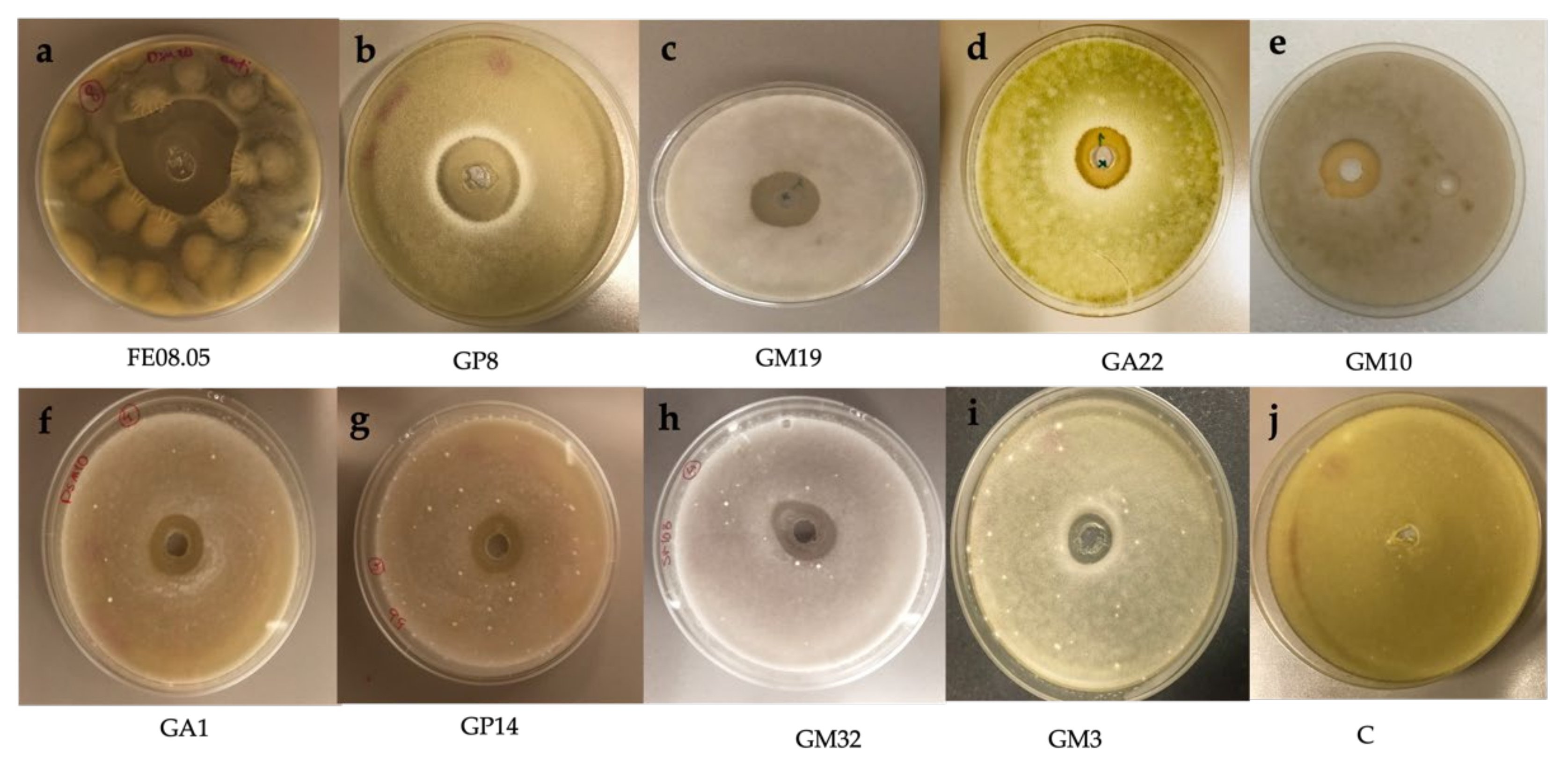
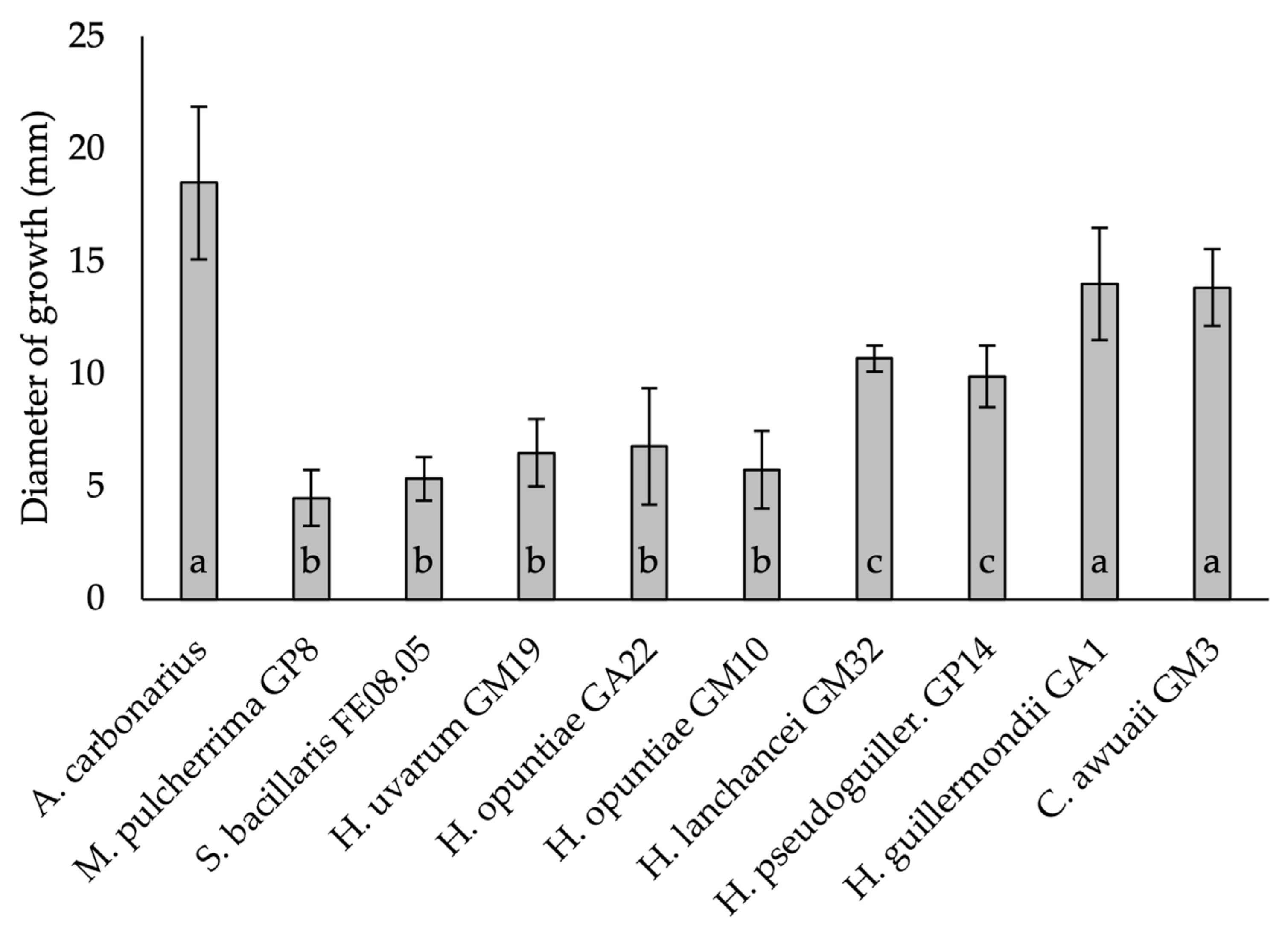
3.2. In-Vitro Antifungal Assays
3.3. Volatile Organic Compound Profiles
3.4. Detached Berry Antifungal Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Alleweldt, G.; Possingham, J.V. Progress in grapevine breeding. Theor. Appl. Genet. 1988, 75, 669–673. [Google Scholar] [CrossRef]
- Gobilik, J.; Enggihon, J. Some aspects on growth, yield, phenology and grape quality of ‘isabella’ grapevine (vitis × labruscana) planted in sandakan, sabah as ornamental plant. Trans. Sci. Technol. 2019, 6, 235–252. [Google Scholar]
- Di Canito, A.; Mateo-Vargas, M.A.; Mazzieri, M.; Cantoral, J.; Foschino, R.; Cordero-Bueso, G.; Vigentini, I. The role of yeasts as biocontrol agents for pathogenic fungi on postharvest grapes: A review. Foods 2021, 10, 1650. [Google Scholar] [CrossRef] [PubMed]
- Rosslenbroich, H.-J.; Stuebler, D. Botrytis cinerea—history of chemical control and novel fungicides for its management. Crop Prot. 2000, 19, 557–561. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Unravelling the diversity of grapevine microbiome. PLoS ONE 2014, 9, e85622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durel, L.; Estrada-Peña, A.; Franc, M.; Mehlhorn, H.; Bouyer, J. Integrated fly management in European ruminant operations from the perspective of directive 2009/128/EC on sustainable use of pesticides. Parasitol. Res. 2015, 114, 379–389. [Google Scholar] [CrossRef]
- Pinto, C.; Gomes, A.C. Vitis vinifera microbiome: From basic research to technological development. BioControl 2016, 61, 243–256. [Google Scholar] [CrossRef]
- Maachia, B.S.; Rafik, E.; Chérif, M.; Nandal, P.; Mohapatra, T.; Bernard, P. Biological Control of the Grapevine Diseases Grey Mold and Powdery Mildew by Bacillus B27 and B29 Strains. 2015. Available online: http://nopr.niscair.res.in/handle/123456789/30442 (accessed on 1 February 2015).
- Pertot, I.; Caffi, T.; Rossi, V.; Mugnai, L.; Hoffmann, C.; Grando, M.; Gary, C.; Lafond, D.; Duso, C.; Thiery, D. A critical review of plant protection tools for reducing pesticide use on grapevine and new perspectives for the implementation of IPM in viticulture. Crop Prot. 2017, 97, 70–84. [Google Scholar] [CrossRef]
- Martins, G.; Lauga, B.; Miot-Sertier, C.; Mercier, A.; Lonvaud, A.; Soulas, M.-L.; Soulas, G.; Masneuf-Pomarède, I. Characterization of epiphytic bacterial communities from grapes, leaves, bark and soil of grapevine plants grown, and their relations. PLoS ONE 2013, 8, e73013. [Google Scholar] [CrossRef] [Green Version]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef]
- Patrignani, F.; Montanari, C.; Serrazanetti, D.I.; Braschi, G.; Vernocchi, P.; Tabanelli, G.; Parpinello, G.P.; Versari, A.; Gardini, F.; Lanciotti, R. Characterisation of yeast microbiota, chemical and sensory properties of organic and biodynamic Sangiovese red wines. Ann. Microbiol. 2017, 67, 99–109. [Google Scholar] [CrossRef]
- Morgan, H.H.; Du Toit, M.; Setati, M.E. The grapevine and wine microbiome: Insights from high-throughput amplicon sequencing. Front. Microbiol. 2017, 8, 820. [Google Scholar] [CrossRef] [Green Version]
- Vivier, M.; Pretorius, I.S. Genetic improvement of grapevine: Tailoring grape varieties for the third millennium. S. Afr. J. Enol. Vitic. 2000, 21, 5–26. [Google Scholar] [CrossRef] [Green Version]
- Ribes, S.; Fuentes, A.; Talens, P.; Barat, J.M. Prevention of fungal spoilage in food products using natural compounds: A review. Crit. Rev. Food Sci. Nutr. 2018, 58, 2002–2016. [Google Scholar] [CrossRef]
- Spadaro, D.; Droby, S. Development of biocontrol products for postharvest diseases of fruit: The importance of elucidating the mechanisms of action of yeast antagonists. Trends Food Sci. Technol. 2016, 47, 39–49. [Google Scholar] [CrossRef]
- Suzzi, G.; Romano, P.; Ponti, I.; Montuschi, C. Natural wine yeasts as biocontrol agents. J. Appl. Bacteriol. 1995, 78, 304–308. [Google Scholar] [CrossRef]
- Freimoser, F.M.; Rueda-Mejia, M.P.; Tilocca, B.; Migheli, Q. Biocontrol yeasts: Mechanisms and applications. World J. Microbiol. Biotechnol. 2019, 35, 154. [Google Scholar] [CrossRef] [Green Version]
- Parafati, L.; Vitale, A.; Restuccia, C.; Cirvilleri, G. Biocontrol ability and action mechanism of food-isolated yeast strains against Botrytis cinerea causing post-harvest bunch rot of table grape. Food Microbiol. 2015, 47, 85–92. [Google Scholar] [CrossRef]
- Cao, X.-H.; Zhao, S.-S.; Liu, D.-Y.; Wang, Z.; Niu, L.-L.; Hou, L.-H.; Wang, C.-L. ROS-Ca2+ is associated with mitochondria permeability transition pore involved in surfactin-induced MCF-7 cells apoptosis. Chem. Biol. Interact. 2011, 190, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Zanzotto, A.; Morroni, M. Major biocontrol studies and measures against fungal and oomycete pathogens of grapevine. In Biocontrol of Major Grapevine Diseases: Leading Research; CABI: Oxford, UK, 2016; pp. 1–34. [Google Scholar]
- Dukare, A.S.; Paul, S.; Nambi, V.E.; Gupta, R.K.; Singh, R.; Sharma, K.; Vishwakarma, R.K. Exploitation of microbial antagonists for the control of postharvest diseases of fruits: A review. Crit. Rev. Food Sci. Nutr. 2019, 59, 1498–1513. [Google Scholar] [CrossRef]
- Pearson, W.R.; Lipman, D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 1988, 85, 2444–2448. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Steel, C.C.; Blackman, J.W.; Schmidtke, L.M. Grapevine bunch rots: Impacts on wine composition, quality, and potential procedures for the removal of wine faults. J. Agric. Food Chem. 2013, 61, 5189–5206. [Google Scholar] [CrossRef] [PubMed]
- Alfonzo, A.; Conigliaro, G.; Torta, L.; Burruano, S.; Moschetti, G. Antagonism of Bacillus subtilis strain AG1 against vine wood fungal pathogens. Phytopathol. Mediterr. 2009, 48, 155–158. [Google Scholar]
- Pantelides, I.S.; Christou, O.; Tsolakidou, M.-D.; Tsaltas, D.; Ioannou, N. Isolation, identification and in vitro screening of grapevine yeasts for the control of black aspergilli on grapes. Biol. Control 2015, 88, 46–53. [Google Scholar] [CrossRef]
- Bleve, G.; Grieco, F.; Cozzi, G.; Logrieco, A.; Visconti, A. Isolation of epiphytic yeasts with potential for biocontrol of Aspergillus carbonarius and Aspergillus niger on grape. Int. J. Food Microbiol. 2006, 108, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Solomon, G.M.; Dodangoda, H.; McCarthy-Walker, T.; Ntim-Gyakari, R.; Newell, P.D. The microbiota of Drosophila suzukii influences the larval development of Drosophila melanogaster. PeerJ 2019, 7, e8097. [Google Scholar] [CrossRef] [Green Version]
- Setati, M.E.; Jacobson, D.; Andong, U.-C.; Bauer, F.F. The vineyard yeast microbiome, a mixed model microbial map. PLoS ONE 2012, 7, e52609, Correction in 2013, 8, doi:10.1371/annotation/b9d307d9-f5c1-4e0d-8945-c5a747b6f58e. [Google Scholar] [CrossRef] [Green Version]
- Nadai, C.; Lemos, W.J.F.; Favaron, F.; Giacomini, A.; Corich, V. Biocontrol activity of Starmerella bacillaris yeast against blue mold disease on apple fruit and its effect on cider fermentation. PLoS ONE 2018, 13, e0204350. [Google Scholar] [CrossRef] [PubMed]
- Junior, W.J.F.L.; Bovo, B.; Nadai, C.; Crosato, G.; Carlot, M.; Favaron, F.; Giacomini, A.; Corich, V. Biocontrol ability and action mechanism of Starmerella bacillaris (Synonym Candida zemplinina) isolated from wine musts against gray mold disease agent botrytis cinerea on grape and their effects on alcoholic fermentation. Front. Microbiol. 2016, 7, 1249, Corrigendum in 2016, 7, 1499. [Google Scholar] [CrossRef]
- Prendes, L.P.; Merín, M.G.; Fontana, A.R.; Bottini, R.A.; Ramirez, M.L.; de Ambrosini, V.I.M. Isolation, identification and selection of antagonistic yeast against Alternaria alternata infection and tenuazonic acid production in wine grapes from Argentina. Int. J. Food Microbiol. 2018, 266, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Guinebretière, M.H.; Nguyen-The, C.; Morrison, N.; Reich, M.; Nicot, P. Isolation and characterization of antagonists for the biocontrol of the postharvest wound pathogen Botrytis cinerea on strawberry fruits. J. Food Prot. 2000, 63, 386–394. [Google Scholar] [CrossRef]
- Stocco, A.F.; Diaz, M.E.; Romera, M.C.R.; Mercado, L.A.; Rivero, M.L.; Ponsone, M.L. Biocontrol of postharvest Alternaria decay in table grapes from Mendoza province. Biol. Control 2019, 134, 114–122. [Google Scholar] [CrossRef]
- Ferreira-Saab, M.; Formey, D.; Torres, M.; Aragón, W.; Padilla, E.A.; Tromas, A.; Sohlenkamp, C.; Schwan-Estrada, K.R.F.; Serrano, M. Compounds released by the biocontrol yeast Hanseniaspora opuntiae protect plants against Corynespora cassiicola and Botrytis cinerea. Front. Microbiol. 2018, 9, 1596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Apaliya, M.T.; Mahunu, G.K.; Chen, L.; Li, W. Control of ochratoxin A-producing fungi in grape berry by microbial antagonists: A review. Trends Food Sci. Technol. 2016, 51, 88–97. [Google Scholar] [CrossRef]
- Türkel, S.; Korukluoğlu, M.; Yavuz, M. Biocontrol activity of the local strain of Metschnikowia pulcherrima on different postharvest pathogens. Biotechnol. Res. Int. 2014, 2014, 397167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, S.S.T.; Beck, J.J.; Sarreal, S.B.L.; Gee, W. The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 2014, 30, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Oro, L.; Feliziani, E.; Ciani, M.; Romanazzi, G.; Comitini, F. Volatile organic compounds from Wickerhamomyces anomalus, Metschnikowia pulcherrima and Saccharomyces cerevisiae inhibit growth of decay causing fungi and control postharvest diseases of strawberries. Int. J. Food Microbiol. 2018, 265, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Hatanaka, K.; Ohata, I.; Yamashita-Kitaguchi, Y.; Kurata, A.; Kishimoto, N. Antifungal activities of volatile substances generated by yeast isolated from Iranian commercial cheese. Food Control 2012, 26, 472–478. [Google Scholar] [CrossRef]
- Pohl, C.H.; Kock, J.L.; Thibane, V.S. Antifungal free fatty acids: A review. Sci. Against Microb. Pathog. Commun. Curr. Res. Technol. Adv. 2011, 3, 61–71. [Google Scholar]
- Cordero-Bueso, G.; Mangieri, N.; Maghradze, D.; Foschino, R.; Valdetara, F.; Cantoral, J.M.; Vigentini, I. Wild grape-associated yeasts as promising biocontrol agents against vitis vinifera fungal pathogens. Front. Microbiol. 2017, 8, 2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
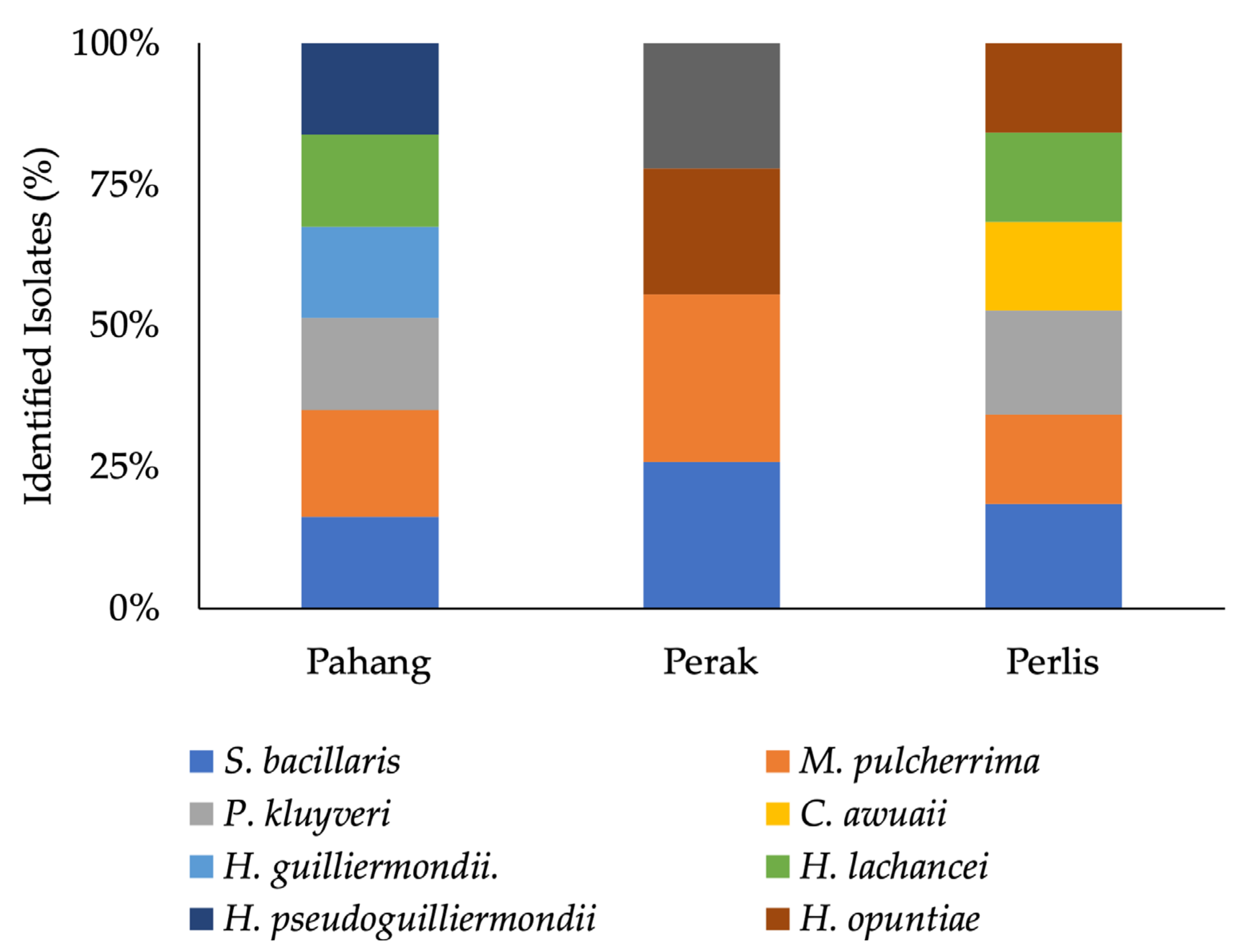
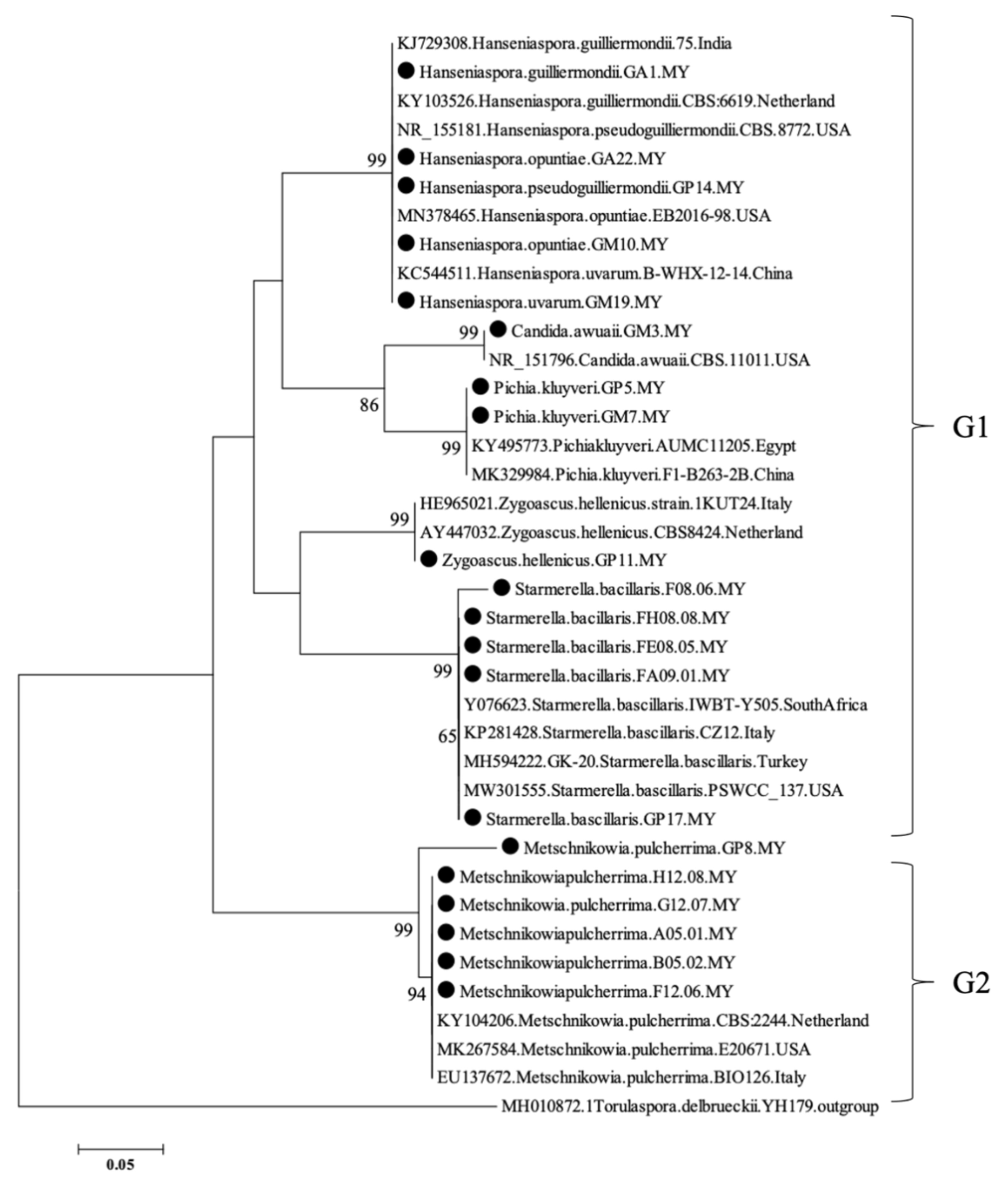

| Isolate | Species Designation | Accession Number/Country/Region | Identity (%) | E-Value |
|---|---|---|---|---|
| F08.06 | Starmerella bacillaris | OK329946/Malaysia/Perak | 99.28 | 0.00 |
| FH08.08 | Starmerella bacillaris | OK329947/Malaysia/Pahang | 99.28 | 0.00 |
| FA09.01 | Starmerella bacillaris | OK329948/Malaysia/Perak | 98.8 | 0.00 |
| FE08.05 | Starmerella bacillaris | OK329949/Malaysia/Pahang | 99.04 | 0.00 |
| GP17 | Starmerella bascillaris | OK329950/Malaysia/Perlis | 99.75 | 0.00 |
| PSWCC_137 | Starmerella bascillaris | MW301555/USA | Ref-isolate | - |
| H12.08 | Metschnikowia pulcherrima | OK329951/Malaysia/Perak | 97.93 | 2 × 10−162 |
| A05.01 | Metschnikowia pulcherrima | OK329952/Malaysia/Perak | 97.93 | 2 × 10−162 |
| B05.02 | Metschnikowia pulcherrima | OK329953/Malaysia/Perak | 97.12 | 2 × 10−162 |
| F12.06 | Metschnikowia pulcherrima | OK329954/Malaysia/Pahang | 97.12 | 2 × 10−162 |
| G12.07 | Metschnikowia pulcherrima | OK329955/Malaysia/Perlis | 97.93 | 2 × 10−162 |
| GP8 | Metschnikowia pulcherrima | OK560819/Malaysia/Perlis | 96.43 | 0.00 |
| E20671 | Metschnikowia pulcherrima | MK267584/USA | Ref-isolate | - |
| GM3 | Candida awuaii | OK329958/Malaysia/Pahang | 87.27 | 1 × 10−94 |
| CBS.11011 | Candida awuaii | NR_151796/USA | Ref-isolate | - |
| GA1 | Hanseniaspora guilliermondii. | OK329959/Malaysia/Perlis | 99.57 | 0.00 |
| CBS:6619 | Hanseniaspora guilliermondii. | KY103526/Netherland | Ref-isolate | - |
| GM10 | Hanseniaspora opuntiae | OK329962/Malaysia/Perak | 100 | 0.00 |
| GA22 | Hanseniaspora opuntiae | OK329963/Malaysia/Pahang | 99.85 | 0.00 |
| EB2016-98 | Hanseniaspora opuntiae | MN378465/USA | Ref-isolate | - |
| GP14 | Hanseniaspora pseudoguilliermondii | OK329961/Malaysia/Perlis | 99.41 | 0.00 |
| CBS.8772 | Hanseniaspora pseudoguilliermondii | NR_155181/USA | Ref-isolate | - |
| GM19 | Hanseniaspora uvarum | OK329964/Malaysia/Pahang | 100 | 0.00 |
| B-WHX | Hanseniaspora uvarum | KC544511/China | Ref-isolate | - |
| GP5 | Pichia kluyveri | OK329956/Malaysia/Perlis | 98.46 | 0.00 |
| GM7 | Pichia kluyveri | OK329957/Malaysia/Pahang | 99.22 | 0.00 |
| F1-B263-2B | Pichi kluyveri | MK329984/China | Ref-isolate | - |
| GP11 | Zygoascus hellenicus | OK329960/Malaysia/Perlis | 99.81 | 0.00 |
| 1KUT24 | Zygoascus hellenicus | HE965021/Italy | Ref-isolate | - |
| Isolate | Designated Species | B. cinerea Antifungal Activity |
|---|---|---|
| FE08.05 | Starmerella bacillaris | + |
| GP8 | Metschnikowia pulcherrima | + |
| GM19 | Hanseniaspora uvarum | + |
| GA22 | Hanseniaspora opuntiae | + |
| GM10 | Hanseniaspora opuntiae | + |
| GA1 | Hanseniaspora guilliermondii | + |
| GP14 | Hanseniaspora pseudoguilliermondii | + |
| GM32 | Hanseniasporalanchancei | + |
| GM3 | Candida awuaii | + |
| Inhibition Ring (mm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| S. bacillaris FE08.05 | M. pulcherrimaGP8 | H. uvarum GM19 | H. opuntiae GA22 | H. opuntiae GM10 | H. guilliermondii GA1 | H. lachancei GM32 | H. pseudoguillier. GP14 | C. awuaii GM3 | |
| B. cinerea | 15.1 a ± 0.4 | 12.4 b ± 0.5 | 10.8 c ± 0.3 | 8.1 d ± 0.3 | 5.8 e ± 0.3 | 3.0 g ± 0.2 | 2.0 h ± 0.2 | 4.4 g ± 0.5 | 6.3 e ± 0.3 |
| A. carbonarius | 14.2 a ± 0.3 | 10.2 b ± 0.3 | 8.2 c ± 0.3 | 6.2 d ± 0.3 | 3.2 e ± 0.3 | 2.2 f ± 0.3 | <1 * | 2.1 f ± 0.1 | 1.2 g ± 0.3 |
| A. ochraceus | 12.2 a ± 0.3 | 8.2 b ± 0.3 | 3.2 e ± 0.3 | 5.9 c ± 0.1 | 3.2 d ± 0.3 | 2.2 f ± 0.3 | <1 | 2.1 f ± 0.1 | 1.2 g ± 0.3 |
| A. alternata | 15.8 a ± 0.2 | 6.3 b ± 0.5 | 8.2 c ± 0.3 | 5.2 d ± 0.3 | 2.1 e ± 0.1 | 1.1 f ± 0.1 | 1.2 f±0.3 | 5.2 d ± 0.3 | 2.2 e ± 0.3 |
| F. oxysporum | 10.5 a ± 0.5 | 5.4 a ± 0.4 | 4.3 c ± 0.2 | 1.2 d ± 0.3 | <1 | <1 | <1 | <1 | <1 |
| P. chlamydospora | <1 | <1 | <1 | <1 | 1.2 a ± 0.3 | <1 | <1 | <1 | <1 |
| Yeast Isolate | ppm eq. | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alcohols | Organic Acids | Esters | |||||||||
| Isobutyl Alcohol | Isoamyl Alcohol | Phenylethyl Alcohol | Isoprenyl Alcohol | Acetic Acid | Isovaleric Acid | n-Caprylic Acid | Pelargonic Acid | n-Capric Acid | Ethyl Propionate | Lauric Acid, Ethyl Ester | |
| M. pulcherrima GP8 | 0.66 a | 8.69 a | 10.91 a | 1.04 a | 0.49 a | 1.30 a | 0.89 a | 0.34 a | 0.77 a | - | 1.80 a |
| S. bacillaris FE08.05 | - * | 8.99 a | 3.16 b | 0.95 a | 0.22 b | 0.17 b | 0.76 a | 0.19 b | 0.88 a | 0.18 | 0.26 b |
| H. opuntiae GA22 | 0.28 b | 2.17 b | 0.17 c | 0.25 b | - | - | - | - | - | - | - |
| H. opuntiae GM10 | 0.16 b | 3.33 c | 1.85 d | 0.21 b | - | - | - | - | 0.30 b | - | - |
| H. uvarum GM19 | 1.60 c | 8.07 a | 2.51 b | 0.58 c | 0.23 b | 0.49 c | 0.23 b | 0.50 c | - | - | 0.86 c |
| H. lanchancei GM32 | - | - | - | - | - | - | - | - | - | - | - |
| H. pseudoguiller GP14 | 0.30 a | 2.50 b | 0.14 c | 0.38 b | - | - | - | - | - | - | - |
| H. guilliermondii GA1 | - | 0.35 d | 0.59 c | 0.25 b | 0.26 b | 0.18 b | 0.45 c | - | - | 0.11 b | |
| C. awuaii GM3 | - | 0.15 d | - | 0.58 c | 0.18 b | - | - | 0.12 b | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabaghian, S.; Braschi, G.; Vannini, L.; Patrignani, F.; Samsulrizal, N.H.; Lanciotti, R. Isolation and Identification of Wild Yeast from Malaysian Grapevine and Evaluation of Their Potential Antimicrobial Activity against Grapevine Fungal Pathogens. Microorganisms 2021, 9, 2582. https://doi.org/10.3390/microorganisms9122582
Sabaghian S, Braschi G, Vannini L, Patrignani F, Samsulrizal NH, Lanciotti R. Isolation and Identification of Wild Yeast from Malaysian Grapevine and Evaluation of Their Potential Antimicrobial Activity against Grapevine Fungal Pathogens. Microorganisms. 2021; 9(12):2582. https://doi.org/10.3390/microorganisms9122582
Chicago/Turabian StyleSabaghian, Simin, Giacomo Braschi, Lucia Vannini, Francesca Patrignani, Nurul Hidayah Samsulrizal, and Rosalba Lanciotti. 2021. "Isolation and Identification of Wild Yeast from Malaysian Grapevine and Evaluation of Their Potential Antimicrobial Activity against Grapevine Fungal Pathogens" Microorganisms 9, no. 12: 2582. https://doi.org/10.3390/microorganisms9122582
APA StyleSabaghian, S., Braschi, G., Vannini, L., Patrignani, F., Samsulrizal, N. H., & Lanciotti, R. (2021). Isolation and Identification of Wild Yeast from Malaysian Grapevine and Evaluation of Their Potential Antimicrobial Activity against Grapevine Fungal Pathogens. Microorganisms, 9(12), 2582. https://doi.org/10.3390/microorganisms9122582









