Microbiome Compositions and Resistome Levels after Antibiotic Treatment of Critically Ill Patients: An Observational Cohort Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Settings
2.2. Ethical Concerns
2.3. Study Participants and Samples
2.4. Antibiotic Regimens
2.5. Laboratory Analyses
2.6. DNA Extraction, Library Preparation and Sequencing
2.7. Sequencing Data Quality Control
2.8. Mapping Reads to Microbial Gene Catalog
2.9. MGS Relative Abundance Calculation
2.10. Resistome Profiling
2.11. Diversity Analyses
2.12. Statistics
3. Results
3.1. Study Group Characterization
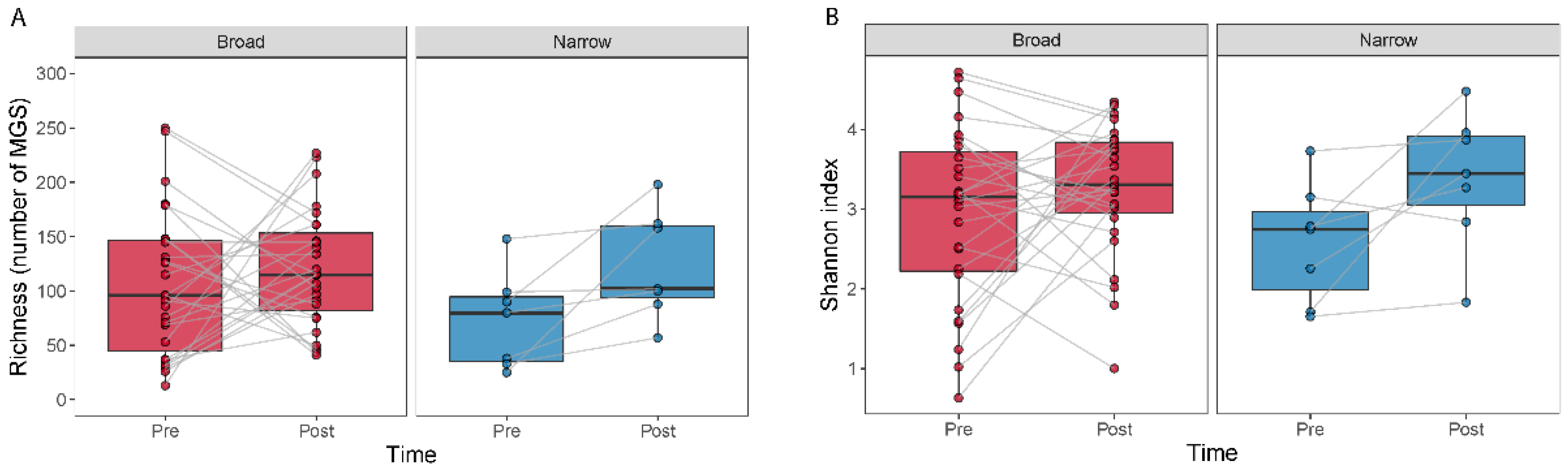
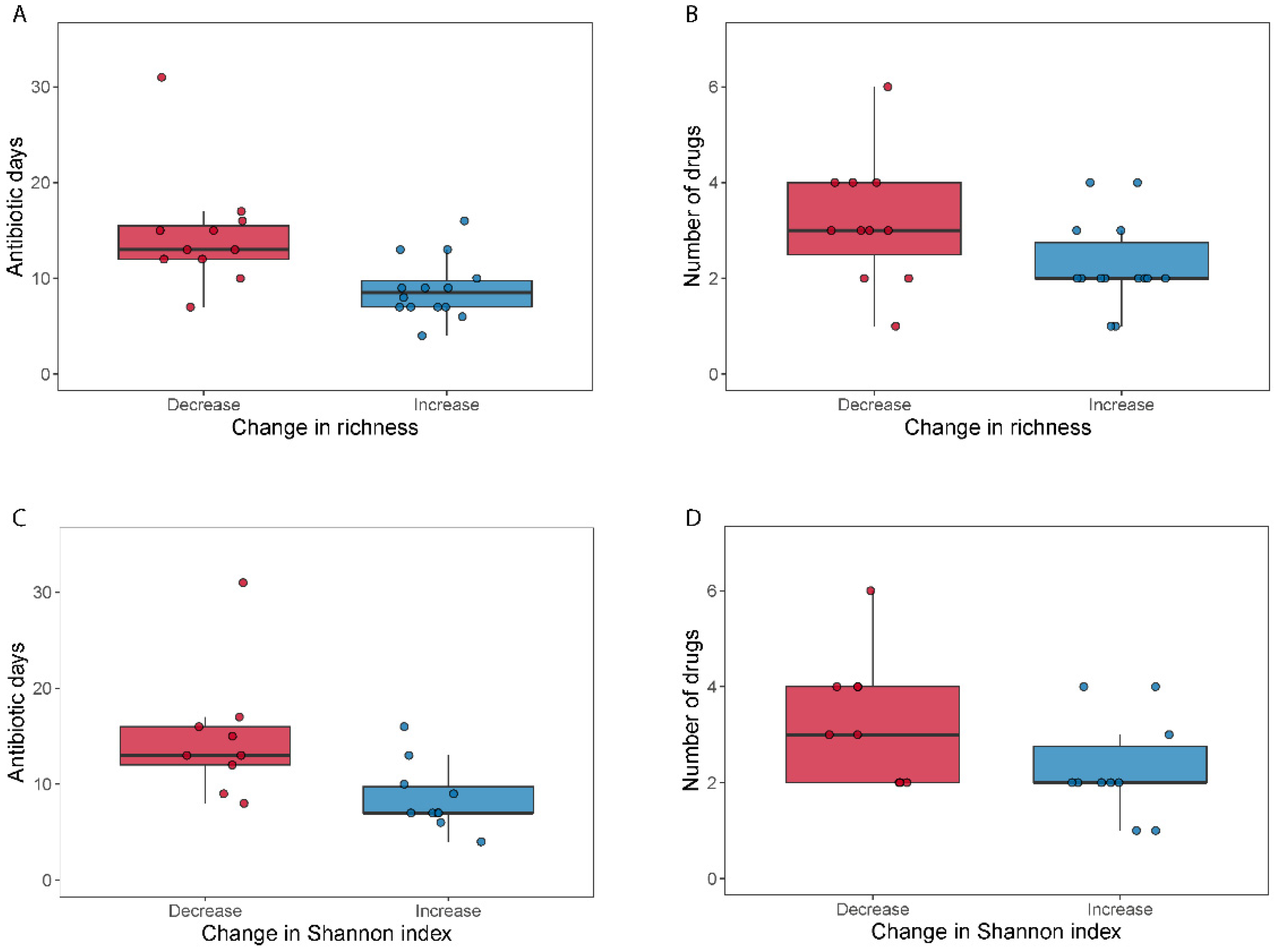
3.2. Higher Number of Drugs and Selection Pressure for Decreased MGS Diversity Samples
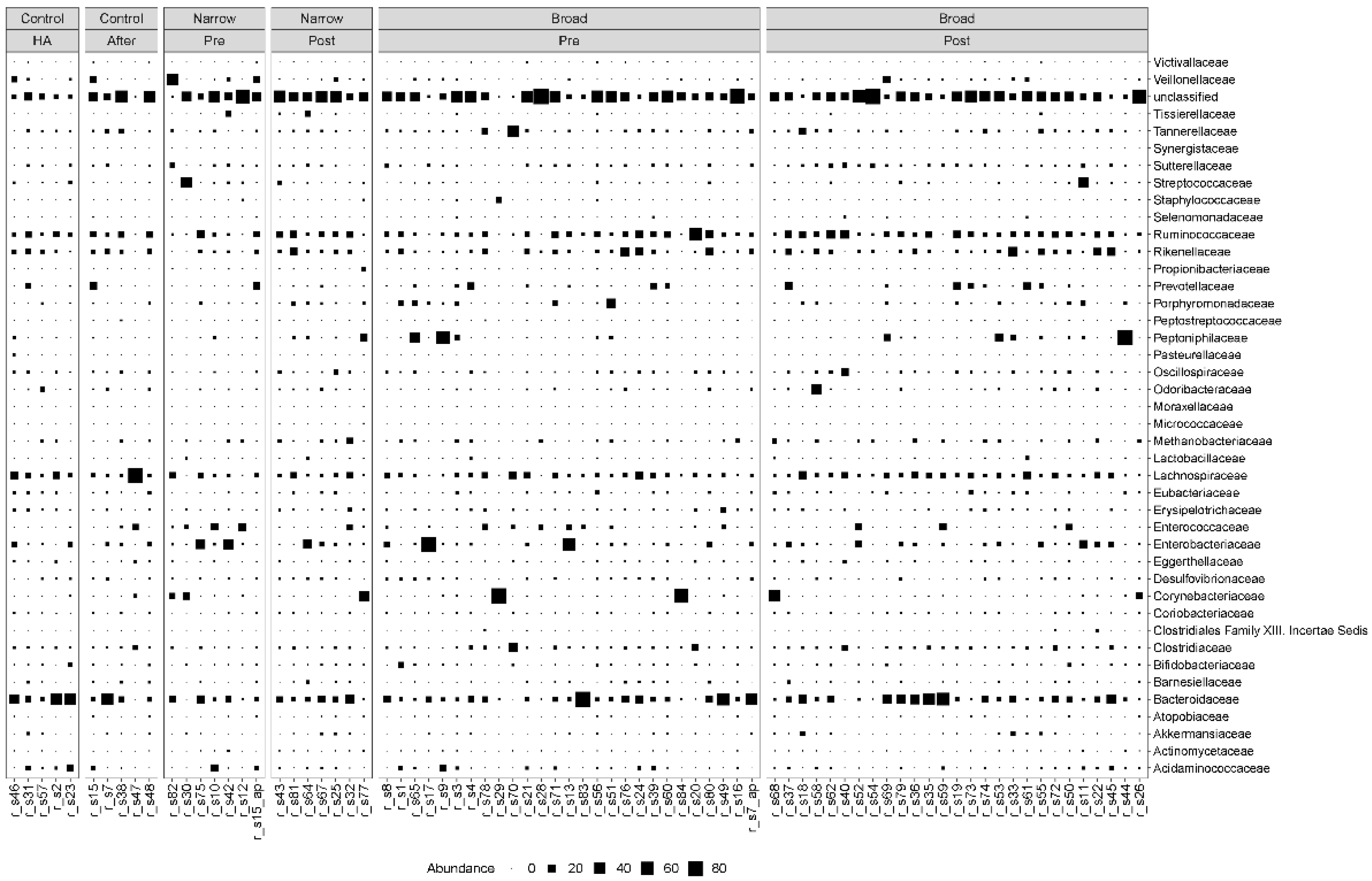
3.3. Clostridiales Species Were Enriched in Post-Treatment Samples
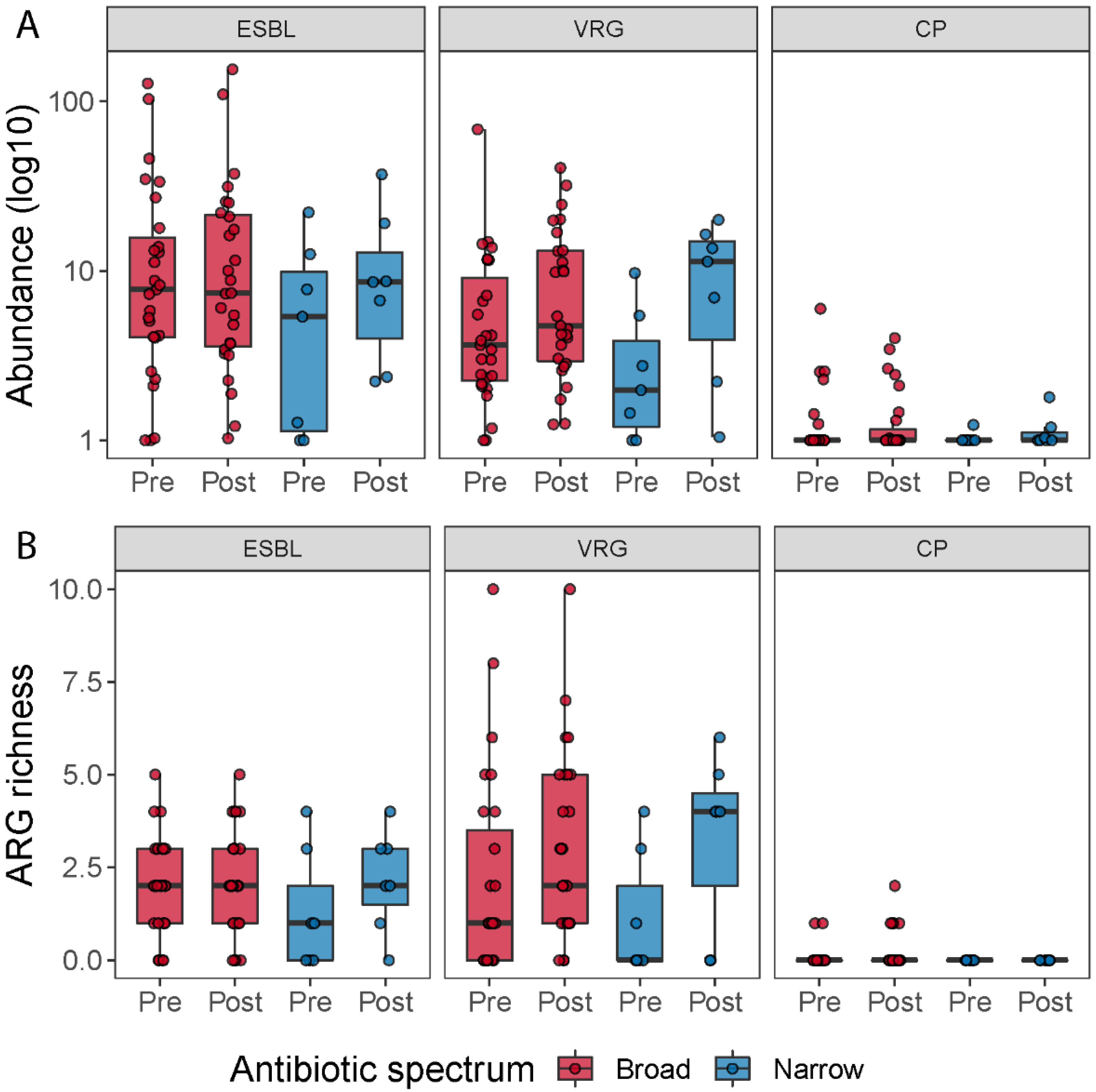
3.4. General Increase of Resistome after Broad-Spectrum Antibiotic
3.5. Vancomycin Resistance Gene Richness Tend to Increase after Antibiotic Treatment
4. Discussion
4.1. Impact of Antibiotic Treatment on Antimicrobial Resistance Genes
4.2. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lawley, T.D.; Walker, A.W. Intestinal colonization resistance. Immunology 2013, 138, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Microbiology: Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Stecher, B. The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection. Microbiol. Spectr. 2015, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Keith, J.W.; Pamer, E.G. Enlisting commensal microbes to resist antibiotic- resistant pathogens. J. Exp. Med. 2019, 216, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Hertz, F.B.; Budding, A.E.; van der Lugt-Degen, M.; Savelkoul, P.H.; Løbner-Olesen, A.; Frimodt-Møller, N. Effects of Antibiotics on the Intestinal Microbiota of Mice. Antibiotics 2020, 9, 191. [Google Scholar] [CrossRef] [PubMed]
- Stacy, A.; Andrade-oliveira, V.; Mcculloch, J.A.; Segre, J.A.; Rehermann, B.; Belkaid, Y.; Stacy, A.; Andrade-oliveira, V.; Mcculloch, J.A.; Hild, B.; et al. Article Infection trains the host for microbiota-enhanced resistance to pathogens ll ll Article Infection trains the host for microbiota-enhanced resistance to pathogens. Cell 2021, 184, 615–627.e17. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; Adler, A.; Abu-Hanna, J.; Percia, S.C.; Matalon, M.K.; Carmeli, Y. Spread of KPC-producing carbapenem-resistant Enterobacteriaceae: The importance of super-spreaders and rectal KPC concentration. Clin. Microbiol. Infect. 2015, 21, 470.e1–470.e7. [Google Scholar] [CrossRef] [PubMed]
- Edlund, C.; Nord, C.E. Effect on the human normal microflora of oral antibiotics for treatment of urinary tract infections. J. Antimicrob. Chemother. 2000, 46, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Ackermann, G.; Khailova, L.; Baird, C.; Heyland, D.; Kozar, R.; Lemieux, M.; Derenski, K.; King, J.; Vis-Kampen, C.; et al. Extreme Dysbiosis of the Microbiome in Critical Illness. mSphere 2018, 1, e00199-16. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.B.; Almeida, M.; Juncker, A.S.; Rasmussen, S.; Li, J.; Sunagawa, S.; Plichta, D.R.; Gautier, L.; Pedersen, A.G.; Le Chatelier, E.; et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 2014, 32, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Boetius Hertz, F.; Lobner-Olesen, A.; Frimodt-Moller, N. Antibiotic Selection of Escherichia coli Sequence Type 131 in a Mouse Intestinal Colonization Model. Antimicrob. Agents Chemother. 2014, 58, 6139–6144. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hertz, F.B.; Schønning, K.; Rasmussen, S.C.; Littauer, P.; Knudsen, J.D.; Løbner-Olesen, A.; Frimodt-Møller, N. Epidemiological factors associated with ESBL- and non ESBL-producing E. coli causing urinary tract infection in general practice. Infect. Dis. 2015, 48, 241–245. [Google Scholar] [CrossRef] [PubMed]
- DANMAP 2019—Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark. Available online: https://backend.orbit.dtu.dk/ws/portalfiles/portal/235092204/DANMAP_2019.pdf (accessed on 1 December 2021).
- Skjøt-Arkil, H.; Mogensen, C.B.; Lassen, A.T.; Johansen, I.S.; Chen, M.; Petersen, P.; Andersen, K.V.; Ellermann-Eriksen, S.; Møller, J.M.; Ludwig, M.; et al. Detection of meticillin-resistant Staphylococcus aureus and carbapenemase-producing Enterobacteriaceae in Danish emergency departments—Evaluation of national screening guidelines. J. Hosp. Infect. 2020, 104, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Nikkels, A.F.; Nikkels-tassoudji, N. Corynebacterium—Associated skin infections. Int. J. Dermatol. 2008, 47, 884–890. [Google Scholar]
- Budding, A.E.; Grasman, M.E.; Eck, A.; Bogaards, J.A.; Vandenbroucke-Grauls, C.M.J.E.; Van Bodegraven, A.A.; Savelkoul, P.H.M. Rectal swabs for analysis of the intestinal microbiota. PLoS ONE 2014, 9, e101344. [Google Scholar] [CrossRef] [PubMed]


| Antimicrobial Treatment | Number of Patients |
|---|---|
| Broad | |
| Cefuroxime, linezolide, meropenem | 1 |
| Piperacillin/Tazobactam | 3 |
| Cefuroxime, Erythromycin, Piperacillin/Tazobactam | 3 |
| Cefuroxime, Piperacillin/Tazobactam | 1 |
| Erythromycin, Piperacillin/Tazobactam, Metronidazole | 1 |
| Dicloxacillin, Piperacillin/Tazobactam | 3 |
| Ciprofloxacin, Piperacillin/Tazobactam | 1 |
| Erythromycin, Piperacillin/Tazobactam | 3 |
| Cefuroxime, Dicloxacillin, Erythromycin, Piperacillin/Tazobactam | 1 |
| Dicloxacillin, Piperacillin/Tazobactam, mero, clari | 1 |
| cipro, genta, Piperacillin/Tazobactam, vanco | 1 |
| Cefuroxime, Erythromycin, Meropenem, Piperacillin/Tazobactam | 1 |
| Erythromycin, Meropenem | 1 |
| Ampicillin, Piperacillin/Tazobactam | 1 |
| Dicloxacillin, Erythromycin, Meropenem, Piperacillin/Tazobactam | 1 |
| Ciprofloxacin, Pivmecillinam, Piperacillin/Tazobactam | 1 |
| Benzylpenicillin, erythromycin, Meropenem, Vancomycin | 1 |
| Erythromycin, Gentamycin, Piperacillin/Tazobactam | 1 |
| Narrow | |
| Cefuroxime, Erythromycin | 1 |
| Cefuroxime, Pivmecillinam, Vancomycin | 1 |
| Pivmecillinam | 1 |
| Ampicillin, Dicloxacillin, Cefuroxime | 1 |
| Benzylpenicillin | 1 |
| Erythromycin, Gentamicin, Meropenem, Piperacillin/Tazobactam, Vancomycin | 1 |
| Cefuroxime | 2 |
| Group | Number of Drugs | Duration of Treatment | Antibiotic Days |
|---|---|---|---|
| All subjects | 0.03 (p = 0.86) | 0.17 (p = 0.32) | 0.20 (p = 0.24) |
| “Narrow” patients | 0.78 (p = 0.038) | 0.32 (p = 0.47) | 0.77 (p = 0.039) |
| “Broad” patients | −0.08 (p = 0.68) | −0.22 (p = 0.25) | 0.037 (p = 0.85) |
| “Broad” Decreased diversity | 0.085 (p = 0.79) | 0.20 (p = 0.54) | 0.14 (p = 0.66) |
| “Broad” Increased diversity | 0.058 (p = 0.83) | −0.070 (p = 0.82) | 0.027 (p = 0.92) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nielsen, K.L.; Olsen, M.H.; Pallejá, A.; Ebdrup, S.R.; Sørensen, N.; Lukjancenko, O.; Marvig, R.L.; Møller, K.; Frimodt-Møller, N.; Hertz, F.B. Microbiome Compositions and Resistome Levels after Antibiotic Treatment of Critically Ill Patients: An Observational Cohort Study. Microorganisms 2021, 9, 2542. https://doi.org/10.3390/microorganisms9122542
Nielsen KL, Olsen MH, Pallejá A, Ebdrup SR, Sørensen N, Lukjancenko O, Marvig RL, Møller K, Frimodt-Møller N, Hertz FB. Microbiome Compositions and Resistome Levels after Antibiotic Treatment of Critically Ill Patients: An Observational Cohort Study. Microorganisms. 2021; 9(12):2542. https://doi.org/10.3390/microorganisms9122542
Chicago/Turabian StyleNielsen, Karen Leth, Markus Harboe Olsen, Albert Pallejá, Søren Røddik Ebdrup, Nikolaj Sørensen, Oksana Lukjancenko, Rasmus L. Marvig, Kirsten Møller, Niels Frimodt-Møller, and Frederik Boëtius Hertz. 2021. "Microbiome Compositions and Resistome Levels after Antibiotic Treatment of Critically Ill Patients: An Observational Cohort Study" Microorganisms 9, no. 12: 2542. https://doi.org/10.3390/microorganisms9122542
APA StyleNielsen, K. L., Olsen, M. H., Pallejá, A., Ebdrup, S. R., Sørensen, N., Lukjancenko, O., Marvig, R. L., Møller, K., Frimodt-Møller, N., & Hertz, F. B. (2021). Microbiome Compositions and Resistome Levels after Antibiotic Treatment of Critically Ill Patients: An Observational Cohort Study. Microorganisms, 9(12), 2542. https://doi.org/10.3390/microorganisms9122542







