Single- and Dual-Species Biofilm Formation by Shiga Toxin-Producing Escherichia coli and Salmonella, and Their Susceptibility to an Engineered Peptide WK2
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation
2.2. Food Contact Surface Materials and Preparation of Beef Juice
2.3. Preparation of Single- and Mixed-Species Cultures
2.4. Single- and Mixed-Species Biofilm Formation on Stainless Steel Surface in Beef Juice
2.5. Enumeration of the Planktonic and Attached Cells
2.6. Peptide Synthesis
2.7. Peptide Testing
2.8. Sensitivity of STEC O113:H21 and S. Choleraesuis ATCC 10708 in Single- and Mixed-Species Biofilms to WK2
2.9. Confocal Laser Scanning Microscopy (CLSM)
2.10. Statistical Analysis
3. Results
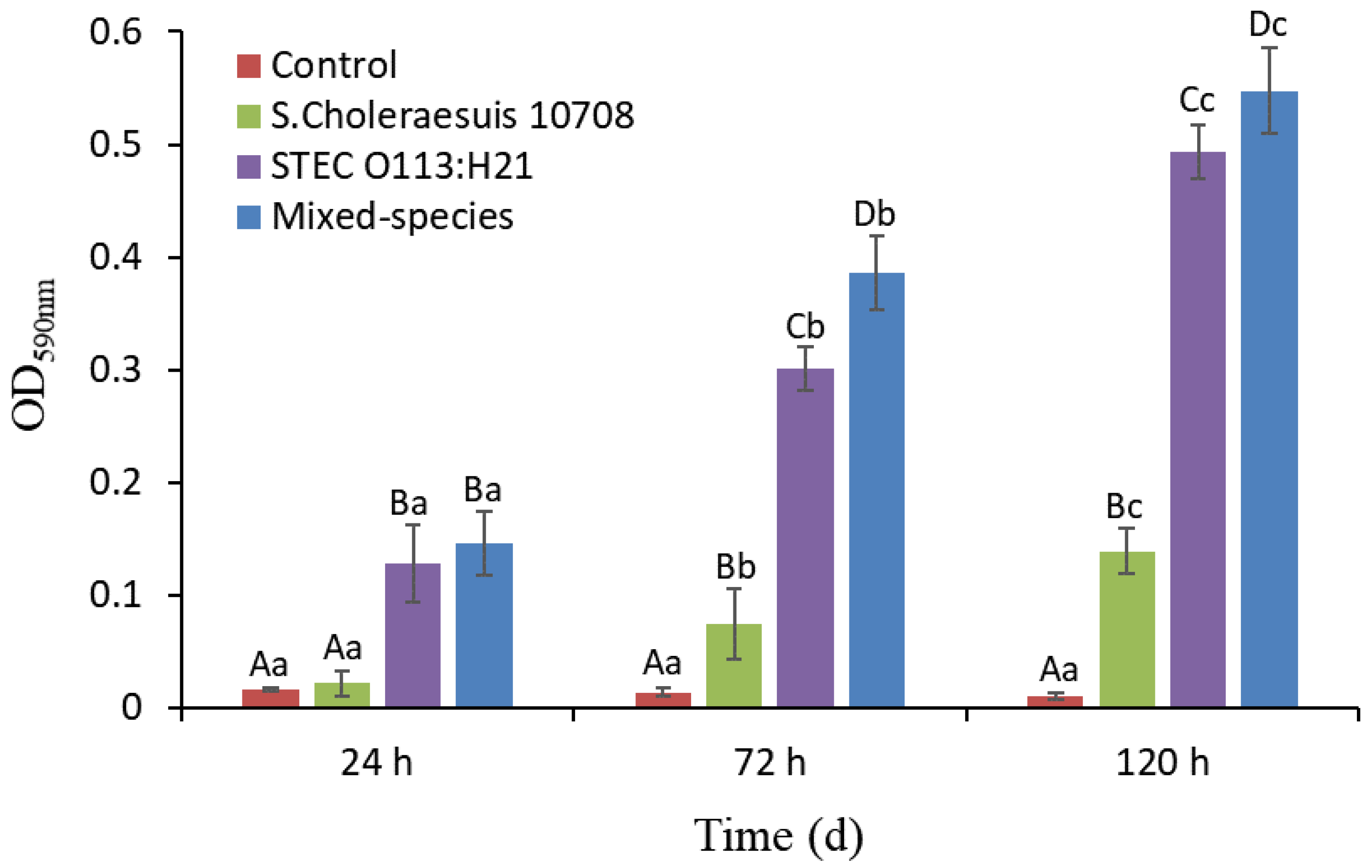
3.1. Single- and Mixed-Species Biofilm Mass Measured by CV Staining
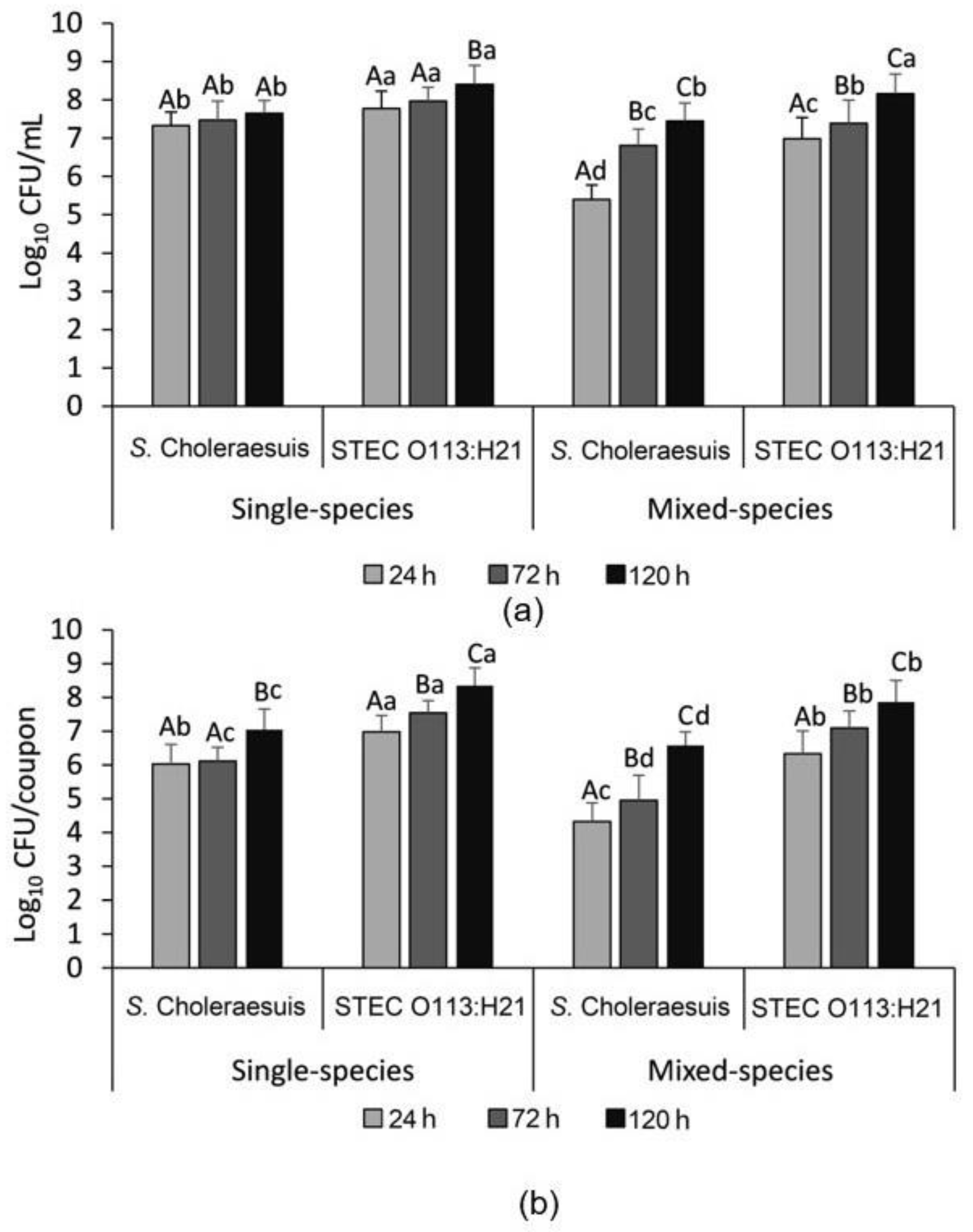
3.2. Enumeration of the Single- and Mixed-Species Planktonic and Attached Bacterial Cells
3.3. The Susceptibility of S. Choleraesuis 10708 and STEC O113:H21 in Single- and Mixed-Species Biofilms to WK2
3.4. Microscopic Examination of Single- and Mixed-Species Biofilms
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Byrne, L.; Adams, N.; Jenkins, C. Association between Shiga toxin-producing Escherichia coli O157: H7 stx gene subtype and disease severity, Rngland, 2009–2019. Emerg. Infect. Dis. 2020, 26, 2394–2400. [Google Scholar] [CrossRef]
- Wang, F.; Yang, Q.; Kase, J.A.; Meng, J.; Clotilde, L.M.; Lin, A.; Ge, B. Current trends in detecting non-O157 Shiga toxin-producing Escherichia coli in food. Foodborne Pathog. Dis. 2013, 10, 665–677. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the united states—major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Giaouris, E.; Simões, M.; Dubois-Brissonnet, F. The role of biofilms in the development and dissemination of microbial resistance within the food industry. Foods 2020, 9, 816. [Google Scholar] [CrossRef]
- Almeida, C.; Azevedo, N.F.; Santos, S.; Keevil, C.W.; Vieira, M.J. Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (pna fish). PLoS ONE 2011, 6, e14786. [Google Scholar] [CrossRef]
- Burmølle, M.; Ren, D.; Bjarnsholt, T.; Sørensen, S.J. Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 2014, 22, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Kalchayanand, N.; Brichta-Harhay, D.M.; Arthur, T.M.; Bosilevac, J.M.; Guerini, M.N.; Wheeler, T.L.; Shackelford, S.D.; Koohmaraie, M. Prevalence rates of Escherichia coli O157: H7 and Salmonella at different sampling sites on cattle hides at a feedlot and processing plant. J. Food Prot. 2009, 72, 1267–1271. [Google Scholar] [CrossRef]
- Stephens, T.; Loneragan, G.; Karunasena, E.; Brashears, M. Reduction of Escherichia coli O157 and Salmonella in feces and on hides of feedlot cattle using various doses of a direct-fed microbial. J. Food Prot. 2007, 70, 2386–2391. [Google Scholar] [CrossRef] [PubMed]
- Callejón, R.M.; Rodriguez-Naranjo, M.I.; Ubeda, C.; Hornedo-Ortega, R.; Garcia-Parrilla, M.C.; Troncoso, A.M. Reported foodborne outbreaks due to fresh produce in the United States and European Union: Trends and causes. Foodborne Pathog. Dis. 2015, 12, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Jahid, I.K.; Ha, S.-D. A review of microbial biofilms of produce: Future challenge to food safety. Food Sci. Biotechnol. 2012, 21, 299–316. [Google Scholar] [CrossRef]
- Uhlich, G.A.; Rogers, D.P.; Mosier, D.A. Escherichia coli serotype O157: H7 retention on solid surfaces and peroxide resistance is enhanced by dual-strain biofilm formation. Foodborne Pathog. Dis. 2010, 7, 935–943. [Google Scholar] [CrossRef]
- Chen, D.; Zhao, T.; Doyle, M.P. Single-and mixed-species biofilm formation by Escherichia coli coli O157: H7 and Salmonella, and their sensitivity to levulinic acid plus sodium dodecyl sulfate. Food Control 2015, 57, 48–53. [Google Scholar] [CrossRef]
- Smet, C.; Govaert, M.; Kyrylenko, A.; Easdani, M.; Walsh, J.L.; Van Impe, J.F. Inactivation of single strains of Listeria monocytogenes and Salmonella Typhimurium planktonic cells biofilms with plasma activated liquids. Front Microbiol. 2019, 10, 1539. [Google Scholar] [CrossRef]
- Pletzer, D.; Hancock, R.E. Antibiofilm peptides: Potential as broad-spectrum agents. J. Bacteriol. 2016, 198, 2572–2578. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Hansen, M.F.; Røder, H.L.; Wang, N.; Burmølle, M.; He, G. Mixed-species biofilms in the food industry: Current knowledge and novel control strategies. Crit. Rev. Food Sci. Nutr. 2020, 60, 2277–2293. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhang, R.; Hai, D.; Lu, Z.; Lv, F.; Zhao, H.; Zhang, C.; McAllister, T.A.; Stanford, K.; Bie, X. Antibiofilm activity and modes of action of a novel β-sheet peptide against multidrug-resistant Salmonella enterica. Food Res. Int. 2019, 125, 108520. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Stanford, K.; Bie, X.M.; Niu, Y.D.; McAllister, T.A. Effects of beef juice on biofilm formation by Shiga toxin-producing Escherichia coli on stainless steel. Foodborne Pathog. Dis. 2019, 17, 235–242. [Google Scholar] [CrossRef]
- Kumar, A.; Alam, A.; Rani, M.; Ehtesham, N.Z.; Hasnain, S.E. Biofilms: Survival and defense strategy for pathogens. Int. J. Med. Microbiol. 2017, 307, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Abebe, G.M. The role of bacterial biofilm in antibiotic resistance and food contamination. Int. J. Microbiol. 2020, 2020, 1705814. [Google Scholar] [CrossRef]
- Wagner, E.M.; Pracser, N.; Thalguter, S.; Fischel, K.; Rychli, K. Identification of biofilm hotspots in a meat processing environment: Detection of spoilage bacteria in multi-species biofilms. Int. J. Food Microbiol. 2020, 328, 108668. [Google Scholar] [CrossRef]
- Lim, E.S.; Lee, J.E.; Kim, J.S.; Koo, O.K. Isolation of indigenous bacteria from a cafeteria kitchen and their biofilm formation and disinfectant susceptibility. LWT 2017, 77, 376–382. [Google Scholar] [CrossRef]
- Sadekuzzaman, M.; Yang, S.; Mizan, M.F.; Ha, S.D. Current and recent advanced strategies for combating biofilms. Compr. Rev. Food Sci. Food Saf. 2015, 14, 491–509. [Google Scholar] [CrossRef]
- Wang, J.; Stanford, K.; McAllister, T.A.; Johnson, R.P.; Chen, J.; Hou, H.; Zhang, G.; Niu, Y.D. Biofilm formation, virulence gene profiles, and antimicrobial resistance of nine serogroups of non-O157 Shiga toxin-producing Escherichia coli. Foodborne Pathog. Dis. 2016, 13, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.L.; Sojka, M.; Dibb-Fuller, M.; Woodward, M.J. Effect of pH, temperature and surface contact on the elaboration of fimbriae and flagella by Salmonella serotype enteritidis. J. Med. Microbiol. 1999, 48, 253–261. [Google Scholar] [CrossRef]
- Mitri, S.; Foster, K.R. The genotypic view of social interactions in microbial communities. Annu. Rev. Genet. 2013, 47, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Lories, B.; Roberfroid, S.; Dieltjens, L.; De Coster, D.; Foster, K.R.; Steenackers, H.P. Biofilm bacteria use stress responses to detect and respond to competitors. Curr. Biol. 2020, 30, 1231–1244. [Google Scholar] [CrossRef] [PubMed]
- Reisner, A.; Höller, B.M.; Molin, S.; Zechner, E.L. Synergistic effects in mixed Escherichia coli biofilms: Conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 2006, 188, 3582–3588. [Google Scholar] [CrossRef]
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immuno Med. Microbiol. 2012, 65, 183–195. [Google Scholar] [CrossRef]
- Lappin-Scott, H.M.; Bass, C. Biofilm formation: Attachment, growth, and detachment of microbes from surfaces. Am. J. Infect. Control 2001, 29, 250–251. [Google Scholar] [CrossRef]
- Wang, R.; Kalchayanand, N.; Schmidt, J.W.; Harhay, D.M. Mixed biofilm formation by Shiga toxin-producing Escherichia coli and Salmonella enterica serovar Typhimurium enhanced bacterial resistance to sanitization due to extracellular polymeric substances. J. Food Prot. 2013, 76, 1513–1522. [Google Scholar] [CrossRef]
- Silva, P.L.; Goulart, L.R.; Reis, T.F.; Mendonça, E.P.; Melo, R.T.; Penha, V.A.; Peres, P.A.; Hoepers, P.G.; Beletti, M.E.; Fonseca, B.B. Biofilm formation in different Salmonella serotypes isolated from poultry. Curr. Microbiol. 2019, 76, 124–129. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Z.; Tang, X.; Stanford, K.; Chen, X.; McAllister, T.A.; Niu, Y.D. Single- and Dual-Species Biofilm Formation by Shiga Toxin-Producing Escherichia coli and Salmonella, and Their Susceptibility to an Engineered Peptide WK2. Microorganisms 2021, 9, 2510. https://doi.org/10.3390/microorganisms9122510
Ma Z, Tang X, Stanford K, Chen X, McAllister TA, Niu YD. Single- and Dual-Species Biofilm Formation by Shiga Toxin-Producing Escherichia coli and Salmonella, and Their Susceptibility to an Engineered Peptide WK2. Microorganisms. 2021; 9(12):2510. https://doi.org/10.3390/microorganisms9122510
Chicago/Turabian StyleMa, Zhi, Xia Tang, Kim Stanford, Xiaolong Chen, Tim A. McAllister, and Yan D. Niu. 2021. "Single- and Dual-Species Biofilm Formation by Shiga Toxin-Producing Escherichia coli and Salmonella, and Their Susceptibility to an Engineered Peptide WK2" Microorganisms 9, no. 12: 2510. https://doi.org/10.3390/microorganisms9122510
APA StyleMa, Z., Tang, X., Stanford, K., Chen, X., McAllister, T. A., & Niu, Y. D. (2021). Single- and Dual-Species Biofilm Formation by Shiga Toxin-Producing Escherichia coli and Salmonella, and Their Susceptibility to an Engineered Peptide WK2. Microorganisms, 9(12), 2510. https://doi.org/10.3390/microorganisms9122510