Mycobacterium abscessus Genetic Determinants Associated with the Intrinsic Resistance to Antibiotics
Abstract
1. Introduction
2. Materials and Methods
2.1. Mycobacterial Strains and Culture Conditions
2.2. Creation of MAB Mutant Library Using MycomarT7 Transduction and Transposon Mutagenesis
2.3. The High-Throughput Screening of the MAB Mutant Library for Antibiotic Susceptibility
2.4. Ligation Mediated PCR (LM-PCR) for Analysis of MAB Gene Knockout Mutants
2.5. Bioinformatic Analysis of DNA Sequencing
2.6. The Antibiotic Susceptibility Testing with the Broth Microdilution Method
2.7. RNA Extraction, cDNA Synthesis, and Real-Time Quantitative PCR
2.8. Biofilm Formation
2.9. MAB Survival Assay in THP-1 Human Macrophages
2.10. The Complementation of MAB Mutants with Function Genes
2.11. Antibiotic Killing Kinetics in Human Macrophages
2.12. Statistics
3. Results
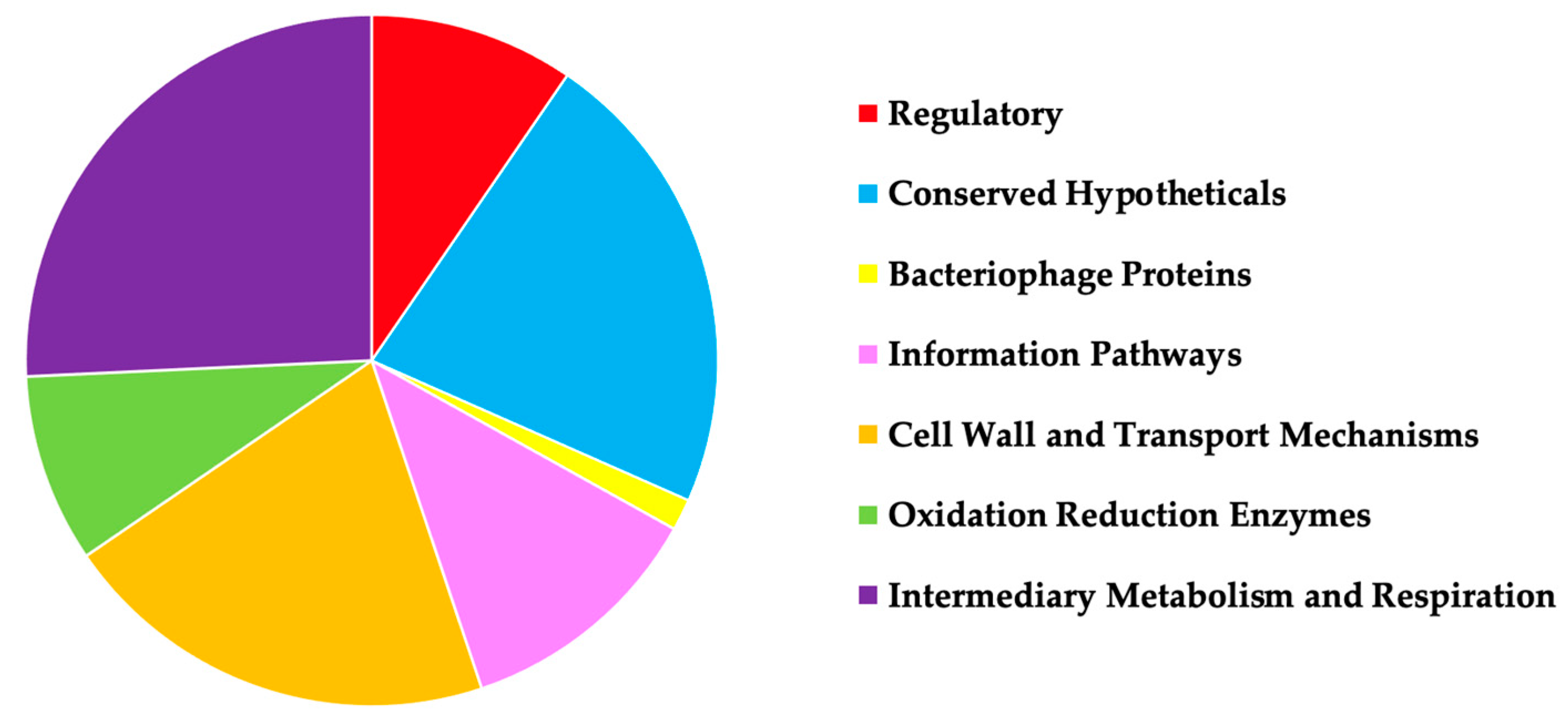
3.1. MAB Gene Knockout Mutants Associated with Increased or Decreased Susceptibility to Antibiotics
3.2. MAB Surface Factors Associated with Differential Antibiotic Susceptibility Patterns
3.3. MAB Surface Factors Associated with the Biofilm Formation
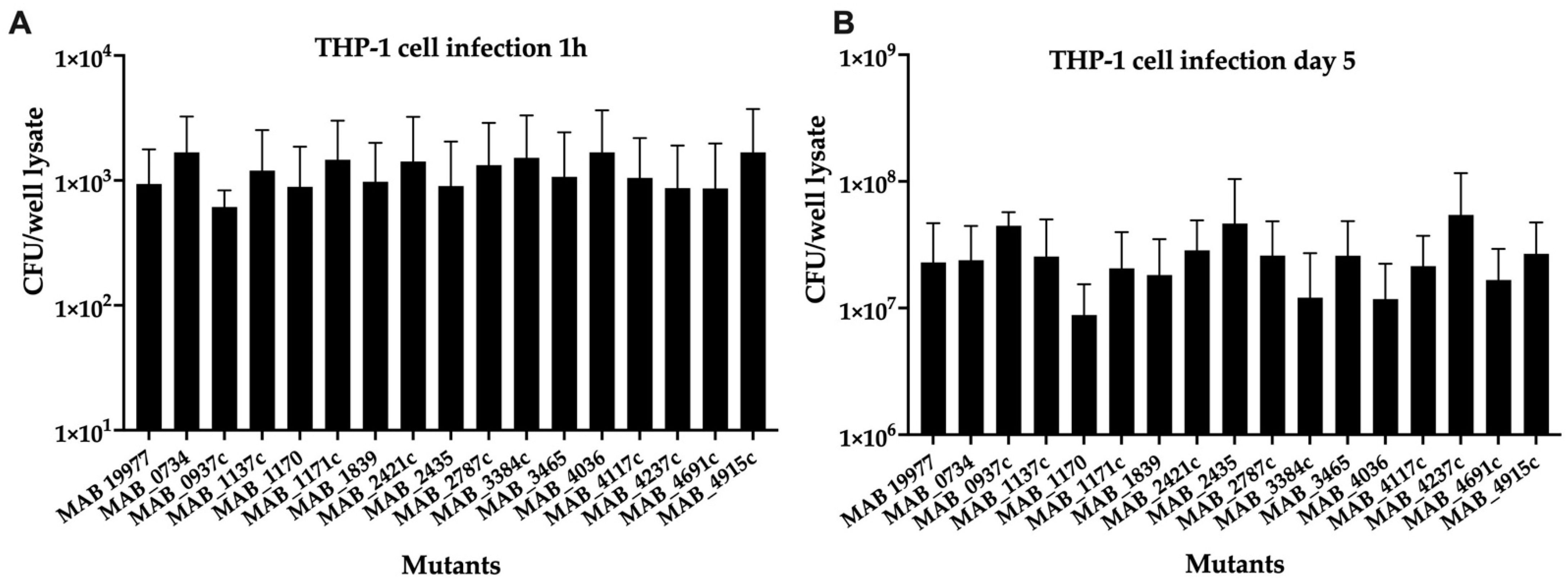
3.4. The Ability of MAB Transport System Mutants to Infect and Grow Intracellularly in THP-1 Human Macrophages
3.5. The Expression of Surface Associated Transport Genes across MAB Clinical Isolates of Cystic Fibrosis Patients
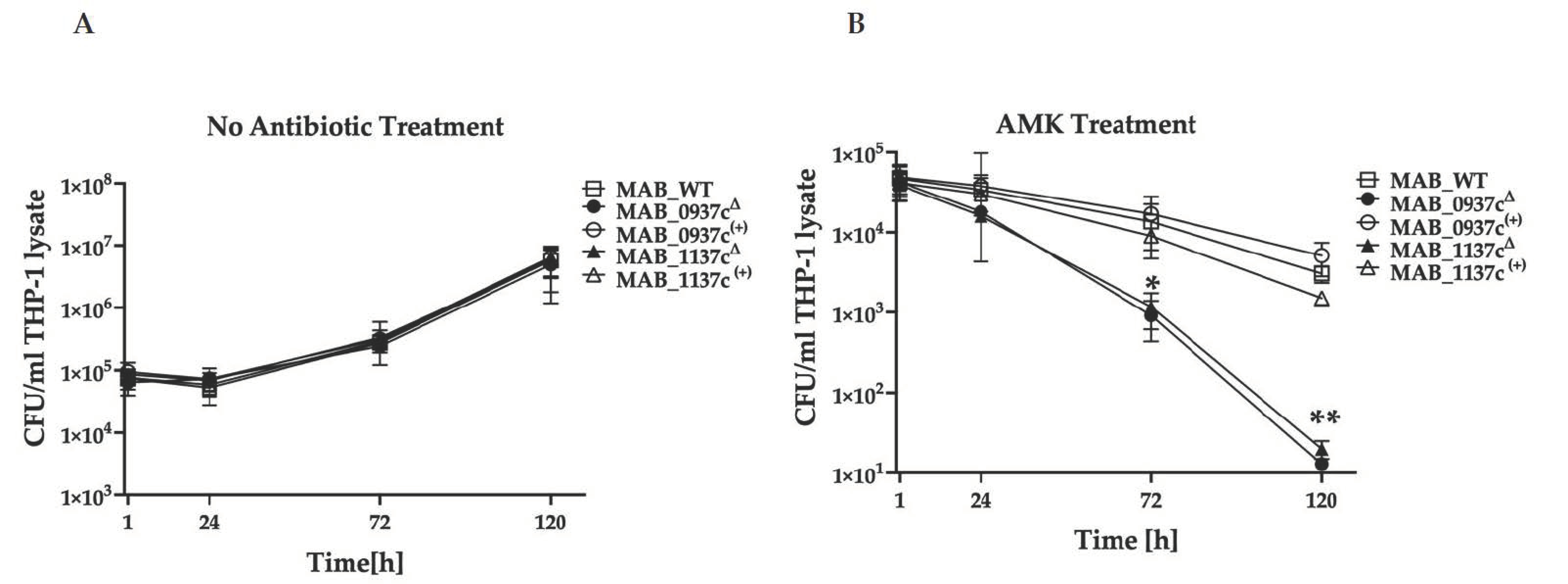
3.6. The Functional Disruption of Efflux Pumps Increases the Antibiotic Efficacy against MAB of the Intracellular Phenotype
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- To, K.; Cao, R.; Yegiazaryan, A.; Owens, J.; Venketaraman, V. General Overview of Nontuberculous Mycobacteria Opportunistic Pathogens: Mycobacterium avium and Mycobacterium abscessus. J. Clin. Med. 2020, 9, 2541. [Google Scholar] [CrossRef] [PubMed]
- Minias, A.; Zukowska, L.; Lach, J.; Jagielski, T.; Strapagiel, D.; Kim, S.Y.; Koh, W.J.; Adam, H.; Bittner, R.; Truden, S.; et al. Subspecies-specific sequence detection for differentiation of Mycobacterium abscessus complex. Sci. Rep. 2020, 10, 16415. [Google Scholar] [CrossRef]
- Prevots, D.R.; Shaw, P.A.; Strickland, D.; Jackson, L.A.; Raebel, M.A.; Blosky, M.A.; Montes de Oca, R.; Shea, Y.R.; Seitz, A.E.; Holland, S.M.; et al. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am. J. Respir. Crit. Care Med. 2010, 182, 970–976. [Google Scholar] [CrossRef]
- Lee, M.R.; Sheng, W.H.; Hung, C.C.; Yu, C.J.; Lee, L.N.; Hsueh, P.R. Mycobacterium abscessus Complex Infections in Humans. Emerg. Infect. Dis. 2015, 21, 1638–1646. [Google Scholar] [CrossRef]
- Nessar, R.; Cambau, E.; Reyrat, J.M.; Murray, A.; Gicquel, B. Mycobacterium abscessus: A new antibiotic nightmare. J. Antimicrob. Chemother. 2012, 67, 810–818. [Google Scholar] [CrossRef]
- Jeon, K.; Kwon, O.J.; Lee, N.Y.; Kim, B.J.; Kook, Y.H.; Lee, S.H.; Park, Y.K.; Kim, C.K.; Koh, W.J. Antibiotic treatment of Mycobacterium abscessus lung disease: A retrospective analysis of 65 patients. Am. J. Respir. Crit. Care Med. 2009, 180, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Ratnatunga, C.N.; Lutzky, V.P.; Kupz, A.; Doolan, D.L.; Reid, D.W.; Field, M.; Bell, S.C.; Thomson, R.M.; Miles, J.J. The Rise of Non-Tuberculosis Mycobacterial Lung Disease. Front. Immunol. 2020, 11, 303. [Google Scholar] [CrossRef]
- Novosad, S.A.; Beekmann, S.E.; Polgreen, P.M.; Mackey, K.; Winthrop, K.L.; Team, M.a.S. Treatment of Mycobacterium abscessus Infection. Emerg. Infect. Dis. 2016, 22, 511–514. [Google Scholar] [CrossRef] [PubMed]
- Vianna, J.S.; Machado, D.; Ramis, I.B.; Silva, F.P.; Bierhals, D.V.; Abril, M.A.; von Groll, A.; Ramos, D.F.; Lourenco, M.C.S.; Viveiros, M.; et al. The Contribution of Efflux Pumps in Mycobacterium abscessus Complex Resistance to Clarithromycin. Antibiotics 2019, 8, 153. [Google Scholar] [CrossRef]
- Bronson, R.A.; Gupta, C.; Manson, A.L.; Nguyen, J.A.; Bahadirli-Talbott, A.; Parrish, N.M.; Earl, A.M.; Cohen, K.A. Global phylogenomic analyses of Mycobacterium abscessus provide context for non cystic fibrosis infections and the evolution of antibiotic resistance. Nat. Commun. 2021, 12, 5145. [Google Scholar] [CrossRef]
- Koh, W.J.; Jeong, B.H.; Kim, S.Y.; Jeon, K.; Park, K.U.; Jhun, B.W.; Lee, H.; Park, H.Y.; Kim, D.H.; Huh, H.J.; et al. Mycobacterial Characteristics and Treatment Outcomes in Mycobacterium abscessus Lung Disease. Clin. Infect. Dis. 2017, 64, 309–316. [Google Scholar] [CrossRef]
- Guo, Q.; Chen, J.; Zhang, S.; Zou, Y.; Zhang, Y.; Huang, D.; Zhang, Z.; Li, B.; Chu, H. Efflux Pumps Contribute to Intrinsic Clarithromycin Resistance in Clinical, Mycobacterium abscessus Isolates. Infect. Drug Resist. 2020, 13, 447–454. [Google Scholar] [CrossRef]
- Richard, M.; Gutierrez, A.V.; Kremer, L. Dissecting erm(41)-Mediated Macrolide-Inducible Resistance in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2020, 64, e01879-19. [Google Scholar] [CrossRef] [PubMed]
- Nasiri, M.J.; Haeili, M.; Ghazi, M.; Goudarzi, H.; Pormohammad, A.; Imani Fooladi, A.A.; Feizabadi, M.M. New Insights in to the Intrinsic and Acquired Drug Resistance Mechanisms in Mycobacteria. Front. Microbiol. 2017, 8, 681. [Google Scholar] [CrossRef]
- Kleinnijenhuis, J.; Oosting, M.; Joosten, L.A.; Netea, M.G.; Van Crevel, R. Innate immune recognition of Mycobacterium tuberculosis. Clin. Dev. Immunol. 2011, 2011, 405310. [Google Scholar] [CrossRef]
- Abrahams, K.A.; Besra, G.S. Mycobacterial cell wall biosynthesis: A multifaceted antibiotic target. Parasitology 2018, 145, 116–133. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Haldimann, K.; Selchow, P.; Reinau, L.M.; Dal Molin, M.; Sander, P. Lipoprotein Glycosylation by Protein-O-Mannosyltransferase (MAB_1122c) Contributes to Low Cell Envelope Permeability and Antibiotic Resistance of Mycobacterium abscessus. Front. Microbiol. 2017, 8, 2123. [Google Scholar] [CrossRef]
- Richard, M.; Gutierrez, A.V.; Viljoen, A.; Rodriguez-Rincon, D.; Roquet-Baneres, F.; Blaise, M.; Everall, I.; Parkhill, J.; Floto, R.A.; Kremer, L. Mutations in the MAB_2299c TetR Regulator Confer Cross-Resistance to Clofazimine and Bedaquiline in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2019, 63, e01316-18. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, A.V.; Richard, M.; Roquet-Baneres, F.; Viljoen, A.; Kremer, L. The TetR Family Transcription Factor MAB_2299c Regulates the Expression of Two Distinct MmpS-MmpL Efflux Pumps Involved in Cross-Resistance to Clofazimine and Bedaquiline in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2019, 63, e01000-19. [Google Scholar] [CrossRef]
- Sarathy, J.P.; Dartois, V.; Lee, E.J. The role of transport mechanisms in mycobacterium tuberculosis drug resistance and tolerance. Pharmaceuticals 2012, 5, 1210. [Google Scholar] [CrossRef]
- Machado, D.; Couto, I.; Perdigao, J.; Rodrigues, L.; Portugal, I.; Baptista, P.; Veigas, B.; Amaral, L.; Viveiros, M. Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS ONE 2012, 7, e34538. [Google Scholar] [CrossRef]
- Dubee, V.; Soroka, D.; Cortes, M.; Lefebvre, A.L.; Gutmann, L.; Hugonnet, J.E.; Arthur, M.; Mainardi, J.L. Impact of beta-lactamase inhibition on the activity of ceftaroline against Mycobacterium tuberculosis and Mycobacterium abscessus. Antimicrob. Agents Chemother. 2015, 59, 2938–2941. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, U.S.; Dartois, V.; Dick, T. Repositioning rifamycins for Mycobacterium abscessus lung disease. Expert Opin. Drug Discov. 2019, 14, 867–878. [Google Scholar] [CrossRef]
- Rudra, P.; Hurst-Hess, K.; Lappierre, P.; Ghosh, P. High Levels of Intrinsic Tetracycline Resistance in Mycobacterium abscessus Are Conferred by a Tetracycline-Modifying Monooxygenase. Antimicrob. Agents Chemother. 2018, 62, e00119-18. [Google Scholar] [CrossRef]
- Rojony, R.; Danelishvili, L.; Campeau, A.; Wozniak, J.M.; Gonzalez, D.J.; Bermudez, L.E. Exposure of Mycobacterium abscessus to Environmental Stress and Clinically Used Antibiotics Reveals Common Proteome Response among Pathogenic Mycobacteria. Microorganisms 2020, 8, 698. [Google Scholar] [CrossRef] [PubMed]
- Rose, S.J.; Bermudez, L.E. Identification of Bicarbonate as a Trigger and Genes Involved with Extracellular DNA Export in Mycobacterial Biofilms. mBio 2016, 7, e01597-16. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Hearn, M.; Eckstein, T.M.; Sommer, S.; Bermudez, L.E. A Mycobacterium avium subsp. paratuberculosis LuxR regulates cell envelope and virulence. Innate Immun. 2010, 16, 235–247. [Google Scholar] [CrossRef]
- Palmer, K.L.; Aye, L.M.; Whiteley, M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 2007, 189, 8079–8087. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Danelishvili, L.; Wagner, D.; Petrofsky, M.; Bermudez, L.E. Identification of virulence determinants of Mycobacterium avium that impact on the ability to resist host killing mechanisms. J. Med. Microbiol. 2010, 59, 8–16. [Google Scholar] [CrossRef]
- Fennelly, K.P.; Ojano-Dirain, C.; Yang, Q.; Liu, L.; Lu, L.; Progulske-Fox, A.; Wang, G.P.; Antonelli, P.; Schultz, G. Biofilm Formation by Mycobacterium abscessus in a Lung Cavity. Am. J. Respir. Crit. Care Med. 2016, 193, 692–693. [Google Scholar] [CrossRef]
- Chakraborty, P.; Kumar, A. The extracellular matrix of mycobacterial biofilms: Could we shorten the treatment of mycobacterial infections? Microb. Cell 2019, 6, 105–122. [Google Scholar] [CrossRef]
- Eladawy, M.; El-Mowafy, M.; El-Sokkary, M.M.A.; Barwa, R. Effects of Lysozyme, Proteinase K, and Cephalosporins on Biofilm Formation by Clinical Isolates of Pseudomonas aeruginosa. Interdiscip. Perspect. Infect. Dis. 2020, 2020, 6156720. [Google Scholar] [CrossRef]
- Viljoen, A.; Dubois, V.; Girard-Misguich, F.; Blaise, M.; Herrmann, J.L.; Kremer, L. The diverse family of MmpL transporters in mycobacteria: From regulation to antimicrobial developments. Mol. Microbiol. 2017, 104, 889–904. [Google Scholar] [CrossRef]
- Ripoll, F.; Pasek, S.; Schenowitz, C.; Dossat, C.; Barbe, V.; Rottman, M.; Macheras, E.; Heym, B.; Herrmann, J.L.; Daffe, M.; et al. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS ONE 2009, 4, e5660. [Google Scholar] [CrossRef]
- Smith, T.; Wolff, K.A.; Nguyen, L. Molecular biology of drug resistance in Mycobacterium tuberculosis. Curr. Top. Microbiol. Immunol. 2013, 374, 53–80. [Google Scholar] [CrossRef] [PubMed]
- Spengler, G.; Kincses, A.; Gajdacs, M.; Amaral, L. New Roads Leading to Old Destinations: Efflux Pumps as Targets to Reverse Multidrug Resistance in Bacteria. Molecules 2017, 22, 468. [Google Scholar] [CrossRef]
- Da Silva, P.E.; Von Groll, A.; Martin, A.; Palomino, J.C. Efflux as a mechanism for drug resistance in Mycobacterium tuberculosis. FEMS Immunol. Med. Microbiol. 2011, 63, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Melly, G.; Purdy, G.E. MmpL Proteins in Physiology and Pathogenesis of M. tuberculosis. Microorganisms 2019, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Lopeman, R.C.; Harrison, J.; Desai, M.; Cox, J.A.G. Mycobacterium abscessus: Environmental Bacterium Turned Clinical Nightmare. Microorganisms 2019, 7, 90. [Google Scholar] [CrossRef]
- De Rossi, E.; Ainsa, J.A.; Riccardi, G. Role of mycobacterial efflux transporters in drug resistance: An unresolved question. FEMS Microbiol. Rev. 2006, 30, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Milano, A.; Pasca, M.R.; Provvedi, R.; Lucarelli, A.P.; Manina, G.; Ribeiro, A.L.; Manganelli, R.; Riccardi, G. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis 2009, 89, 84–90. [Google Scholar] [CrossRef]
- Hartkoorn, R.C.; Uplekar, S.; Cole, S.T. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 2979–2981. [Google Scholar] [CrossRef]
- Louw, G.E.; Warren, R.M.; Gey van Pittius, N.C.; McEvoy, C.R.; Van Helden, P.D.; Victor, T.C. A balancing act: Efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 2009, 53, 3181–3189. [Google Scholar] [CrossRef]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef]
- Rismondo, J.; Schulz, L.M. Not Just Transporters: Alternative Functions of ABC Transporters in Bacillus subtilis and Listeria monocytogenes. Microorganisms 2021, 9, 163. [Google Scholar] [CrossRef]
- Rojony, R.; Martin, M.; Campeau, A.; Wozniak, J.M.; Gonzalez, D.J.; Jaiswal, P.; Danelishvili, L.; Bermudez, L.E. Quantitative analysis of Mycobacterium avium subsp hominissuis proteome in response to antibiotics and during exposure to different environmental conditions. Clin. Proteom. 2019, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Mailaender, C.; Reiling, N.; Engelhardt, H.; Bossmann, S.; Ehlers, S.; Niederweis, M. The MspA porin promotes growth and increases antibiotic susceptibility of both Mycobacterium bovis BCG and Mycobacterium tuberculosis. Microbiol. (Read.) 2004, 150, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. The Citric Acid Cycle Oxidizes Two-Carbon Units, 5th ed.; W. H. Freeman: New York, NY, USA, 2002. [Google Scholar]
- Wagner, T.; Bellinzoni, M.; Wehenkel, A.; O’Hare, H.M.; Alzari, P.M. Functional plasticity and allosteric regulation of alpha-ketoglutarate decarboxylase in central mycobacterial metabolism. Chem. Biol. 2011, 18, 1011–1020. [Google Scholar] [CrossRef]
- Daniel, J.; Sirakova, T.; Kolattukudy, P. An acyl-CoA synthetase in Mycobacterium tuberculosis involved in triacylglycerol accumulation during dormancy. PLoS ONE 2014, 9, e114877. [Google Scholar] [CrossRef]
- Weissbach, H.; Etienne, F.; Hoshi, T.; Heinemann, S.H.; Lowther, W.T.; Matthews, B.; St John, G.; Nathan, C.; Brot, N. Peptide methionine sulfoxide reductase: Structure, mechanism of action, and biological function. Arch. Biochem. Biophys. 2002, 397, 172–178. [Google Scholar] [CrossRef]
- Deng, W.; Li, C.; Xie, J. The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell Signal. 2013, 25, 1608–1613. [Google Scholar] [CrossRef]
- Housseini, B.I.K.; Phan, G.; Broutin, I. Functional Mechanism of the Efflux Pumps Transcription Regulators From Pseudomonas aeruginosa Based on 3D Structures. Front. Mol. Biosci. 2018, 5, 57. [Google Scholar] [CrossRef]
- Li, Y.; He, Z.G. The mycobacterial LysR-type regulator OxyS responds to oxidative stress and negatively regulates expression of the catalase-peroxidase gene. PLoS ONE 2012, 7, e30186. [Google Scholar] [CrossRef][Green Version]
- Liu, H.; Yang, M.; He, Z.G. Novel TetR family transcriptional factor regulates expression of multiple transport-related genes and affects rifampicin resistance in Mycobacterium smegmatis. Sci. Rep. 2016, 6, 27489. [Google Scholar] [CrossRef]
- Hu, J.; Zhao, L.; Yang, M. A GntR family transcription factor positively regulates mycobacterial isoniazid resistance by controlling the expression of a putative permease. BMC Microbiol. 2015, 15, 214. [Google Scholar] [CrossRef]
- Zeng, J.; Deng, W.; Yang, W.; Luo, H.; Duan, X.; Xie, L.; Li, P.; Wang, R.; Fu, T.; Abdalla, A.E.; et al. Mycobacterium tuberculosis Rv1152 is a Novel GntR Family Transcriptional Regulator Involved in Intrinsic Vancomycin Resistance and is a Potential Vancomycin Adjuvant Target. Sci. Rep. 2016, 6, 28002. [Google Scholar] [CrossRef]
- Routh, M.D.; Su, C.C.; Zhang, Q.; Yu, E.W. Structures of AcrR and CmeR: Insight into the mechanisms of transcriptional repression and multi-drug recognition in the TetR family of regulators. Biochim. Biophys. Acta 2009, 1794, 844–851. [Google Scholar] [CrossRef]
- Grkovic, S.; Hardie, K.M.; Brown, M.H.; Skurray, R.A. Interactions of the QacR multidrug-binding protein with structurally diverse ligands: Implications for the evolution of the binding pocket. Biochemistry 2003, 42, 15226–15236. [Google Scholar] [CrossRef]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef]
- Pasqua, M.; Grossi, M.; Zennaro, A.; Fanelli, G.; Micheli, G.; Barras, F.; Colonna, B.; Prosseda, G. The Varied Role of Efflux Pumps of the MFS Family in the Interplay of Bacteria with Animal and Plant Cells. Microorganisms 2019, 7, 285. [Google Scholar] [CrossRef]
- Johansen, M.D.; Herrmann, J.L.; Kremer, L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol. 2020, 18, 392–407. [Google Scholar] [CrossRef]
- Danelishvili, L.; Shulzhenko, N.; Chinison, J.J.J.; Babrak, L.; Hu, J.; Morgun, A.; Burrows, G.; Bermudez, L.E. Mycobacterium tuberculosis Proteome Response to Antituberculosis Compounds Reveals Metabolic “Escape” Pathways That Prolong Bacterial Survival. Antimicrob. Agents Chemother. 2017, 61, e00430-17. [Google Scholar] [CrossRef]


| Antibiotic | MAB 19977 | |
|---|---|---|
| MIC (μg/mL) | BC (μg/mL) | |
| AMK | <16 | 32 |
| CLA | <2 | 8 |
| FOX | <16 | 128 |
| MAB Gene | Sequence (5′-3′) Forward | Sequence (5′-3′) Reverse |
|---|---|---|
| MAB_0734 | CTCGCCAACGCGAGTAATTC | CAGCCCAGCTGATAACCCAT |
| MAB_0937c | ATCAGTGATGTCGCCAAGGG | AGCGTCAACAGAACGACGAT |
| MAB_1137c | GTGATATCGCTGCTCGGTGA | CGCGACGATGAAAATCGCAT |
| MAB_1170 | CTGTGATTGGGGCGCTATTG | TTCTTCGTCTTAGCCGCCG |
| MAB_1171c | GCCAACGCATCCAAACCTAC | GATTCTGCGGTAGAGCCGAT |
| MAB_1839 | TGGGGAATTCGTTACGCGAG | CCACGCCAACTTCACAAAGG |
| MAB_2421c | CTCGACTACGAGGGGAAAGC | GGTCGTCCATTTGTCACCGA |
| MAB_2435 | GAGATTCGTCTCGGCGATCA | GTCAGATGGGGAAACAGCGA |
| MAB_2787c | GGGCTCACCTACTCCTCGTA | TTGGTTCTCCGACTTGGTGG |
| MAB_3384c | AGGGTTCAGTCACGCATTGT | GTAACGCCCGTCGATCATCT |
| MAB_3465 | GGAGCTACGCACACCAAAGA | GCGATTCAGCACTTCCCTGA |
| MAB_4036 | GACCACCCGCAATGTACAGA | TGCATGATGGTTCCGGTTGA |
| MAB_4117c | CTCCACGGCGTATTCGGTAA | ACAGCGGATCACCATTGACA |
| MAB_4237c | AATCTGACTGTGCACCAGGG | ATCAGCTCACCGTCGATACG |
| MAB_4691c | TGCGATAGACGACGTACTGC | CAATGGACCGGTGATGGTGA |
| MAB_4915c | ATGTCGAAATGCGATGGGGT | GCCGATAACACTACGGGTCC |
| 16S | GGCTAACCATCCGTCTCTGG | CGGAAGAAAGTCGTCGGTCA |
| Gene | Function | Conserved Domain/Notes | MIC/BC |
|---|---|---|---|
| MAB_0734 | MspA membrane porin | Contains a signal peptide | BC |
| MAB_0937c | MmpL membrane drug exporter protein | Transport protein with a role in drug resistance | MIC |
| MAB_1137c | Putative MmpL membrane protein | Possible role in drug resistance | MIC |
| MAB_1170 | Putative membrane transporter protein. | TauE, sulfite exporter. Integral membrane protein involved in the transport of anions across the cytoplasmic membrane during taurine metabolism as an exporter of sulfoacetate. | MIC |
| MAB_1171c | Conserved hypothetical protein | Integral membrane transporter protein | MIC |
| MAB_1839 | Diguanylate cyclase/phosphodiesterase | GGDEF Diguanylate-cyclase/ABC2_membrane superfamily regulates cell surface adhesion in bacteria/ABC-2-type transporter | MIC |
| MAB_2421c | Hypothetical protein | PknH_C-like extracellular domain | BC |
| MAB_2435 | Molybdenum ABC transporter ModC, ATP-binding protein | P-loop_NTPase superfamily | MIC |
| MAB_2787c | Probable molybdenum ABC transporter, periplasmic | Periplasmic-binding protein type 2 superfamily | MIC |
| MAB_3384c | Putative ABC transporter, ATP-binding protein | ModF ABC-type molybdenum transport system | MIC |
| MAB_3465 | Putative sulfate transporter/anti-Sigma factor | MIC | |
| MAB_4036 | Conserved hypothetical protein | ABC_6TM_exporters superfamily: Six-transmembrane helical domain of an uncharacterized ABC exporter | MIC |
| MAB_4117c | Putative membrane protein | MmpS family protein with a potential role in the transport of MmpL substrates | BC |
| MAB_4237c | Putative amino acid ABC transporter, ATP-binding protein | GlnQ | MIC |
| MAB_4691c | Probable non-ribosomal peptide synthetase PstA | Sulfate Transporter and anti-Sigma factor antagonist | MIC |
| MAB_4915c | Hypothetical protein | PknH_C-like extracellular domain | BC |
| Bacterial ID | MIC for Broth Dilution in 7H9 (μg/mL) | |||
|---|---|---|---|---|
| AMK | CLA | FOX | ||
| MIC mutants | MAB_0937c | 6 | 0.5 | 4 |
| MAB_1137c | 6 | 0.5 | 4 | |
| MAB_1170 | 12 | 0.5 | 16 | |
| MAB_1171c | 16 | 0.5 | 4 | |
| MAB_1839 | 12 | 0.5 | 8 | |
| MAB_2435 | 10 | 0.5 | 16 | |
| MAB_2787c | 10 | 2 | 4 | |
| MAB_3384c | 10 | 0.5 | 16 | |
| MAB_3465 | 10 | 2 | 16 | |
| MAB_4036 | 10 | 0.5 | 4 | |
| MAB_4237c | 10 | 2 | 8 | |
| MAB_4691 | 10 | 0.5 | 4 | |
| BC mutants | MAB_0734 | 128 | 8 | 32 |
| MAB_2421c | 128 | 4 | 32 | |
| MAB_4117c | 64 | 4 | 32 | |
| MAB_4915c | 128 | 8 | 32 | |
| Clinical isolates | MAB 19977 | 16 | 2 | 16 |
| NR 49093 strain DJO 44274 | >256 | >32 | 16 | |
| NR 44273 strain 4529 | >128 | >32 | 16 | |
| NR 44273 strain 4529 | 16 | 4 | 16 | |
| DNA00703 | 16 | 4 | 32 | |
| DNA01163 | 16 | 4 | 32 | |
| DNA01627 | 16 | 16 | 32 | |
| DNA01715 | 16 | 4 | 32 | |
| DNA01639 | 16 | 2 | 32 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorzynski, M.; Week, T.; Jaramillo, T.; Dzalamidze, E.; Danelishvili, L. Mycobacterium abscessus Genetic Determinants Associated with the Intrinsic Resistance to Antibiotics. Microorganisms 2021, 9, 2527. https://doi.org/10.3390/microorganisms9122527
Gorzynski M, Week T, Jaramillo T, Dzalamidze E, Danelishvili L. Mycobacterium abscessus Genetic Determinants Associated with the Intrinsic Resistance to Antibiotics. Microorganisms. 2021; 9(12):2527. https://doi.org/10.3390/microorganisms9122527
Chicago/Turabian StyleGorzynski, Mylene, Tiana Week, Tiana Jaramillo, Elizaveta Dzalamidze, and Lia Danelishvili. 2021. "Mycobacterium abscessus Genetic Determinants Associated with the Intrinsic Resistance to Antibiotics" Microorganisms 9, no. 12: 2527. https://doi.org/10.3390/microorganisms9122527
APA StyleGorzynski, M., Week, T., Jaramillo, T., Dzalamidze, E., & Danelishvili, L. (2021). Mycobacterium abscessus Genetic Determinants Associated with the Intrinsic Resistance to Antibiotics. Microorganisms, 9(12), 2527. https://doi.org/10.3390/microorganisms9122527






