Developing a Live Probiotic Vaccine Based on the Enterococcus faecium L3 Strain Expressing Influenza Neuraminidase
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Analysis
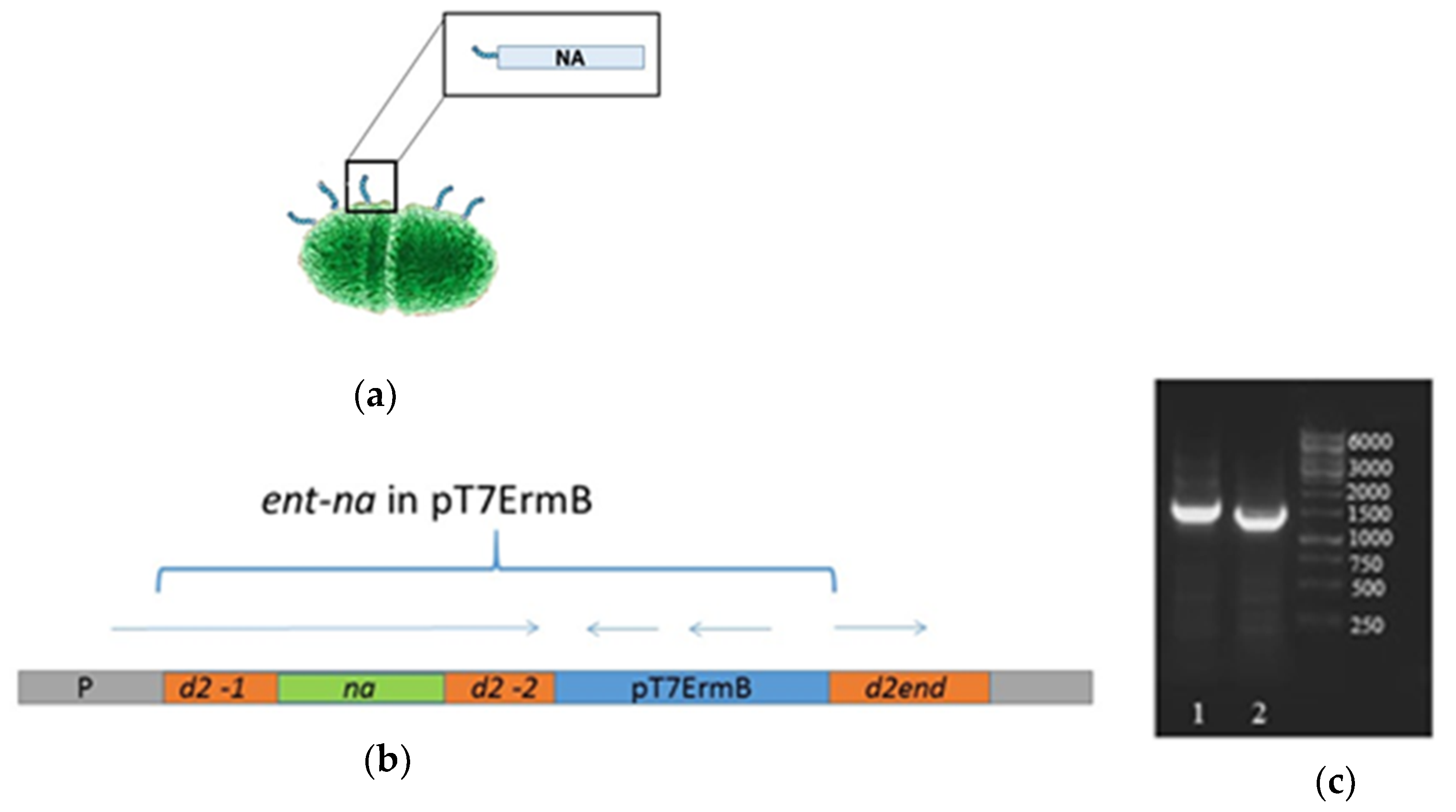
2.2. Obtaining a Chimeric Protein from E. faecium L3 d2 Gene and a Fragment of the NA Gene
2.3. Real-Time Reverse Transcriptase PCR (rRT-PCR)
2.4. Study of Expression of NA Protein on the Surface of E. faecium L3 Using ELISA
2.5. Immunoelectron Microscopy to Study the Structure of E. faecium L3 Pili with Expression of Viral Proteins
2.6. Oral Immunization of Mice with Live Probiotic Vaccine
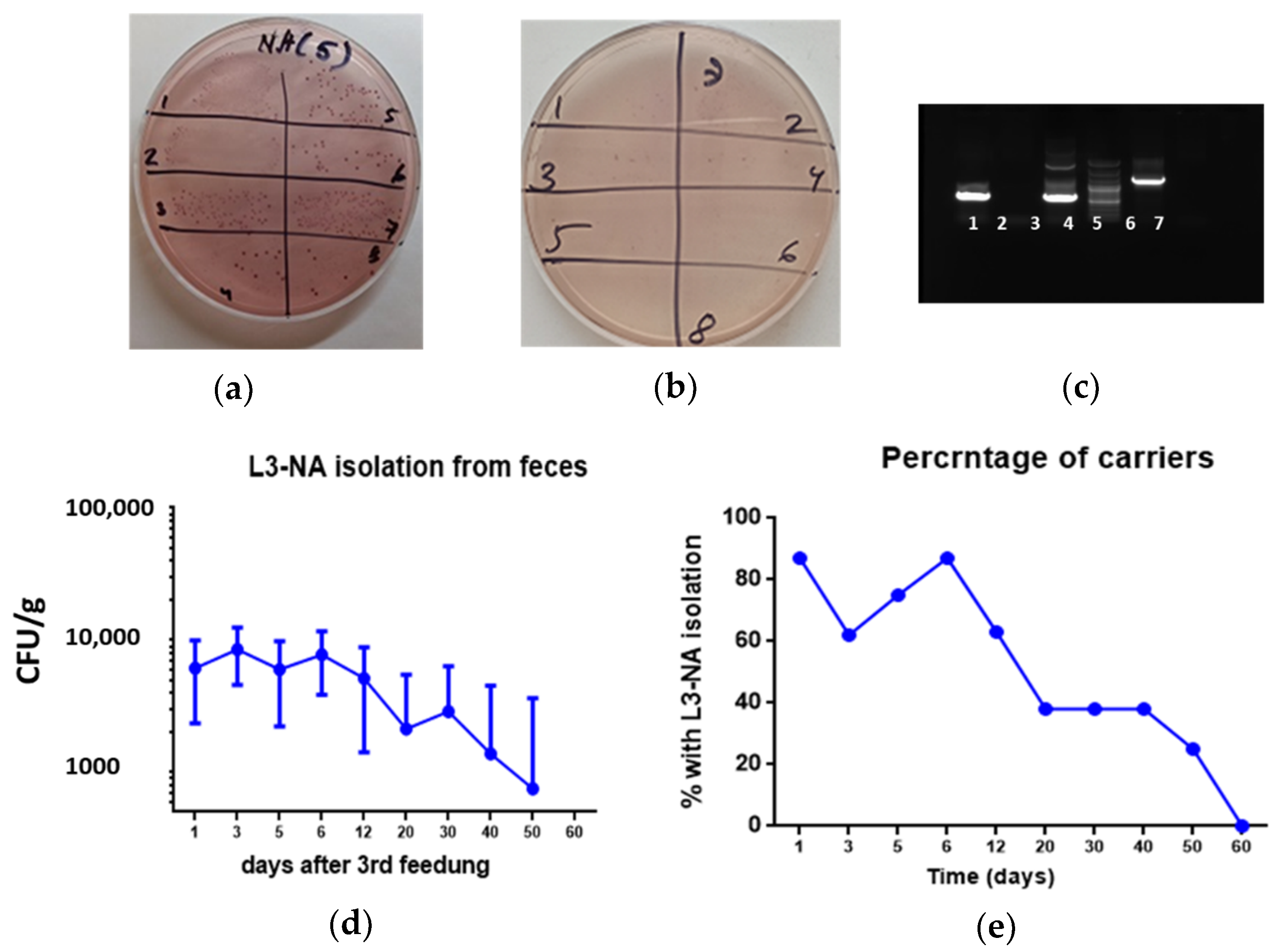
2.7. Evaluation of L3-NA Persistence in the Gastrointestinal Tract of Vaccinated Mice
2.8. Ethic Statement
2.9. Determination of NA-Specific IgG and IgA
2.10. Study of Protection against Influenza Infection after Vaccination with Live Probiotic Vaccine
2.11. Statistics
3. Results
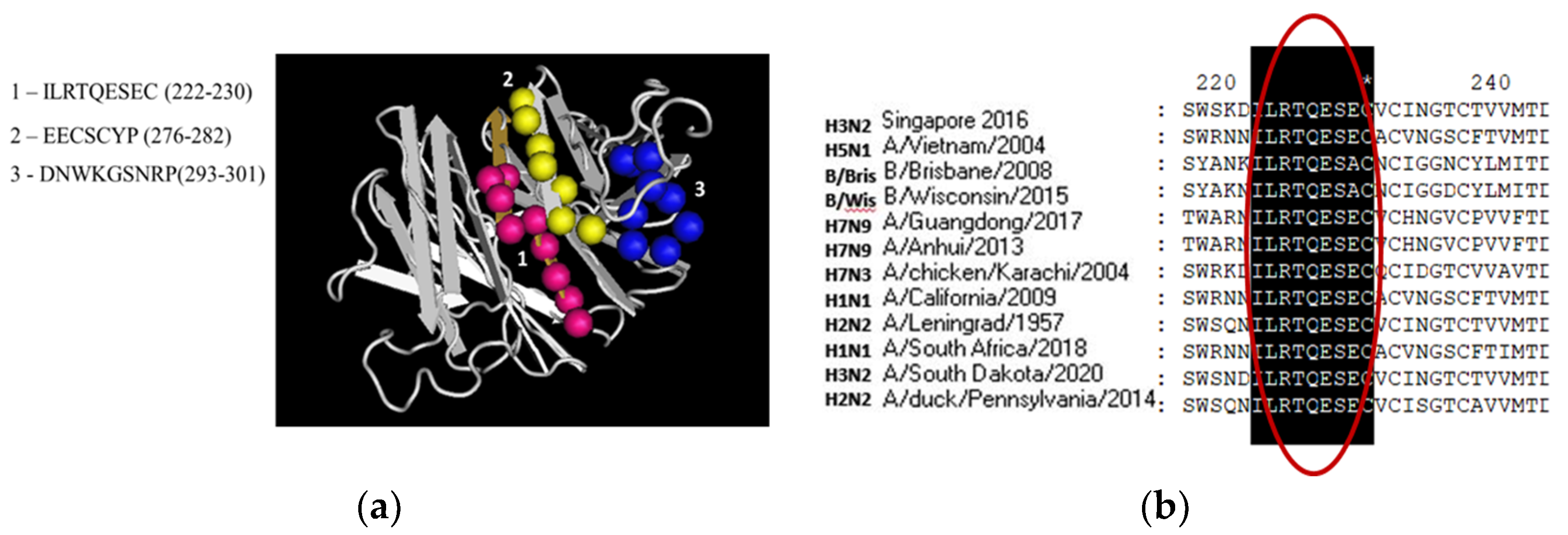
3.1. Molecular Analysis of NA Insertion
3.2. Generation of the Live Probiotic Influenza Vaccine Candidate
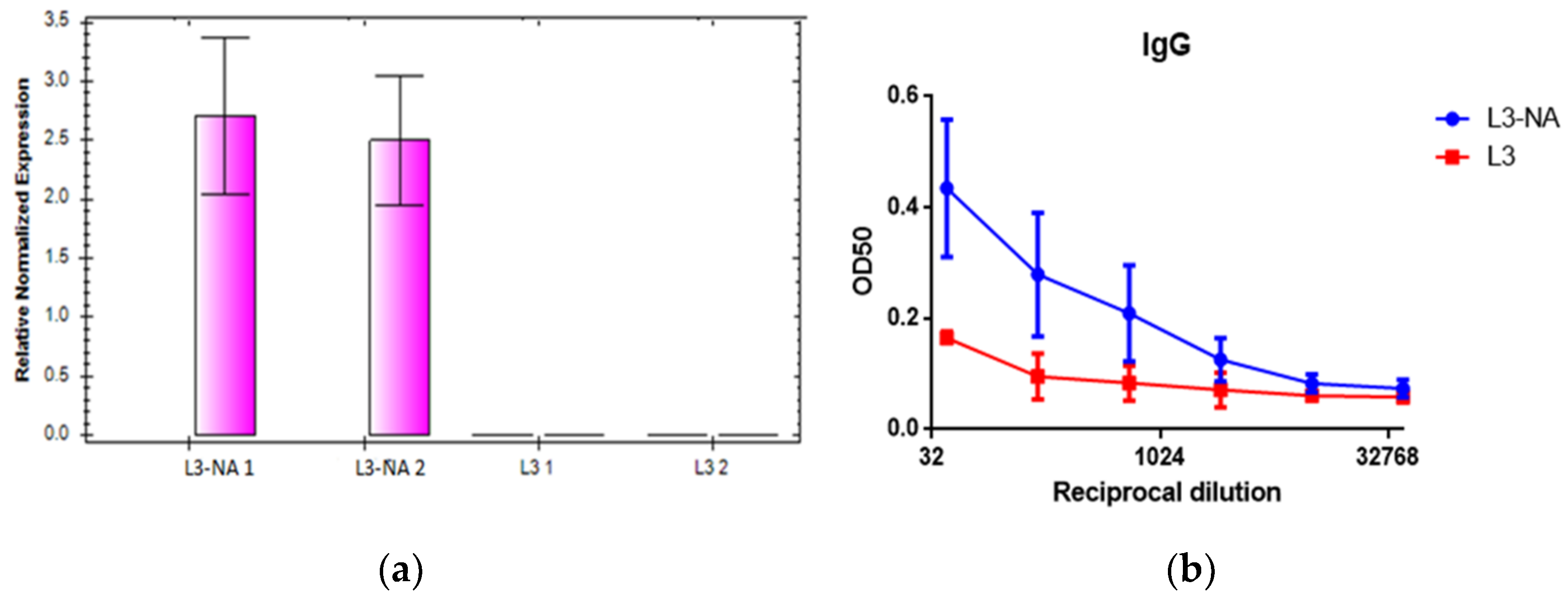
3.3. Expression Levels of the NA mRNA in the Bacteria
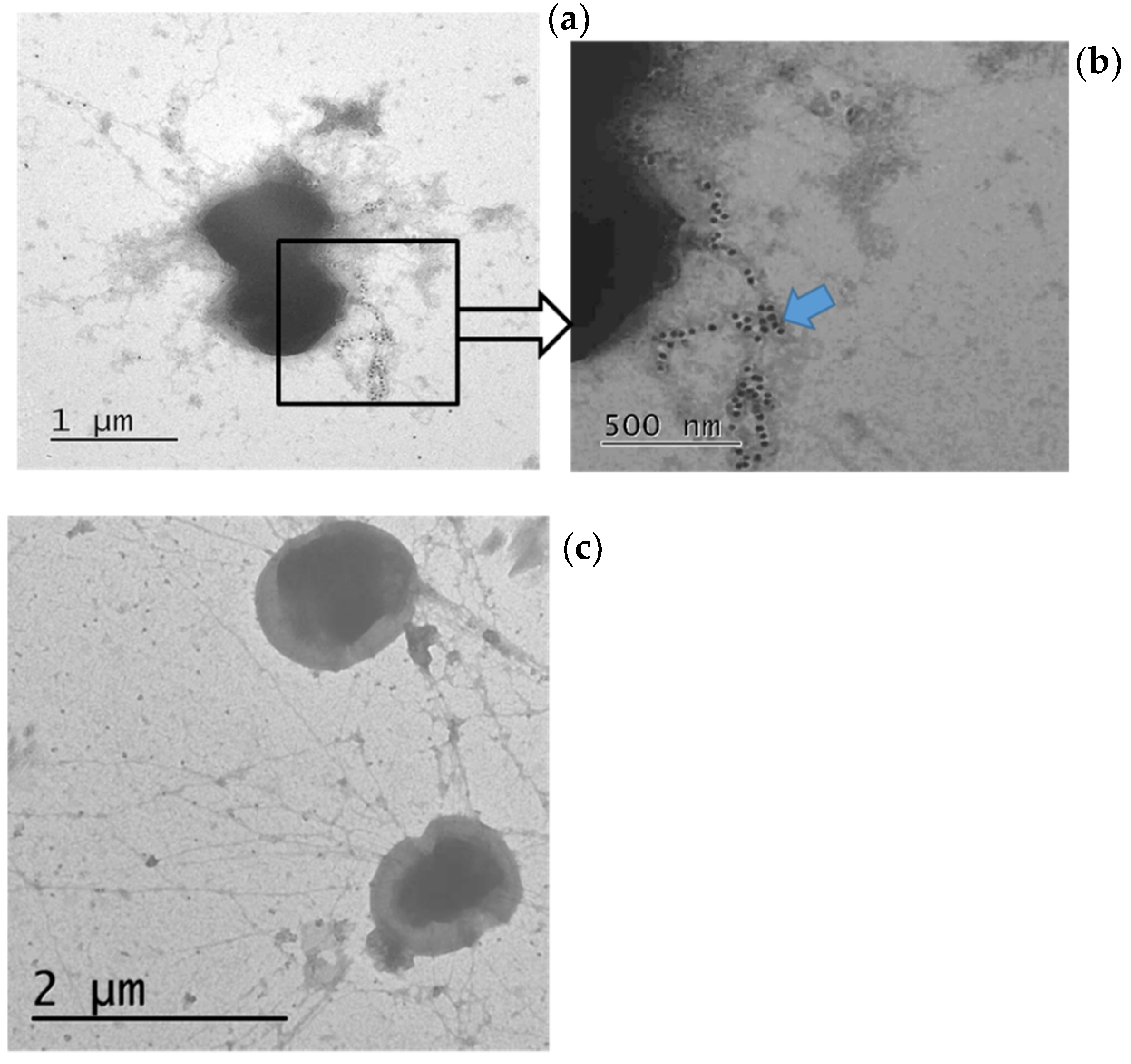
3.4. Immunoelectron Microscopy
3.5. Bacterial Persistence in the Intestines of CBA Mice
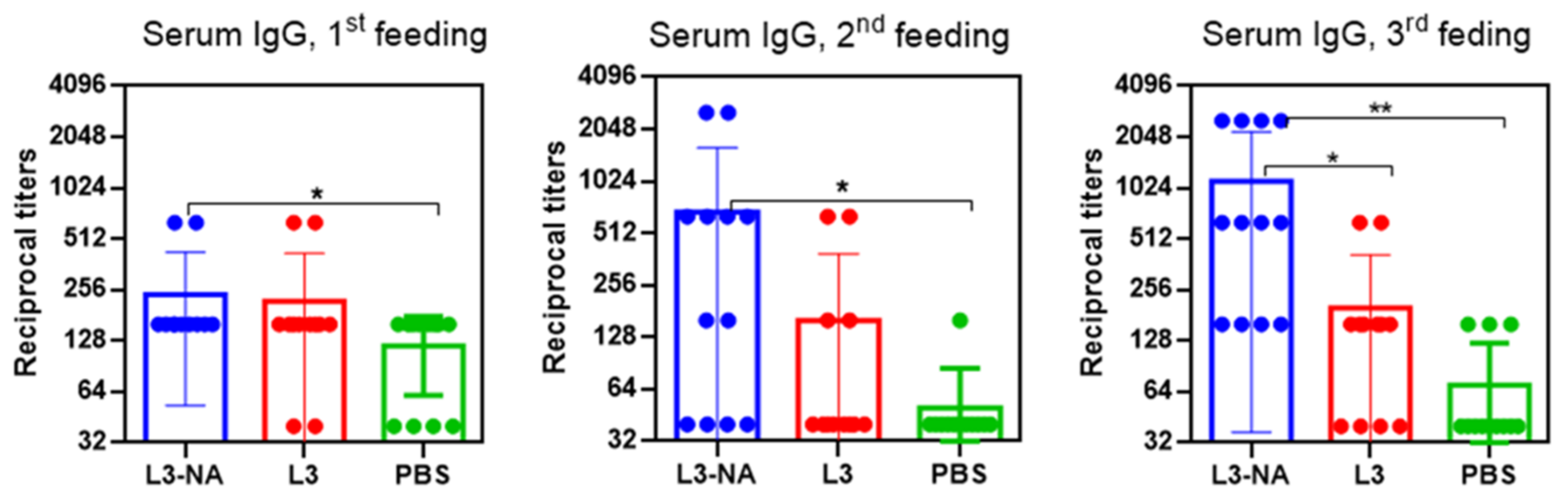
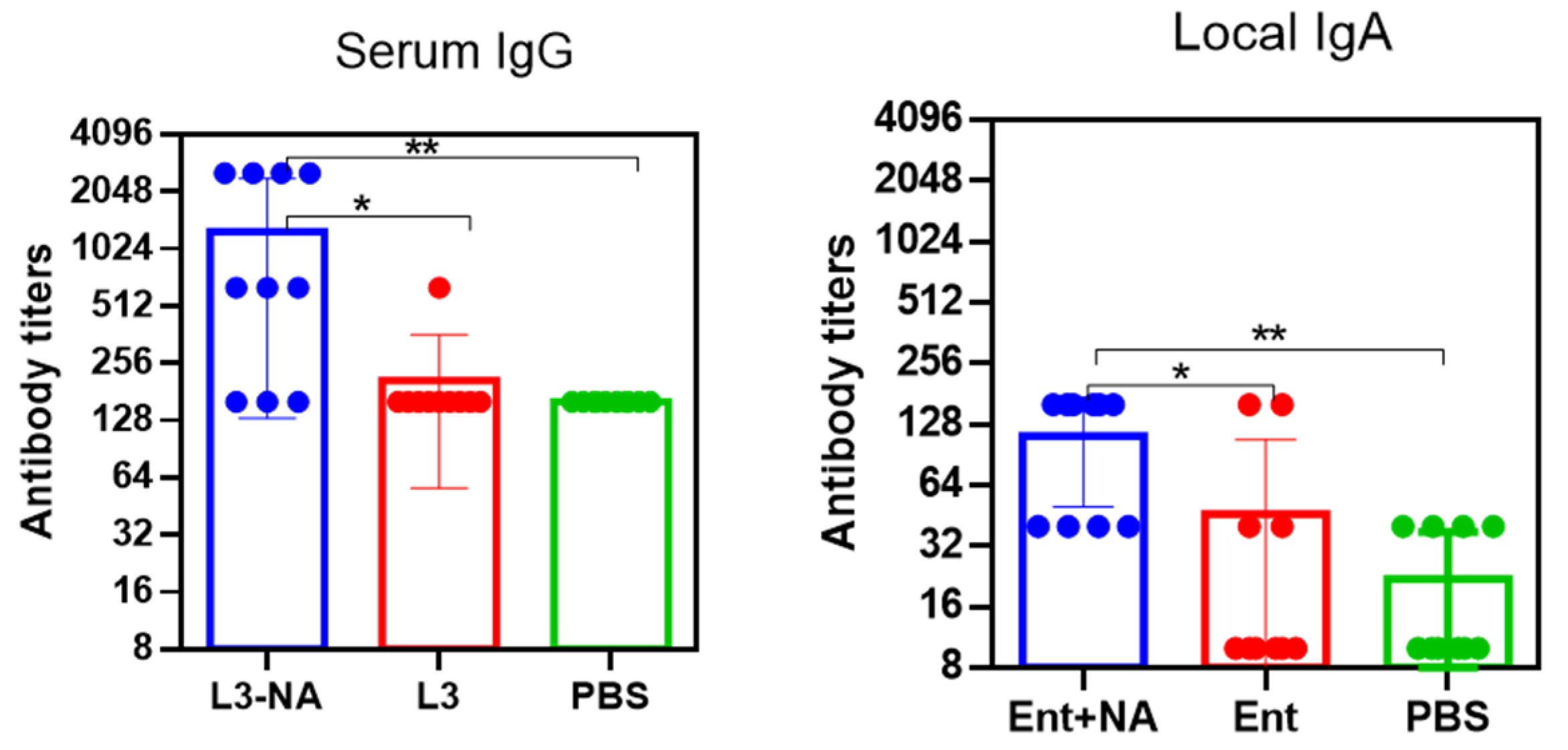
3.6. Immunogenicity in CBA Mice
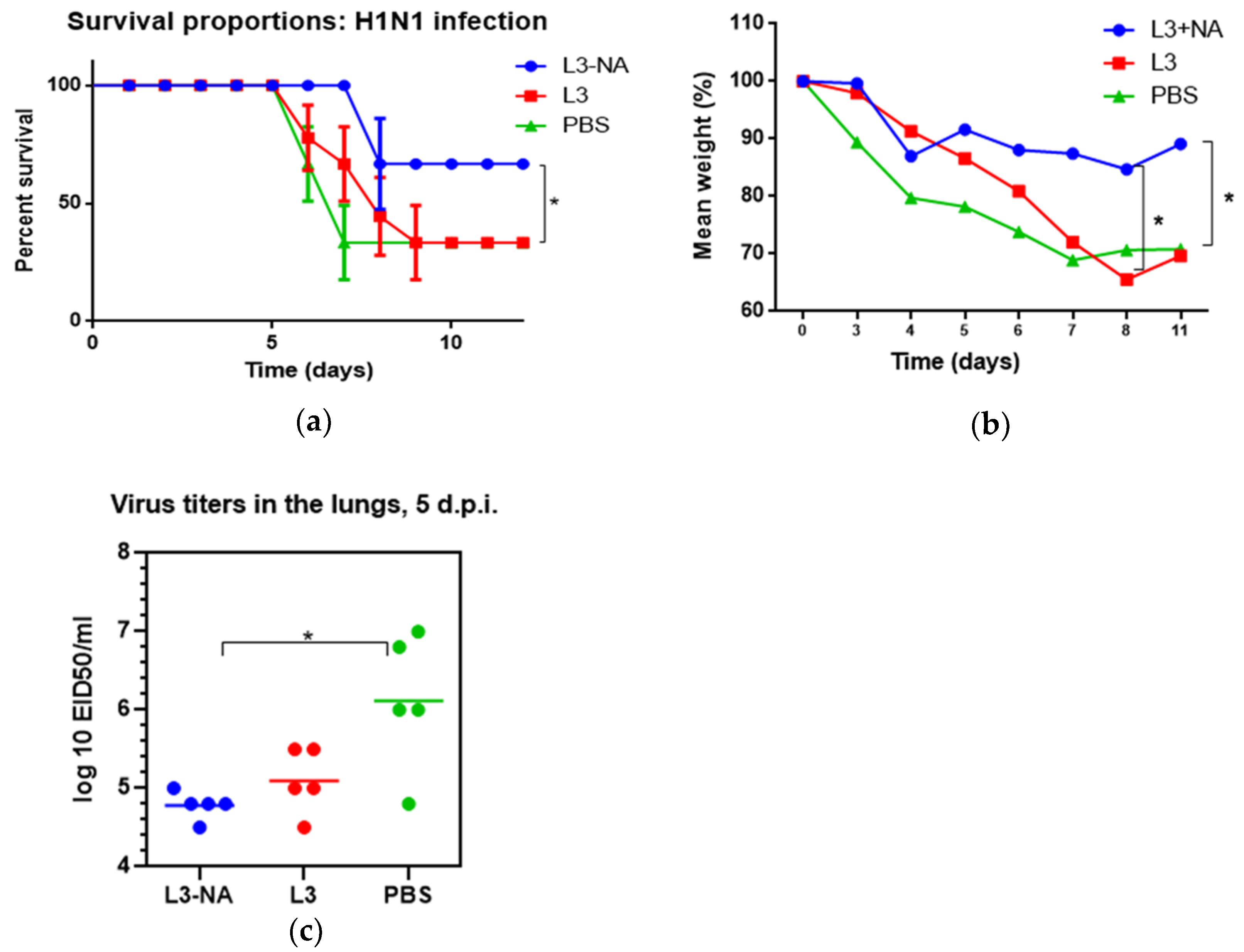
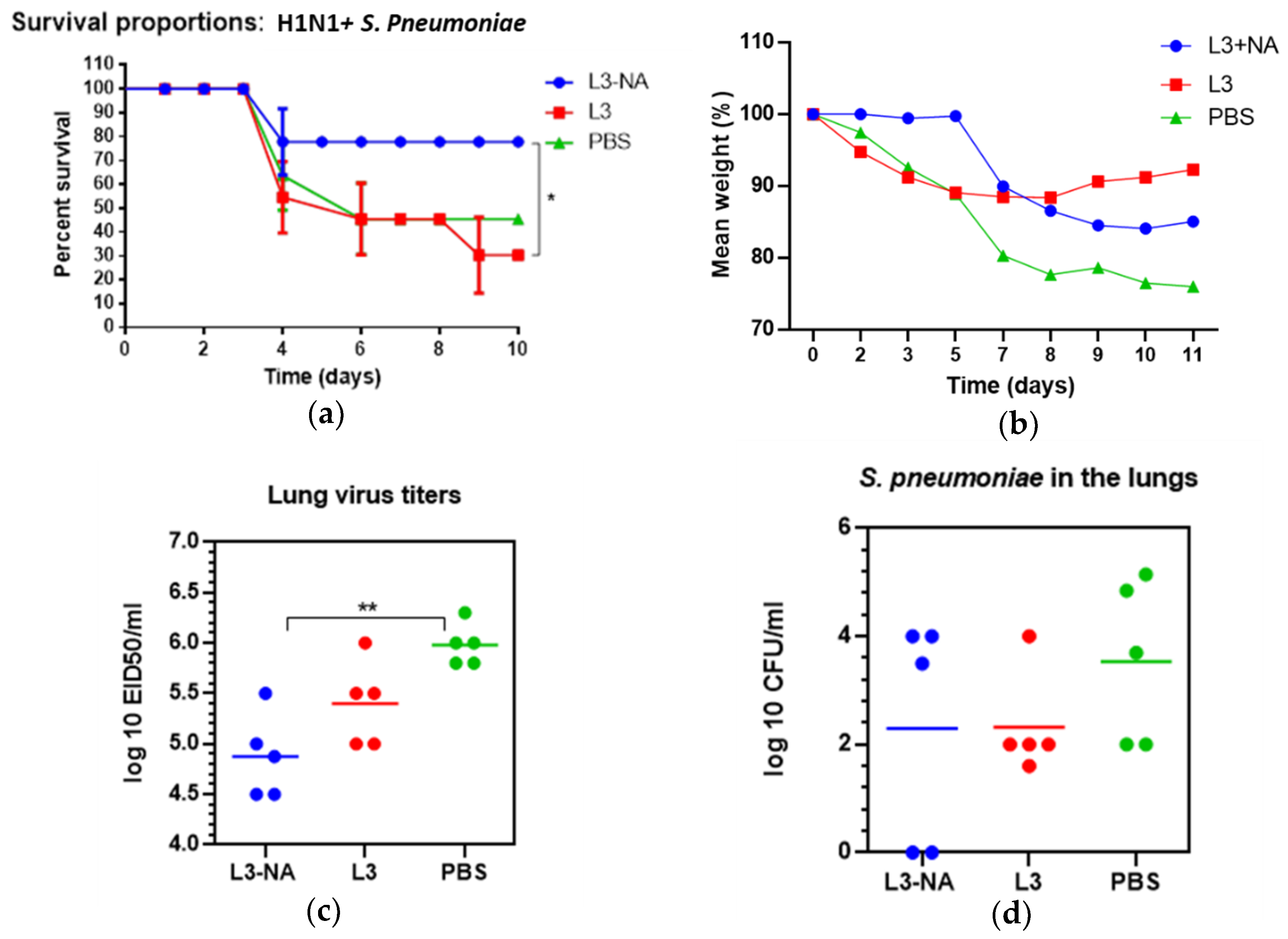
3.7. Protection against Lethal Influenza Infection
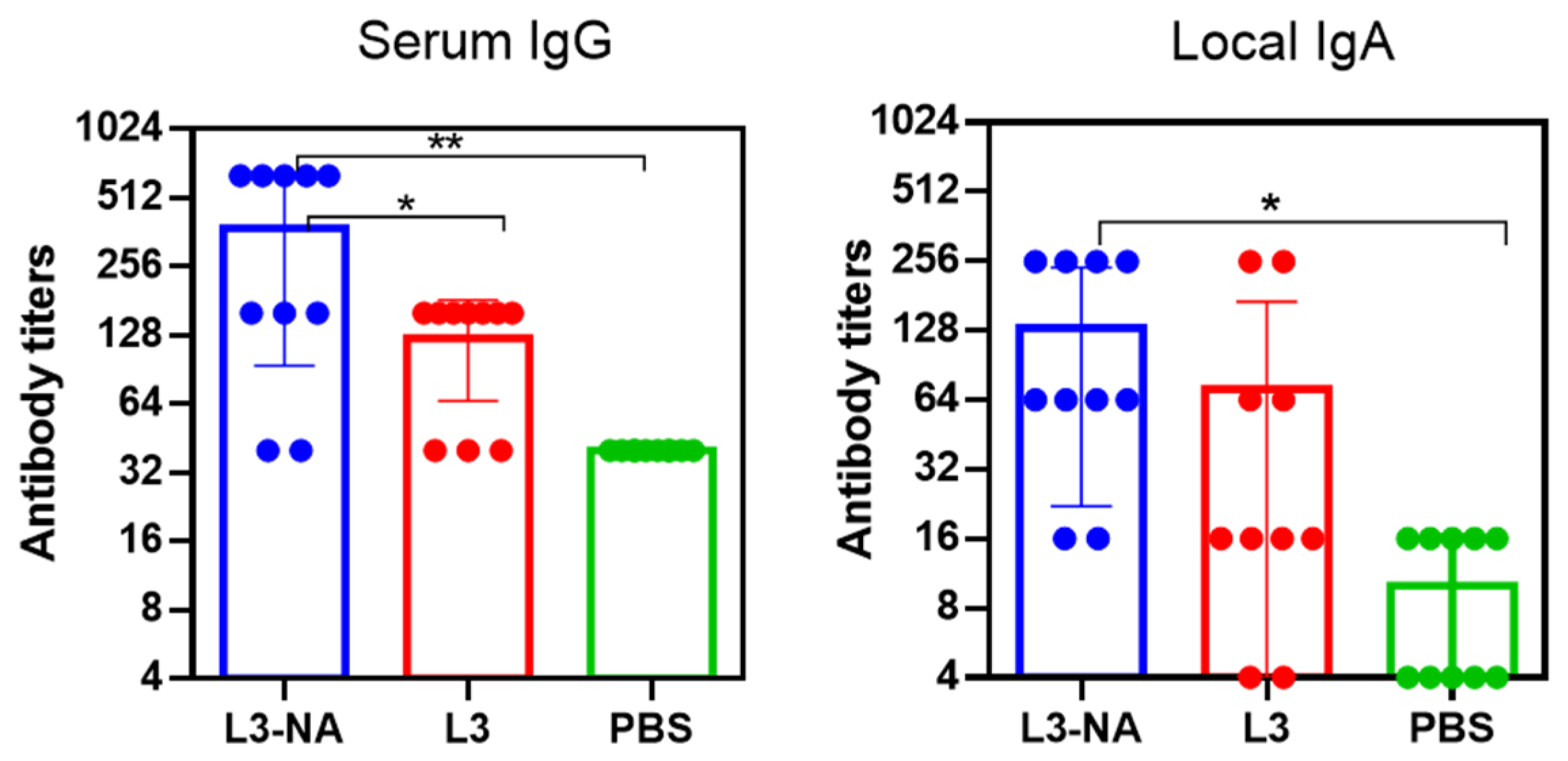
3.8. Immunogenicity and Protection against Virus-Bacterial Infection in Balb/c Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ma, Y.; Luo, Y.; Huang, X.; Song, F.; Liu, G. Construction of Bifidobacterium infantis as a live oral vaccine that expresses antigens of the major fimbrial subunit (CfaB) and the B subunit of heat-labile enterotoxin (LTB) from enterotoxigenic Escherichia coli. Microbiology 2012, 158, 498–504. [Google Scholar] [CrossRef]
- Lei, H.; Peng, X.; Zhao, D.; Ouyang, J.; Jiao, H.; Shu, H.; Ge, X. Lactococcus lactis displayed neuraminidase confers cross protective immunity against influenza A viruses in mice. Virology 2015, 476, 189–195. [Google Scholar] [CrossRef]
- Pebody, R.; McMenamin, J.; Nohynek, H. Live attenuated influenza vaccine (LAIV): Recent effectiveness results from the USA and implications for LAIV programmes elsewhere. Arch. Dis. Child. 2017, 103, 101–105. [Google Scholar] [CrossRef]
- Chung, J.R.; Flannery, B.; Ambrose, C.S.; Bégué, R.E.; Caspard, H.; DeMarcus, L.; Fowlkes, A.L.; Kersellius, G.; Steffens, A.; Fry, A.M.; et al. Live Attenuated and Inactivated Influenza Vaccine Effectiveness. Pediatrics 2019, 143, e20182094. [Google Scholar] [CrossRef]
- Ermolenko, E.I.; Desheva, Y.A.; Kolobov, A.A.; Kotyleva, M.P.; Sychev, I.A.; Suvorov, A.N. Anti–Influenza Activity of Enterocin B In vitro and Protective Effect of Bacteriocinogenic Enterococcal Probiotic Strain on Influenza Infection in Mouse Model. Probiotics Antimicrob. Proteins 2018, 11, 705–712. [Google Scholar] [CrossRef]
- Yermolenko, E.; Chernish, A.; Aleshina, G.; Cvetkova, E.; Martsinkovskaya, I.; Kolodjieva, V.; Suvorov, A. Antagonistic activity of Enterococcus faecium L3 against different groups of pathogenic streptococci. Int. Congr. Ser. 2006, 1289, 363–366. [Google Scholar] [CrossRef]
- Tarasova, E.; Yermolenko, E.; Donets, V.; Sundukova, Z.; Bochkareva, A.; Borschev, I.; Suvorova, M.; Ilyasov, I.; Simanenkov, V.; Suvorov, A. The influence of probiotic Enterococcus faecium strain L5 on the microbiota and cytokines expression in rats with dysbiosis induced by antibiotics. Benef. Microbes 2010, 1, 265–270. [Google Scholar] [CrossRef]
- Karaseva, A.; Tsapieva, A.; Pachebat, J.; Suvorov, A. Draft Genome Sequence of Probiotic Enterococcus faecium Strain L-3. Genome Announc. 2016, 4, e01622-15. [Google Scholar] [CrossRef]
- Telford, J.L.; Barocchi, M.A.; Margarit, I.; Rappuoli, R.; Grandi, G. Pili in Gram-positive pathogens. Nat. Rev. Genet. 2006, 4, 509–519. [Google Scholar] [CrossRef]
- Danne, C.; Dramsi, S. Pili of Gram-positive bacteria: Roles in host colonization. Res. Microbiol. 2012, 163, 645–658. [Google Scholar] [CrossRef]
- Lu, X.; Edwards, L.E.; Desheva, Y.; Nguyen, D.C.; Rekstin, A.; Stephenson, I.; Szretter, K.; Cox, N.J.; Rudenko, L.; Klimov, A.; et al. Cross-protective immunity in mice induced by live-attenuated or inactivated vaccines against highly pathogenic influenza A (H5N1) viruses. Vaccine 2006, 24, 6588–6593. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Couzens, L.; Burke, D.F.; Wan, H.; Wilson, P.; Memoli, M.J.; Eichelberger, M.C. Antigenic drift of the influenza A (H1N1) pdm09 virus neuraminidase results in reduced effectiveness of A/California/7/2009 (H1N1pdm09)-specific antibodies. Mbio 2019, 10, e00307-19. [Google Scholar] [CrossRef] [PubMed]
- Wohlbold, T.J.; Krammer, F. In the shadow of hemagglutinin: A growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses 2014, 6, 2465–2494. [Google Scholar] [CrossRef]
- Giurgea, L.T.; Morens, D.M.; Taubenberger, J.K.; Memoli, M.J. Influenza Neuraminidase: A Neglected Protein and Its Potential for a Better Influenza Vaccine. Vaccines 2020, 8, 409. [Google Scholar] [CrossRef]
- Eichelberger, M.C.; Monto, A.S. Neuraminidase, the Forgotten Surface Antigen, Emerges as an Influenza Vaccine Target for Broadened Protection. J. Infect. Dis. 2019, 219, S75–S80. [Google Scholar] [CrossRef]
- Krammer, F.; Fouchier, R.A.M.; Eichelberger, M.C.; Webby, R.J.; Shaw-Saliba, K.; Wan, H.; Wilson, P.C.; Compans, R.W.; Skountzou, I.; Monto, A.S. NAction! How Can Neuraminidase-Based Immunity Contribute to Better Influenza Virus Vaccines? MBio 2018, 9, e02332-17. [Google Scholar] [CrossRef]
- Eichelberger, M.C.; Morens, D.M.; Taubenberger, J.K. Neuraminidase as an influenza vaccine antigen: A low hanging fruit, ready for picking to improve vaccine effectiveness. Curr. Opin. Immunol. 2018, 53, 38–44. [Google Scholar] [CrossRef]
- Quan, F.-S.; Kim, M.-C.; Lee, B.-J.; Song, J.-M.; Compans, R.; Kang, S.-M. Influenza M1 VLPs containing neuraminidase induce heterosubtypic cross-protection. Virology 2012, 430, 127–135. [Google Scholar] [CrossRef]
- Bui, C.; Bethmont, A.; Chughtai, A.; Gardner, L.; Sarkar, S.; Hassan, S.; Seale, H.; MacIntyre, C.R. A Systematic Review of the Comparative Epidemiology of Avian and Human Influenza A H5N1 and H7N9—Lessons and Unanswered Questions. Transbound. Emerg. Dis. 2015, 63, 602–620. [Google Scholar] [CrossRef]
- Wang, Y.; Geer, L.; Chappey, C.; Kans, J.A.; Bryant, S.H. Cn3D: Sequence and structure views for Entrez. Trends Biochem. Sci. 2000, 25, 300–302. [Google Scholar] [CrossRef]
- Chen, J.; Liu, H.; Yang, J.; Chou, K.-C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 2007, 33, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Raghava, G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins: Struct. Funct. Bioinform. 2006, 65, 40–48. [Google Scholar] [CrossRef]
- Larsen, J.E.P.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sweredoski, M.J.; Baldi, P. COBEpro: A novel system for predicting continuous B-cell epitopes. Protein Eng. Des. Sel. 2008, 22, 113–120. [Google Scholar] [CrossRef]
- Ponomarenko, J.V.; Bui, H.-H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef]
- Rubinstein, N.D.; Mayrose, I.; Martz, E.; Pupko, T. Epitopia: A web-server for predicting B-cell epitopes. BMC Bioinform. 2009, 10, 287. [Google Scholar] [CrossRef]
- Gupalova, T.; Leontieva, G.; Kramskaya, T.; Grabovskaya, K.; Kuleshevich, E.; Suvorov, A. Development of experimental pneumococcal vaccine for mucosal immunization. PLoS ONE 2019, 14, e0218679. [Google Scholar] [CrossRef]
- Waterborg, J.H. The Lowry method for protein quantitation. In The Protein Protocols Handbook; Humana Press: Totowa, NJ, USA, 2009; pp. 7–10. [Google Scholar]
- Desheva, Y.; Smolonogina, T.; Donina, S.; Rudenko, L. Study of Neuraminidase-Inhibiting Antibodies in Clinical Trials of Live Influenza Vaccines. Antibodies 2020, 9, 20. [Google Scholar] [CrossRef]
- Silver, L.M. Mouse Genetics: Concepts and Applications; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Huang, P.; Xu, Y.; Ni, H.; Zhong, J.; Zhang, X.; Tan, S.; Wu, D.; Qiu, B.; Guan, D.; Wen, M.; et al. Linear B-cell epitope mapping of neuraminidases of the 2009 A H1N1 viruses based on immunoinformatics. Vaccine 2011, 29, 1278–1282. [Google Scholar] [CrossRef]
- Melzig, M.F.; Schwerdtfeger, S.M. Sialidases in biological systems. Die Pharm.-Int. J. Pharm. Sci. 2010, 65, 551–561. [Google Scholar]
- Regev-Yochay, G.; Raz, M.; Dagan, R.; Porat, N.; Shainberg, B.; Pinco, E.; Keller, N.; Rubinstein, E. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin. Infect. Dis. 2004, 38, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.M.; Hashem, A.M.; Li, C.; Van Domselaar, G.; Larocque, L.; Wang, J.; Smith, D.; Cyr, T.; Farnsworth, A.; He, R.; et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antivir. Res. 2013, 100, 567–574. [Google Scholar] [CrossRef]
- Gottlieb, T.; Ben-Yedidia, T. Epitope-based approaches to a universal influenza vaccine. J. Autoimmun. 2014, 54, 15–20. [Google Scholar] [CrossRef]
- Herrera-Rodriguez, J.; Meijerhof, T.; Niesters, H.G.; Stjernholm, G.; Hovden, A.-O.; Sørensen, B.; Ökvist, M.; Sommerfelt, M.A.; Huckriede, A. A novel peptide-based vaccine candidate with protective efficacy against influenza A in a mouse model. Virology 2018, 515, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Soema, P.C.; Huber, S.K.R.; Willems, G.-J.; Jacobi, R.; Hendriks, M.; Soethout, E.; Jiskoot, W.; de Jonge, J.; Van Beek, J.; Kersten, G.F.A.; et al. Whole-Inactivated Influenza Virus Is a Potent Adjuvant for Influenza Peptides Containing CD8+ T Cell Epitopes. Front. Immunol. 2018, 9, 525. [Google Scholar] [CrossRef]
- Bermúdez-Humarán, L.G.; Kharrat, P.; Chatel, J.-M.; Langella, P. Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb. Cell Factories 2011, 10, S4. [Google Scholar] [CrossRef]
- De Azevedo, M.; Karczewski, J.; Lefévre, F.; Azevedo, V.; Miyoshi, A.; Wells, J.M.; Langella, P.; Chatel, J.-M. In vitro and in vivo characterization of DNA delivery using recombinant Lactococcus lactis expressing a mutated form of L. monocytogenes Internalin A. BMC Microbiol. 2012, 12, 299. [Google Scholar] [CrossRef] [PubMed]
- Gupalova, T.; Leontieva, G.; Kramskaya, T.; Grabovskaya, K.; Bormotova, E.; Korjevski, D.; Suvorov, A. Development of experimental GBS vaccine for mucosal immunization. PLoS ONE 2018, 13, e0196564. [Google Scholar] [CrossRef]
- Smith, G.E.; Sun, X.; Bai, Y.; Liu, Y.V.; Massare, M.J.; Pearce, M.B.; Belser, J.A.; Maines, T.R.; Creager, H.; Glenn, G.M.; et al. Neuraminidase-based recombinant virus-like particles protect against lethal avian influenza A(H5N1) virus infection in ferrets. Virology 2017, 509, 90–97. [Google Scholar] [CrossRef]
- Kramskaya, T.; Leontieva, G.; Desheva, Y.; Grabovskaya, K.; Gupalova, T.; Rudenko, L.; Suvorov, A. Combined immunization with attenuated live influenza vaccine and chimeric pneumococcal recombinant protein improves the outcome of virus-bacterial infection in mice. PLoS ONE 2019, 14, e0222148. [Google Scholar] [CrossRef]
- Taghinezhad, S.S.; Mohseni, A.H.; Bermúdez-Humarán, L.G.; Casolaro, V.; Cortes-Perez, N.G.; Keyvani, H.; Simal-Gandara, J. Probiotic-Based Vaccines May Provide Effective Protection against COVID-19 Acute Respiratory Disease. Vaccines 2021, 9, 466. [Google Scholar] [CrossRef] [PubMed]
| Subtype/Lineage | Name | Sequence Number |
|---|---|---|
| H2N2 | A/Leningrad/134/17/1957 | L37330 |
| H2N2 | A/duck/Pennsylvania/14-030488-005/2014 | QHQ82622 |
| H3N2 | A/17/Singapore/2016/3571 | EPI1313135 |
| H3N2 | A/South Dakota/12/2020 | MT467157 |
| H1N1 | A/California/07/2009 | HM138502 |
| H1N1 | A/South Africa/A201-004-041/2018 | MN716456 |
| H5N1 | A/Viet Nam/1203/2004 | ABP52008 |
| H7N3 | A/chicken/Karachi/NARC-100/2004 | FJ577544 |
| H7N9 | A/Anhui/1-DEWH730/2013 | CY187620 |
| H7N9 | A/Guangdong/HP001/2017 | AQY18948 |
| B/Victoria | B/Brisbane/60/2008 | AFH57913 |
| B/Yamagata | B/Wisconsin/04/2015 | ALH38476 |
| Primers | Direction | Nucleotide Sequence from 5′ to 3′ |
|---|---|---|
| EV | forward | GCATATGGCGATCCTGGCAAGTGTTAT 1 |
| FV | reverse | GGAATTCCTAGAGCTGTCGTCGTTCC |
| A1 | forward | GCTCTAGAGCCGATGAGAGCAGCTGGTATTG |
| D1 | reverse | CAACAGGATCCAAAGCATCGTTGG |
| B1 | forward | TGAGTGAACCACAGCCAGAA |
| Seq F | forward | GGACACCACAACCATCGAAG |
| Seq R | reverse | AGCTGGACCATGCTACACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desheva, Y.; Leontieva, G.; Kramskaya, T.; Gupalova, T.; Losev, I.; Kuleshevich, E.; Bormotova, E.; Kopteva, O.; Kudar, P.; Suvorov, A. Developing a Live Probiotic Vaccine Based on the Enterococcus faecium L3 Strain Expressing Influenza Neuraminidase. Microorganisms 2021, 9, 2446. https://doi.org/10.3390/microorganisms9122446
Desheva Y, Leontieva G, Kramskaya T, Gupalova T, Losev I, Kuleshevich E, Bormotova E, Kopteva O, Kudar P, Suvorov A. Developing a Live Probiotic Vaccine Based on the Enterococcus faecium L3 Strain Expressing Influenza Neuraminidase. Microorganisms. 2021; 9(12):2446. https://doi.org/10.3390/microorganisms9122446
Chicago/Turabian StyleDesheva, Yulia, Galina Leontieva, Tatiana Kramskaya, Tatiana Gupalova, Igor Losev, Eugenia Kuleshevich, Elena Bormotova, Olga Kopteva, Polina Kudar, and Alexander Suvorov. 2021. "Developing a Live Probiotic Vaccine Based on the Enterococcus faecium L3 Strain Expressing Influenza Neuraminidase" Microorganisms 9, no. 12: 2446. https://doi.org/10.3390/microorganisms9122446
APA StyleDesheva, Y., Leontieva, G., Kramskaya, T., Gupalova, T., Losev, I., Kuleshevich, E., Bormotova, E., Kopteva, O., Kudar, P., & Suvorov, A. (2021). Developing a Live Probiotic Vaccine Based on the Enterococcus faecium L3 Strain Expressing Influenza Neuraminidase. Microorganisms, 9(12), 2446. https://doi.org/10.3390/microorganisms9122446







