Role of Bifidobacteria on Infant Health
Abstract
1. The Genus Bifidobacterium: A Landmark of the Healthy Breastfed Infant
2. The Genus Bifidobacterium and Neonatal Health
2.1. Starting Foundation of Health: Bifidobacteria in the Dyad Mother-Infant
2.2. Bifidobacterial Features Contributing to Beneficial Action in the Early Gut
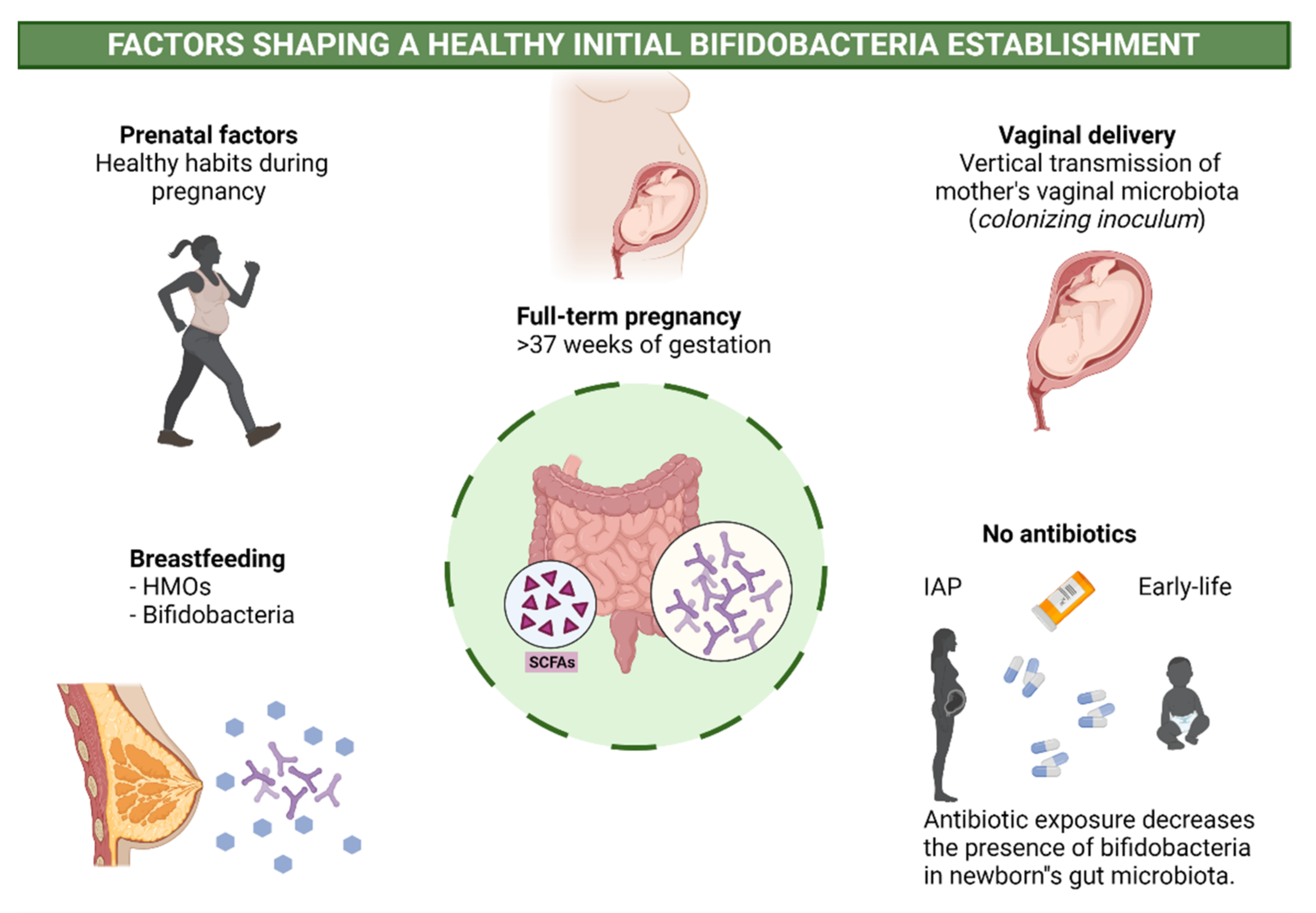
2.3. Perinatal Factors Affecting Bifidobacteria and Health
3. The Genus Bifidobacterium and Child Development
3.1. Bifidobacterium Genus as a Biomarker of Metabolic Diseases in Infanthood
3.2. Maturation of the Immune System: Early Bifidobacterial Alterations Leading to Atopy
3.3. Brain Development and Social Behavior: New Target to Study the Significance of Bifidobacterium in Infant Health
4. The Genus Bifidobacterium as a Probiotic for Infants
4.1. Animal Models
4.2. Clinical Trials
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Palacio, S.D.; Montes, S.A.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Tissier, H. Recherchers Sur La Flora Intestinale Normale Et Pathologique Du Nourisson. Ph.D. Thesis, University of Paris, Paris, France, 1900. [Google Scholar]
- Scardovi, V. Genus Bifidobacterium Orla-Jensen 1924, 472al; Williams and Wilkins: Baltimore, MD, USA, 1986. [Google Scholar]
- Alessandri, G.; van Sinderen, D.; Ventura, M. The genus Bifidobacterium: From genomics to functionality of an important component of the mammalian gut microbiota. Comput. Struct. Biotechnol. J. 2021, 19, 1472–1487. [Google Scholar] [CrossRef]
- Ahn, J.B.; Hwang, H.J.; Park, J.H. Physiological responses of oxygen-tolerant anaerobic bifidobacterium longum under oxygen. J. Microbiol. Biotechnol. 2001, 11, 443–451. [Google Scholar]
- Mattarelli, B. The Family Bifidobacteriaceae, 3rd ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Picard, C.; Fioramonti, J.; Francois, A.; Robinson, T.; Neant, F.; Matuchansky, C. Review article: Bifidobacteria as probiotic agents–physiological effects and clinical benefits. Aliment. Pharmacol. Ther. 2005, 22, 495–512. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M.M. Chapter 12—Bifidobacteria as probiotic organisms: An introduction. In The Microbiota in Gastrointestinal Pathophysiology; Floch, M.H., Ringel, Y., Allan Walker, W., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 125–126. [Google Scholar]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef] [PubMed]
- Lewis, Z.T.; Mills, D.A. Differential Establishment of Bifidobacteria in the Breastfed Infant Gut; Karger, A.G., Ed.; Karger Publishers: Basel, Switzerland, 2017; pp. 149–160. [Google Scholar]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Vaiserman, A.M.; Koliada, A.; Marotta, F. Gut microbiota: A player in aging and a target for anti-aging intervention. Ageing Res. Rev. 2017, 35, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Salazar, N.; Arboleya, S.; Fernández-Navarro, T.; Reyes-Gavilán, C.G.D.L.; Gonzalez, S.; Gueimonde, M. Age-Associated Changes in Gut Microbiota and Dietary Components Related with the Immune System in Adulthood and Old Age: A Cross-Sectional Study. Nutrients 2019, 11, 1765. [Google Scholar] [CrossRef] [PubMed]
- Gueimonde, M.; Ouwehand, A.; Pitkälä, K.; Strandberg, T.; Finne-Soveri, H.; Salminen, S. Fecal Bifidobacterium Levels in Elderly Nursing Home Patients. Biosci. Microflora 2010, 29, 111–113. [Google Scholar] [CrossRef]
- Karlsson, C.L.J.; Molin, G.; Cilio, C.M.; Ahrné, S. The Pioneer Gut Microbiota in Human Neonates Vaginally Born at Term—A Pilot Study. Pediatr. Res. 2011, 70, 282–286. [Google Scholar] [CrossRef]
- Aggett, P.J.; Agostoni, C.; Axelsson, I.; Edwards, C.A.; Goulet, O.; Hernell, O.; Koletzko, B.; Lafeber, H.N.; Micheli, J.-L.; Michaelsen, K.F.; et al. Nondigestible Carbohydrates in the Diets of Infants and Young Children: A Commentary by the ESPGHAN Committee on Nutrition. J. Pediatr. Gastroenterol. Nutr. 2003, 36, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, M.; Wu, S.; Lebrilla, C.B.; Chapkin, R.; Ivanov, I.; Donovan, S.M. Fecal Microbiota Composition of Breast-Fed Infants Is Correlated With Human Milk Oligosaccharides Consumed. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 825–833. [Google Scholar] [CrossRef]
- Solís, G.; Reyes-Gavilan, C.D.L.; Fernández, N.; Margolles, A.; Gueimonde, M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 2010, 16, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Langa, S.; Reviriego, C.; Jiménez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 754–758. [Google Scholar] [CrossRef]
- Collado, M.; Delgado, S.; Maldonado, A.; Rodríguez, J. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett. Appl. Microbiol. 2009, 48, 523–528. [Google Scholar] [CrossRef]
- Gueimonde, M.; Laitinen, K.; Salminen, S.; Isolauri, E. Breast Milk: A Source of Bifidobacteria for Infant Gut Development and Maturation? Neonatology 2007, 92, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Lyons, K.E.; Ryan, C.A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients 2020, 12, 1039. [Google Scholar] [CrossRef]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z.; et al. Bifidobacteria-mediated immune system imprinting early in life. Cell 2021, 184, 3884–3898. [Google Scholar] [CrossRef]
- Victora, C.G.; Bahl, R.; Barros, A.J.D.; França, G.V.A.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef]
- Lapidaire, W.; Lucas, A.; Clayden, J.D.; Clark, C.; Fewtrell, M.S. Human milk feeding and cognitive outcome in preterm infants: The role of infection and NEC reduction. Pediatr. Res. 2021, 1–8. [Google Scholar] [CrossRef]
- Tojo, R.; Suárez, A.; Clemente, M.G.; de los Reyes-Gavilán, C.G.; Margolles, A.; Gueimonde, M.; Ruas-Madiedo, P. Intestinal microbiota in health and disease: Role of bifidobacteria in gut homeostasis. World J. Gastroenterol. 2014, 20, 15163–15176. [Google Scholar] [CrossRef]
- Kalliomäki, M.; Collado, M.C.; Salminen, S.; Isolauri, E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; LeVan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Capilla, A.; Nova, E.; Castillejo, G.; Varea, V.; Pozo, T.; Garrote, J.A.; Polanco, I.; López, A.; Ribes-Koninckx, C.; et al. Influence of Milk-Feeding Type and Genetic Risk of Developing Coeliac Disease on Intestinal Microbiota of Infants: The PROFICEL Study. PLoS ONE 2012, 7, e30791. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.; Isolauri, E.; He, F.; Hashimoto, H.; Benno, Y.; Salminen, S. Differences in Bifidobacterium flora composition in allergic and healthy infants. J. Allergy Clin. Immunol. 2001, 108, 144–145. [Google Scholar] [CrossRef] [PubMed]
- Saturio, S.; Nogacka, A.; Suárez, M.; Fernández, N.; Mantecón, L.; Mancabelli, L.; Milani, C.; Ventura, M.; Reyes-Gavilán, C.D.L.; Solís, G.; et al. Early-Life Development of the Bifidobacterial Community in the Infant Gut. Int. J. Mol. Sci. 2021, 22, 3382. [Google Scholar] [CrossRef]
- Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA—Opinion of the Scientific Committee. Eur. Food Saf. Auth. J. 2007, 5. [CrossRef]
- Gueimonde, M.; Arboleya, S. Resistance of Bifidobacteria toward Antibiotics; Springer: Berlin, Gemany, 2021; Volume 2278, pp. 195–208. [Google Scholar] [CrossRef]
- Turroni, F.; Marchesi, J.R.; Foroni, E.; Gueimonde, M.; Shanahan, F.; Margolles, A.; van Sinderen, D.; Ventura, M. Microbiomic analysis of the bifidobacterial population in the human distal gut. Multidiscip. J. Microb. Ecol. 2009, 3, 745–751. [Google Scholar] [CrossRef]
- Masco, L.; Huys, G.; De Brandt, E.; Temmerman, R.; Swings, J. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 2005, 102, 221–230. [Google Scholar] [CrossRef]
- Turroni, F.; Foroni, E.; Pizzetti, P.; Giubellini, V.; Ribbera, A.; Merusi, P.; Cagnasso, P.; Bizzarri, B.; De’Angelis, G.L.; Shanahan, F.; et al. Exploring the Diversity of the Bifidobacterial Population in the Human Intestinal Tract. Appl. Environ. Microbiol. 2009, 75, 1534–1545. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, E.; Matsuki, T.; Kubota, H.; Makino, H.; Sakai, T.; Oishi, K.; Kushiro, A.; Fujimoto, J.; Watanabe, K.; Watanuki, M.; et al. Ethnic diversity of gut microbiota: Species characterization of Bacteroides fragilis group and genus Bifidobacterium in healthy Belgian adults, and comparison with data from Japanese subjects. J. Biosci. Bioeng. 2013, 116, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.; Sugahara, H.; Odamaki, T.; Xiao, J. Different physiological properties of human-residential and non-human-residential bifidobacteria in human health. Benef. Microbes 2018, 9, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of Bifidobacteria within the Infant Gut Microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef] [PubMed]
- Odamaki, T.; Bottacini, F.; Kato, K.; Mitsuyama, E.; Yoshida, K.; Horigome, A.; Xiao, J.-Z.; Van Sinderen, D. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci. Rep. 2018, 8, 85. [Google Scholar] [CrossRef]
- Reuter, G. The Lactobacillus and Bifidobacterium microflora of the human intestine: Composition and succession. Curr. Issues Intest. Microbiol. 2001, 2, 43–53. [Google Scholar]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The Placenta Harbors a Unique Microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef]
- De Goffau, M.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G. Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar] [CrossRef]
- Perez-Muñoz, M.E.; Arrieta, M.-C.; Ramer-Tait, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 2017, 5, 48. [Google Scholar] [CrossRef]
- Makino, H. Bifidobacterial strains in the intestines of newborns originate from their mothers. Biosci. Microbiota Food Health 2018, 37, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Makino, H.; Kushiro, A.; Ishikawa, E.; Kubota, H.; Gawad, A.; Sakai, T.; Oishi, K.; Martin, R.; Ben-Amor, K.; Knol, J.; et al. Mother-to-Infant Transmission of Intestinal Bifidobacterial Strains Has an Impact on the Early Development of Vaginally Delivered Infant’s Microbiota. PLoS ONE 2013, 8, e78331. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Mancabelli, L.; Lugli, G.A.; Duranti, S.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Ferretti, P.; Gorfer, V.; et al. Exploring Vertical Transmission of Bifidobacteria from Mother to Child. Appl. Environ. Microbiol. 2015, 81, 7078–7087. [Google Scholar] [CrossRef]
- Kumar, H.; Collado, M.C.; Wopereis, H.; Salminen, S.; Knol, J.; Roeselers, G. The Bifidogenic Effect Revisited—Ecology and Health Perspectives of Bifidobacterial Colonization in Early Life. Microorganisms 2020, 8, 1855. [Google Scholar] [CrossRef] [PubMed]
- Butel, M.-J.; Waligora-Dupriet, A.-J.; Wydau-Dematteis, S. The developing gut microbiota and its consequences for health. J. Dev. Orig. Health Dis. 2018, 9, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Mikami, K.; Takahashi, H.; Kimura, M.; Isozaki, M.; Izuchi, K.; Shibata, R.; Sudo, N.; Matsumoto, H.; Koga, Y. Influence of Maternal Bifidobacteria on the Establishment of Bifidobacteria Colonizing the Gut in Infants. Pediatr. Res. 2009, 65, 669–674. [Google Scholar] [CrossRef]
- Albesharat, R.; Ehrmann, M.A.; Korakli, M.; Yazaji, S.; Vogel, R.F. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 2011, 34, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Matsumiya, Y.; Kato, N.; Watanabe, K.; Kato, H. Molecular epidemiological study of vertical transmission of vaginal Lactobacillus species from mothers to newborn infants in Japanese, by arbitrarily primed polymerase chain reaction. J. Infect. Chemother. 2002, 8, 43–49. [Google Scholar] [CrossRef]
- Makino, H.; Kushiro, A.; Ishikawa, E.; Muylaert, D.; Kubota, H.; Sakai, T.; Oishi, K.; Martin, R.; Ben Amor, K.; Oozeer, R.; et al. Transmission of Intestinal Bifidobacterium longum subsp.longumStrains from Mother to Infant, Determined by Multilocus Sequencing Typing and Amplified Fragment Length Polymorphism. Appl. Environ. Microbiol. 2011, 77, 6788–6793. [Google Scholar] [CrossRef]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Armanini, F.; Turroni, F.; James, K.; Ferretti, P.; Gorfer, V.; Ferrario, C.; Milani, C.; et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 2017, 5, 66. [Google Scholar] [CrossRef]
- Asnicar, F.; Manara, S.; Zolfo, M.; Truong, D.T.; Scholz, M.; Armanini, F.; Ferretti, P.; Gorfer, V.; Pedrotti, A.; Tett, A.; et al. Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. MSystems 2017, 2, e00164-16. [Google Scholar] [CrossRef]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe 2018, 24, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Biagi, E.; Quercia, S.; Aceti, A.; Beghetti, I.; Rampelli, S.; Turroni, S.; Faldella, G.; Candela, M.; Brigidi, P.; Corvaglia, L. The Bacterial Ecosystem of Mother’s Milk and Infant’s Mouth and Gut. Front. Microbiol. 2017, 8, 1214. [Google Scholar] [CrossRef] [PubMed]
- Selma-Royo, M.; Lerma, J.C.; Cortés-Macías, E.; Collado, M.C. Human milk microbiome: From actual knowledge to future perspective. Semin. Perinatol. 2021, 45, 151450. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Macías, E.; Selma-Royo, M.; Martínez-Costa, C.; Collado, M. Breastfeeding Practices Influence the Breast Milk Microbiota Depending on Pre-Gestational Maternal BMI and Weight Gain over Pregnancy. Nutrients 2021, 13, 1518. [Google Scholar] [CrossRef]
- Cortes-Macías, E.; Selma-Royo, M.; García-Mantrana, I.; Calatayud, M.; González, S.; Martínez-Costa, C.; Collado, M.C. Maternal Diet Shapes the Breast Milk Microbiota Composition and Diversity: Impact of Mode of Delivery and Antibiotic Exposure. J. Nutr. 2020, 151, 330–340. [Google Scholar] [CrossRef]
- Bode, L. The functional biology of human milk oligosaccharides. Early Hum. Dev. 2015, 91, 619–622. [Google Scholar] [CrossRef]
- Wiciński, M.; Sawicka, E.; Gębalski, J.; Kubiak, K.; Malinowski, B. Human Milk Oligosaccharides: Health Benefits, Potential Applications in Infant Formulas, and Pharmacology. Nutrients 2020, 12, 266. [Google Scholar] [CrossRef]
- Sakanaka, M.; Gotoh, A.; Yoshida, K.; Odamaki, T.; Koguchi, H.; Xiao, J.-Z.; Kitaoka, M.; Katayama, T. Varied Pathways of Infant Gut-Associated Bifidobacterium to Assimilate Human Milk Oligosaccharides: Prevalence of the Gene Set and Its Correlation with Bifidobacteria-Rich Microbiota Formation. Nutrients 2019, 12, 71. [Google Scholar] [CrossRef]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; De Los Reyes-Gavilán, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef]
- Leblanc, J.G.; Chain, F.; Martín, R.; Humaran, L.G.B.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Factories 2017, 16, 79. [Google Scholar] [CrossRef] [PubMed]
- Zabel, B.; Yde, C.C.; Roos, P.; Marcussen, J.; Jensen, H.M.; Salli, K.; Hirvonen, J.; Ouwehand, A.C.; Morovic, W. Novel Genes and Metabolite Trends in Bifidobacterium longum subsp. infantis Bi-26 Metabolism of Human Milk Oligosaccharide 2′-fucosyllactose. Sci. Rep. 2019, 9, 7983. [Google Scholar] [CrossRef]
- Alessandri, G.; Ossiprandi, M.C.; Mac Sharry, J.; Van Sinderen, D.; Ventura, M. Bifidobacterial Dialogue With Its Human Host and Consequent Modulation of the Immune System. Front. Immunol. 2019, 10, 2348. [Google Scholar] [CrossRef] [PubMed]
- Motherway, M.O.; Houston, A.; O’Callaghan, G.; Reunanen, J.; O’Brien, F.; O’Driscoll, T.; Casey, P.G.; De Vos, W.M.; Van Sinderen, D.; Shanahan, F. A Bifidobacterial pilus-associated protein promotes colonic epithelial proliferation. Mol. Microbiol. 2018, 111, 287–301. [Google Scholar] [CrossRef]
- Ewaschuk, J.B.; Diaz, H.; Meddings, L.; Diederichs, B.; Dmytrash, A.; Backer, J.; Langen, M.L.-V.; Madsen, K.L. Secreted bioactive factors fromBifidobacterium infantisenhance epithelial cell barrier function. Am. J. Physiol. Liver Physiol. 2008, 295, G1025–G1034. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Ouwehand, A.C.; Isolauri, E.; Hashimoto, H.; Benno, Y.; Salminen, S. Comparison of mucosal adhesion and species identification of bifidobacteria isolated from healthy and allergic infants. FEMS Immunol. Med. Microbiol. 2001, 30, 43–47. [Google Scholar] [CrossRef]
- Martinez, F.A.C.; Balciunas, E.M.; Converti, A.; Cotter, P.D.; de Souza Oliveira, R.P. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol. Adv. 2013, 31, 482–488. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Sugahara, H.; Odamaki, T.; Hashikura, N.; Abe, F.; Xiao, J.-Z. Differences in folate production by bifidobacteria of different origins. Biosci. Microbiota Food Health 2015, 34, 87–93. [Google Scholar] [CrossRef]
- Huda, M.N.; Lewis, Z.; Kalanetra, K.M.; Rashid, M.; Ahmad, S.M.; Raqib, R.; Qadri, F.; Underwood, M.A.; Mills, D.A.; Stephensen, C.B. Stool Microbiota and Vaccine Responses of Infants. Pediatrics 2014, 134, e362–e372. [Google Scholar] [CrossRef]
- Arboleya, S.; Binetti, A.; Salazar, N.; Fernández, N.; Solís, G.; Barranco, A.H.; Margolles, A.; Reyes-Gavilan, C.D.L.; Gueimonde, M. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 2011, 79, 763–772. [Google Scholar] [CrossRef]
- Arboleya, S.; Sánchez, G.S.; Milani, C.; Duranti, S.; Solís, G.; Fernández, N.; Reyes-Gavilan, C.D.L.; Ventura, M.; Margolles, A.; Gueimonde, M. Intestinal Microbiota Development in Preterm Neonates and Effect of Perinatal Antibiotics. J. Pediatr. 2014, 166, 538–544. [Google Scholar] [CrossRef]
- Barrett, E.; Kerr, C.; Murphy, K.; O’Sullivan, O.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.P.; O’Toole, P.W.; Cotter, P.; Fitzgerald, G.F.; et al. The individual-specific and diverse nature of the preterm infant microbiota. Arch. Dis. Child. Fetal Neonatal Ed. 2013, 98, F334–F340. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; Blakstad, E.W.; Moltu, S.J.; Strømmen, K.; Nakstad, B.; Rønnestad, A.E.; Brække, K.; Iversen, P.O.; Drevon, C.A.; De Vos, W. Intestinal microbiota development and gestational age in preterm neonates. Sci. Rep. 2018, 8, 2453. [Google Scholar] [CrossRef]
- Forsgren, M.; Isolauri, E.; Salminen, S.; Rautava, S. Late preterm birth has direct and indirect effects on infant gut microbiota development during the first six months of life. Acta Paediatr. 2017, 106, 1103–1109. [Google Scholar] [CrossRef]
- Hill, C.J.; Lynch, D.B.; Murphy, K.; Ulaszewska, M.; Jeffery, I.; O’Shea, C.A.; Watkins, C.; Dempsey, E.M.; Mattivi, F.; Tuohy, K.; et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome 2017, 5, 4. [Google Scholar] [CrossRef]
- Masi, A.C.; Stewart, C.J. The role of the preterm intestinal microbiome in sepsis and necrotising enterocolitis. Early Hum. Dev. 2019, 138, 104854. [Google Scholar] [CrossRef]
- Plummer, E.L.; for the ProPrems Study Group; Bulach, D.M.; Murray, G.L.; Jacobs, S.E.; Tabrizi, S.N.; Garland, S.M. Gut microbiota of preterm infants supplemented with probiotics: Sub-study of the ProPrems trial. BMC Microbiol. 2018, 18, 184. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Embleton, N.; Marrs, E.C.L.; Smith, D.P.; Nelson, A.; Abdulkadir, B.; Skeath, T.; Petrosino, J.F.; Perry, J.D.; Berrington, J.E.; et al. Temporal bacterial and metabolic development of the preterm gut reveals specific signatures in health and disease. Microbiome 2016, 4, 67. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Embleton, N.; Marrs, E.C.L.; Smith, D.P.; Fofanova, T.; Nelson, A.; Skeath, T.; Perry, J.D.; Petrosino, J.F.; Berrington, J.E.; et al. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome 2017, 5, 75. [Google Scholar] [CrossRef]
- De Jong, J.C.; Ijssennagger, N.; van Mil, S.W. Breast milk nutrients driving intestinal epithelial layer maturation via Wnt and Notch signaling: Implications for necrotizing enterocolitis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2021, 1867, 166229. [Google Scholar] [CrossRef] [PubMed]
- Qazi, S.A.; Stoll, B.J. Neonatal Sepsis. Pediatr. Infect. Dis. J. 2009, 28, S1–S2. [Google Scholar] [CrossRef] [PubMed]
- Carl, M.A.; Ndao, I.M.; Springman, A.C.; Manning, S.; Johnson, J.R.; Johnston, B.D.; Burnham, C.-A.D.; Weinstock, E.S.; Weinstock, G.; Wylie, T.N.; et al. Sepsis From the Gut: The Enteric Habitat of Bacteria That Cause Late-Onset Neonatal Bloodstream Infections. Clin. Infect. Dis. 2014, 58, 1211–1218. [Google Scholar] [CrossRef]
- Ye, Q.; Yu, J. A Study on Fucosyltransferase 2 Gene Polymorphism and Secretion Status Related to Neonatal Necrotizing Enterocolitis. J. Healthc. Eng. 2021, 2021, 7219850. [Google Scholar] [CrossRef] [PubMed]
- Maynard, C.L.; Elson, C.O.; Hatton, R.; Weaver, C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Brennan, A.-M.; Murphy, B.P.; Kiely, M.E. Optimising preterm nutrition: Present and future. Proc. Nutr. Soc. 2016, 75, 154–161. [Google Scholar] [CrossRef]
- Younge, N.E.; Newgard, C.B.; Cotten, C.M.; Goldberg, R.N.; Muehlbauer, M.J.; Bain, J.R.; Stevens, R.D.; O’Connell, T.M.; Rawls, J.F.; Seed, P.C.; et al. Disrupted Maturation of the Microbiota and Metabolome among Extremely Preterm Infants with Postnatal Growth Failure. Sci. Rep. 2019, 9, 8167. [Google Scholar] [CrossRef]
- Arboleya, S.; Camblor, P.M.; Solís, G.; Suárez, M.; Fernández, N.; de Los Reyes-Gavilan, C.D.L.; Gueimonde, M. Intestinal Microbiota and Weight-Gain in Preterm Neonates. Front. Microbiol. 2017, 8, 183. [Google Scholar] [CrossRef]
- Blanton, L.V.; Charbonneau, M.R.; Salih, T.; Barratt, M.J.; Venkatesh, S.; Ilkaveya, O.; Subramanian, S.; Manary, M.J.; Trehan, I.; Jorgensen, J.M.; et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 2016, 351, aad3311. [Google Scholar] [CrossRef]
- Gehrig, J.L.; Venkatesh, S.; Chang, H.-W.; Hibberd, M.C.; Kung, V.L.; Cheng, J.; Chen, R.Y.; Subramanian, S.; Cowardin, C.A.; Meier, M.; et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science 2019, 365, 6449. [Google Scholar] [CrossRef]
- Raman, A.S.; Gehrig, J.L.; Venkatesh, S.; Chang, H.-W.; Hibberd, M.C.; Subramanian, S.; Kang, G.; Bessong, P.O.; Lima, A.A.; Kosek, M.N.; et al. A sparse covarying unit that describes healthy and impaired human gut microbiota development. Science 2019, 365, 6449. [Google Scholar] [CrossRef]
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; Van den Brandt, P.A.; Stobberingh, E.E. Factors Influencing the Composition of the Intestinal Microbiota in Early Infancy. Pediatrics 2006, 118, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed]
- Rutayisire, E.; Huang, K.; Liu, Y.; Tao, F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: A systematic review. BMC Gastroenterol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Werlang, I.C.R.; Mueller, N.T.; Pizoni, A.; Wisintainer, H.; Matte, U.; Costa, S.H.D.A.M.; Ramos, J.G.L.; Goldani, M.Z.; Dominguez-Bello, M.G.; Goldani, H.A.S. Associations of birth mode with cord blood cytokines, white blood cells, and newborn intestinal bifidobacteria. PLoS ONE 2018, 13, e0205962. [Google Scholar] [CrossRef]
- Morais, L.H.; Golubeva, A.V.; Moloney, G.M.; Moya-Pérez, A.; Ventura-Silva, A.P.; Arboleya, S.; Bastiaanssen, T.F.; O’Sullivan, O.; Rea, K.; Borre, Y.; et al. Enduring Behavioral Effects Induced by Birth by Caesarean Section in the Mouse. Curr. Biol. 2020, 30, 3761–3774.e6. [Google Scholar] [CrossRef]
- Reyman, M.; Van Houten, M.A.; Van Baarle, D.; Bosch, A.A.T.M.; Man, W.H.; Chu, M.L.J.N.; Arp, K.; Watson, R.L.; Sanders, E.A.M.; Fuentes, S.; et al. Author Correction: Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019, 10, 5352. [Google Scholar] [CrossRef]
- Nogacka, A.; Salazar, N.; Suárez, M.; Milani, C.; Arboleya, S.; Solís, G.; Fernández, N.; Alaez, L.; Hernández-Barranco, A.M.; Reyes-Gavilan, C.D.L.; et al. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome 2017, 5, 93. [Google Scholar] [CrossRef]
- Imoto, N.; Morita, H.; Amanuma, F.; Maruyama, H.; Watanabe, S.; Hashiguchi, N. Maternal antimicrobial use at delivery has a stronger impact than mode of delivery on bifidobacterial colonization in infants: A pilot study. J. Perinatol. 2018, 38, 1174–1181. [Google Scholar] [CrossRef]
- Saturio, S.; Suárez, M.; Mancabelli, L.; Fernández, N.; Mantecón, L.; Reyes-Gavilán, C.G.D.L.; Ventura, M.; Gueimonde, M.; Arboleya, S.; Solís, G. Effect of Intrapartum Antibiotics Prophylaxis on the Bifidobacterial Establishment within the Neonatal Gut. Microorganisms 2021, 9, 1867. [Google Scholar] [CrossRef]
- Fouhy, F.; Guinane, C.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.; Fitzgerald, G.F.; Stanton, C.; et al. High-Throughput Sequencing Reveals the Incomplete, Short-Term Recovery of Infant Gut Microbiota following Parenteral Antibiotic Treatment with Ampicillin and Gentamicin. Antimicrob. Agents Chemother. 2012, 56, 5811–5820. [Google Scholar] [CrossRef] [PubMed]
- Uzan-Yulzari, A.; Turta, O.; Belogolovski, A.; Ziv, O.; Kunz, C.; Perschbacher, S.; Neuman, H.; Pasolli, E.; Oz, A.; Ben-Amram, H.; et al. Neonatal antibiotic exposure impairs child growth during the first six years of life by perturbing intestinal microbial colonization. Nat. Commun. 2021, 12, 443. [Google Scholar] [CrossRef] [PubMed]
- Turta, O.; Rautava, S. Antibiotics, obesity and the link to microbes—What are we doing to our children? BMC Med. 2016, 14, 57. [Google Scholar] [CrossRef] [PubMed]
- Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; Naghavi, M.; et al. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [CrossRef]
- Soderborg, T.K.; Clark, S.; Mulligan, C.E.; Janssen, R.C.; Babcock, L.; Ir, D.; Young, B.; Krebs, N.; Lemas, D.J.; Johnson, L.K.; et al. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat. Commun. 2018, 9, 4462, Correction in 2019, 10, 2965. [Google Scholar] [CrossRef]
- Collado, M.C.; Cernada, M.; Bäuerl, C.; Vento, M.; Pérez-Martínez, G. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes 2012, 3, 352–365. [Google Scholar] [CrossRef]
- Dreisbach, C.; Prescott, S.; Alhusen, J. Influence of Maternal Prepregnancy Obesity and Excessive Gestational Weight Gain on Maternal and Child Gastrointestinal Microbiome Composition: A Systematic Review. Biol. Res. Nurs. 2019, 22, 114–125. [Google Scholar] [CrossRef]
- Kim, H.; Sitarik, A.R.; Woodcroft, K.; Johnson, C.C.; Zoratti, E. Birth Mode, Breastfeeding, Pet Exposure, and Antibiotic Use: Associations With the Gut Microbiome and Sensitization in Children. Curr. Allergy Asthma Rep. 2019, 19, 22. [Google Scholar] [CrossRef]
- Tun, H.; Bridgman, S.L.; Chari, R.; Field, C.; Guttman, D.S.; Becker, A.B.; Mandhane, P.J.; Turvey, S.; Subbarao, P.; Sears, M.R.; et al. Roles of Birth Mode and Infant Gut Microbiota in Intergenerational Transmission of Overweight and Obesity From Mother to Offspring. JAMA Pediatr. 2018, 172, 368–377. [Google Scholar] [CrossRef]
- Sutharsan, R.; Mannan, M.; Doi, A.S.; Al Mamun, A. Caesarean delivery and the risk of offspring overweight and obesity over the life course: A systematic review and bias-adjusted meta-analysis. Clin. Obes. 2015, 5, 293–301. [Google Scholar] [CrossRef]
- Vu, K.; Lou, W.; Tun, H.M.; Konya, T.B.; Morales-Lizcano, N.; Chari, R.S.; Field, C.J.; Guttman, D.S.; Mandal, R.; Wishart, D.S.; et al. From Birth to Overweight and Atopic Disease: Multiple and Common Pathways of the Infant Gut Microbiome. Gastroenterology 2021, 160, 128–144.e10. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; Zijlmans, M.A.C.; Kuitunen, M.; Kukkonen, K.; Savilahti, E.; Salonen, A.; De Weerth, C.; De Vos, W.M. Childhood BMI in relation to microbiota in infancy and lifetime antibiotic use. Microbiome 2017, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Murri, M.; Leiva, I.; Gomez-Zumaquero, J.M.; Tinahones, F.J.; Cardona, F.; Soriguer, F.; Queipo-Ortuño, M.I. Gut microbiota in children with type 1 diabetes differs from that in healthy children: A case-control study. BMC Med. 2013, 11, 46. [Google Scholar] [CrossRef]
- Isolauri, E. Development of healthy gut microbiota early in life. J. Paediatr. Child Health 2012, 48, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Asher, M.I.; Montefort, S.; Björkstén, B.; Lai, C.K.; Strachan, D.P.; Weiland, S.K.; Williams, H. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet 2006, 368, 733–743. [Google Scholar] [CrossRef]
- Wold, A.E. The hygiene hypotheslis revised: Is the rising frequency of allergy due to changes in the intestinal flora? Allergy 1998, 53, 20–25. [Google Scholar] [CrossRef]
- Alfven, T.; Braun-Fahrlander, C.; Brunekreef, B.; von Mutius, E.; Riedler, J.; Scheynius, A.; van Hage, M.; Wickman, M.; Benz, M.R.; Budde, J.; et al. Allergic diseases and atopic sensitization in children related to farming and anthroposophic lifestyle—The PARSIFAL study. Allergy 2006, 61, 414–421. [Google Scholar] [CrossRef]
- Fieten, K.B.; Totté, J.E.; Levin, E.; Reyman, M.; Meijer, Y.; Knulst, A.; Schuren, F.; Pasmans, S.G. Fecal Microbiome and Food Allergy in Pediatric Atopic Dermatitis: A Cross-Sectional Pilot Study. Int. Arch. Allergy Immunol. 2018, 175, 77–84. [Google Scholar] [CrossRef]
- Ruohtula, T.; de Goffau, M.; Nieminen, J.K.; Honkanen, J.; Siljander, H.; Hämäläinen, A.-M.; Peet, A.; Tillmann, V.; Ilonen, J.; Niemelä, O.; et al. Maturation of Gut Microbiota and Circulating Regulatory T Cells and Development of IgE Sensitization in Early Life. Front. Immunol. 2019, 10, 2494. [Google Scholar] [CrossRef]
- Low, J.S.Y.; Soh, E.S.; Lee, Y.K.; Kwek, K.Y.C.; Holbrook, J.D.; Van Der Beek, E.M.; Shek, L.; Goh, A.E.N.; Teoh, O.H.; Godfrey, K.M.; et al. Ratio of Klebsiella/Bifidobacterium in early life correlates with later development of paediatric allergy. Benef. Microbes 2017, 8, 681–695. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, S.; Wang, J.; Zhang, L.; Mu, Y.; Huang, K.; Zhao, B.; Zhang, K.; Cui, Y.; Li, S. Variations in early gut microbiome are associated with childhood eczema. FEMS Microbiol. Lett. 2019, 366, fnz020. [Google Scholar] [CrossRef]
- Sjödin, K.S.; Hammarström, M.-L.; Rydén, P.; Sjödin, A.; Hernell, O.; Engstrand, L.; West, C.E. Temporal and long-term gut microbiota variation in allergic disease: A prospective study from infancy to school age. Allergy 2018, 74, 176–185. [Google Scholar] [CrossRef]
- Melli, L.C.F.L.; Carmo-Rodrigues, M.S.D.; Araújo-Filho, H.B.; Mello, C.; Tahan, S.; Pignatari, A.C.C.; Solé, D.; de Morais, M.B. Gut microbiota of children with atopic dermatitis: Controlled study in the metropolitan region of São Paulo, Brazil. Allergol. Immunopathol. 2020, 48, 107–115. [Google Scholar] [CrossRef]
- Guo, L.; Bai, H.; Dong, Y.; Huang, D.X.; Zhang, X.; Gong, S.; Zhao, X.; Fei, P. Comparative Analysis of Fecal Microbiota in 5–8-Year-old Children with and without Cow Milk Protein Allergy. Iran. J. Pediatr. 2016, 26, e6397. [Google Scholar] [CrossRef]
- Melli, L.; Carmo-Rodrigues, M.D.; Araújo-Filho, H.; Solé, D.; de Morais, M.B. Intestinal microbiota and allergic diseases: A systematic review. Allergol. Immunopathol. 2016, 44, 177–188. [Google Scholar] [CrossRef]
- Mah, K.; Björkstén, B.; Lee, B.; Van Bever, H.; Shek, L.; Tan, T.; Lee, Y.; Chua, K. Distinct Pattern of Commensal Gut Microbiota in Toddlers with Eczema. Int. Arch. Allergy Immunol. 2006, 140, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Narisawa, Y.; Arase, S.; Okamatsu, H.; Ikenaga, T.; Tajiri, Y.; Kumemura, M. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J. Allergy Clin. Immunol. 2003, 111, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Barbi, J.; Pan, F. The regulation of immune tolerance by FOXP. Nat. Rev. Immunol. 2017, 17, 703–717. [Google Scholar] [CrossRef]
- Zuo, L.; Yuan, K.-T.; Yu, L.; Meng, Q.-H.; Chung, P.C.-K.; Yang, D.-H. Bifidobacterium infantisattenuates colitis by regulating T cell subset responses. World J. Gastroenterol. 2014, 20, 18316–18329. [Google Scholar] [CrossRef]
- Ashraf, R.; Vasiljevic, T.; Day, S.; Smith, S.; Donkor, O. Lactic acid bacteria and probiotic organisms induce different cytokine profile and regulatory T cells mechanisms. J. Funct. Foods 2014, 6, 395–409. [Google Scholar] [CrossRef]
- Verma, R.; Lee, C.; Jeun, E.-J.; Yi, J.; Kim, K.S.; Ghosh, A.; Byun, S.; Lee, C.-G.; Kang, H.-J.; Kim, G.-C.; et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3 + regulatory T cells. Sci. Immunol. 2018, 3, eaat6975. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Carrera-Bastos, P.; Schmitz, G. Milk: A postnatal imprinting system stabilizing FoxP3 expression and regulatory T cell differentiation. Clin. Transl. Allergy 2016, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Sayal, K.; Heron, J.; Maughan, B.; Rowe, R.; Ramchandani, P. Infant temperament and childhood psychiatric disorder: Longitudinal study. Child Care Health Dev. 2013, 40, 292–297. [Google Scholar] [CrossRef]
- Sampson, T.R.; Mazmanian, S.K. Control of Brain Development, Function, and Behavior by the Microbiome. Cell Host Microbe 2015, 17, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Aatsinki, A.-K.; Lahti, L.; Uusitupa, H.-M.; Munukka, E.; Keskitalo, A.; Nolvi, S.; O’Mahony, S.; Pietilä, S.; Elo, L.L.; Eerola, E.; et al. Gut microbiota composition is associated with temperament traits in infants. Brain Behav. Immun. 2019, 80, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, X.; Yu, Y.; Liu, Y.; Zhang, Q.; Bai, J. Association between Gut Microbiota and Infant’s Temperament in the First Year of Life in a Chinese Birth Cohort. Microorganisms 2020, 8, 753. [Google Scholar] [CrossRef]
- Kelsey, C.M.; Prescott, S.; McCulloch, J.A.; Trinchieri, G.; Valladares, T.L.; Dreisbach, C.; Alhusen, J.; Grossmann, T. Gut microbiota composition is associated with newborn functional brain connectivity and behavioral temperament. Brain Behav. Immun. 2020, 91, 472–486. [Google Scholar] [CrossRef] [PubMed]
- Coretti, L.; Paparo, L.; Riccio, M.P.; Amato, F.; Cuomo, M.; Natale, A.; Borrelli, L.; Corrado, G.; De Caro, C.; Comegna, M.; et al. Gut Microbiota Features in Young Children with Autism Spectrum Disorders. Front. Microbiol. 2018, 9, 3146. [Google Scholar] [CrossRef]
- Bojović, K.; Ignjatović, Đ.; Bajic, S.S.; Milutinović, D.V.; Tomić, M.; Golić, N.; Tolinački, M. Gut Microbiota Dysbiosis Associated With Altered Production of Short Chain Fatty Acids in Children With Neurodevelopmental Disorders. Front. Cell. Infect. Microbiol. 2020, 10, 223. [Google Scholar] [CrossRef]
- Flannery, J.E.; Stagaman, K.; Burns, A.R.; Hickey, R.J.; Roos, L.E.; Giuliano, R.J.; Fisher, P.A.; Sharpton, T.J. Gut Feelings Begin in Childhood: The Gut Metagenome Correlates with Early Environment, Caregiving, and Behavior. MBio 2020, 11, e02780-19. [Google Scholar] [CrossRef]
- Golubeva, A.V.; Joyce, A.S.; Moloney, G.; Burokas, A.; Sherwin, E.; Arboleya, S.; Flynn, I.; Khochanskiy, D.; Moya-Pérez, A.; Peterson, V.; et al. Microbiota-related Changes in Bile Acid & Tryptophan Metabolism are Associated with Gastrointestinal Dysfunction in a Mouse Model of Autism. EBioMedicine 2017, 24, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Luk, B.; Veeraragavan, S.; Engevik, M.; Balderas, M.; Major, A.; Runge, J.; Luna, R.A.; Versalovic, J. Postnatal colonization with human “infant-type” Bifidobacterium species alters behavior of adult gnotobiotic mice. PLoS ONE 2018, 13, e0196510. [Google Scholar] [CrossRef]
- Ehrlich, A.M.; Henrick, B.; Pacheco, A.; Taft, D.; Xu, G.; Huda, N.; Lozada-Contreras, M.; Goodson, M.; Slupsky, C.; Mills, D.; et al. Bifidobacterium grown on human milk oligosaccharides produce tryptophan metabolite Indole-3-lactic acid that significantly decreases inflammation in intestinal cells In Vitro. FASEB J. 2018, 32, lb359. [Google Scholar] [CrossRef]
- Sakurai, T.; Odamaki, T.; Xiao, J.-Z. Production of Indole-3-Lactic Acid by Bifidobacterium Strains Isolated from Human Infants. Microorganisms 2019, 7, 340. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.B.; Tanaka, A.; Kuhara, T.; Xiao, J.-Z. Potential Effects of Indole-3-Lactic Acid, a Metabolite of Human Bifidobacteria, on NGF-Induced Neurite Outgrowth in PC12 Cells. Microorganisms 2020, 8, 398. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, A.M.; Pacheco, A.R.; Henrick, B.M.; Taft, D.; Xu, G.; Huda, M.N.; Mishchuk, D.; Goodson, M.L.; Slupsky, C.; Barile, D.; et al. Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells. BMC Microbiol. 2020, 20, 357. [Google Scholar] [CrossRef]
- Wang, B.; Brand-Miller, J.; McNeil, Y.; McVeagh, P. Sialic Acid Concentration of Brain Gangliosides: Variation among Eight Mammalian Species. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 1998, 119, 435–439. [Google Scholar] [CrossRef]
- Wang, B.; McVeagh, P.; Petocz, P.; Brand-Miller, J. Brain ganglioside and glycoprotein sialic acid in breastfed compared with formula-fed infants. Am. J. Clin. Nutr. 2003, 78, 1024–1029. [Google Scholar] [CrossRef]
- Tannock, G.W.; Lee, P.S.; Wong, K.H.; Lawley, B. Why Don’t All Infants Have Bifidobacteria in Their Stool? Front. Microbiol. 2016, 7, 834. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Arboleya, S.; Salazar, N.; Solís, G.; Fernández, N.; Gueimonde, M.; Reyes-Gavilán, C.G.D.L. In Vitro evaluation of the impact of human background microbiota on the response to Bifidobacterium strains and fructo-oligosaccharides. Br. J. Nutr. 2013, 110, 2030–2036. [Google Scholar] [CrossRef]
- El Aidy, S.; Hooiveld, G.; Tremaroli, V.; Bäckhed, F.; Kleerebezem, M. The gut microbiota and mucosal homeostasis. Gut Microbes 2013, 4, 118–124. [Google Scholar] [CrossRef]
- Kiu, R.; Treveil, A.; Harnisch, L.C.; Caim, S.; Leclaire, C.; van Sinderen, D.; Korcsmaros, T.; Hall, L.J. Bifidobacterium breve UCC2003 Induces a Distinct Global Transcriptomic Program in Neonatal Murine Intestinal Epithelial Cells. IScience 2020, 23, 101336. [Google Scholar] [CrossRef]
- Rigo-Adrover, M.D.M.; Franch, A.; Castell, M.; Pérez-Cano, F.J. Preclinical Immunomodulation by the Probiotic Bifidobacterium breve M-16V in Early Life. PLoS ONE 2016, 11, e0166082. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, Y.; Ikegami, T.; Izumi, H.; Namura, M.; Ikeda, T.; Ikuse, T.; Baba, Y.; Kudo, T.; Suzuki, R.; Shimizu, T. Effects of Bifidobacterium breve on inflammatory gene expression in neonatal and weaning rat intestine. Pediatr. Res. 2011, 71, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Ehara, T.; Izumi, H.; Tsuda, M.; Nakazato, Y.; Iwamoto, H.; Namba, K.; Takeda, Y. Combinational effects of prebiotic oli-gosaccharides on bifidobacterial growth and host gene expression in a simplified mixed culture model and neonatal mice. Br. J. Nutr. 2016, 116, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Ehara, T.; Sugahara, H.; Matsubara, T.; Mitsuyama, E.; Nakazato, Y.; Tsuda, M.; Shimizu, T.; Odamaki, T.; Xiao, J.-Z.; et al. The Combination of Bifidobacterium breve and Three Prebiotic Oligosaccharides Modifies Gut Immune and Endocrine Functions in Neonatal Mice. J. Nutr. 2019, 149, 344–353. [Google Scholar] [CrossRef]
- Lyons, A.; O’Mahony, D.; O’Brien, F.; Mac Sharry, J.; Sheil, B.; Ceddia, M.; Russell, W.M.; Forsythe, P.; Bienenstock, J.; Kiely, B.; et al. Bacterial strain-specific induction of Foxp3+T regulatory cells is protective in murine allergy models. Clin. Exp. Allergy 2010, 40, 811–819. [Google Scholar] [CrossRef]
- Sudo, N.; Sawamura, S.A.; Tanaka, K.; Aiba, Y.; Kubo, C.; Koga, Y. The requirement of intestinal bacterial flora for the de-velopment of an ige production system fully susceptible to oral tolerance induction. J. Immunol. 1997, 159, 1739–1745. [Google Scholar]
- Tanaka, K.; Ishikawa, H. Role of intestinal bacterial flora in oral tolerance induction. Histol. Histopathol. 2004, 19, 907–914. [Google Scholar] [CrossRef]
- Cheng, R.; Yao, J.; Wan, Q.; Guo, J.; Pu, F.; Shi, L.; Hu, W.; Yang, Y.; Li, L.; Li, M.; et al. Oral administration of Bifidobacterium bifidum TMC3115 to neonatal mice may alleviate IgE-mediated allergic risk in adulthood. Benef. Microbes 2018, 9, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Feleszko, W.; Jaworska, J.; Rha, R.-D.; Steinhausen, S.; Avagyan, A.; Jaudszus, A.; Ahrens, B.; Groneberg, D.A.; Wahn, U.; Hamelmann, E. Probiotic-induced suppression of allergic sensitization and airway inflammation is associated with an increase of T regulatory-dependent mechanisms in a murine model of asthma. Clin. Exp. Allergy 2006, 37, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Terada-Ikeda, C.; Kitabatake, M.; Hiraku, A.; Kato, K.; Yasui, S.; Imakita, N.; Ouji-Sageshima, N.; Iwabuchi, N.; Hamada, K.; Ito, T. Maternal supplementation with Bifidobacterium breve M-16V prevents their offspring from allergic airway inflammation accelerated by the prenatal exposure to an air pollutant aerosol. PLoS ONE 2020, 15, e0238923. [Google Scholar] [CrossRef]
- Schwarzer, M.; Srutkova, D.; Schabussova, I.; Hudcovic, T.; Akgün, J.; Wiedermann, U.; Kozakova, H. Neonatal colonization of germ-free mice with Bifidobacterium longum prevents allergic sensitization to major birch pollen allergen Bet v1. Vaccine 2013, 31, 5405–5412. [Google Scholar] [CrossRef]
- Kostadinova, A.I.; Meulenbroek, L.A.P.M.; Van Esch, B.C.A.M.; Hofman, G.A.; Garssen, J.; Willemsen, L.E.M.; Knippels, L.M.J. A Specific Mixture of Fructo-Oligosaccharides and Bifidobacterium breve M-16V Facilitates Partial Non-Responsiveness to Whey Protein in Mice Orally exposed to β-Lactoglobulin-Derived Peptides. Front. Immunol. 2017, 7, 673. [Google Scholar] [CrossRef]
- Schouten, B.; Van Esch, B.C.A.M.; Hofman, G.A.; Van Doorn, S.A.C.M.; Knol, J.; Nauta, A.J.; Garssen, J.; Willemsen, L.E.M.; Knippels, L.M.J. Cow Milk Allergy Symptoms Are Reduced in Mice Fed Dietary Synbiotics during Oral Sensitization with Whey. J. Nutr. 2009, 139, 1398–1403. [Google Scholar] [CrossRef]
- Sasajima, N.; Ogasawara, T.; Takemura, N.; Fujiwara, R.; Watanabe, J.; Sonoyama, K. Role of intestinal bifidobacterium pseudolongum in dietary fructo-oligosaccharide inhibition of 2,4-dinitrofluorobenzene-induced contact hypersensitivity in mice. Br. J. Nutr. 2010, 103, 539–548. [Google Scholar] [CrossRef]
- Cheng, R.; Guo, J.; Pu, F.; Wan, C.; Shi, L.; Li, H.; Yang, Y.; Huang, C.; Li, M.; He, F. Loading ceftriaxone, vancomycin, and Bifidobacteria bifidum TMC3115 to neonatal mice could differently and consequently affect intestinal microbiota and immunity in adulthood. Sci. Rep. 2019, 9, 3254. [Google Scholar] [CrossRef]
- Luck, B.; Engevik, M.A.; Ganesh, B.; Lackey, E.P.; Lin, T.; Balderas, M.; Major, A.; Runge, J.; Luna, R.A.; Sillitoe, R.V.; et al. Bifidobacteria shape host neural circuits during postnatal development by promoting synapse formation and microglial function. Sci. Rep. 2020, 10, 7737. [Google Scholar] [CrossRef]
- Moya-Pérez, A.; Perez-Villalba, A.; Benitez-Paez, A.; Campillo, I.; Sanz, Y. Bifidobacterium CECT 7765 modulates early stress-induced immune, neuroendocrine and behavioral alterations in mice. Brain Behav. Immun. 2017, 65, 43–56. [Google Scholar] [CrossRef]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.-N.; Kubo, C.; Koga, Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Caplan, M.S.; Miller–Catchpole, R.; Kaup§, S.; Russell, T.; Lickerman, M.; Amer, M.; Xiao, Y.; Thomson, R. Bifidobacterial supplementation reduces the incidence of necrotizing enterocolitis in a neonatal rat model. Gastroenterology 1999, 117, 577–583. [Google Scholar] [CrossRef]
- Bergmann, K.R.; Liu, S.X.; Tian, R.; Kushnir, A.; Turner, J.R.; Li, H.-L.; Chou, P.M.; Weber, C.; De Plaen, I.G. Bifidobacteria Stabilize Claudins at Tight Junctions and Prevent Intestinal Barrier Dysfunction in Mouse Necrotizing Enterocolitis. Am. J. Pathol. 2013, 182, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Lv, H.; Li, M.; Su, H.; Huang, L.; Li, J.; Yuan, W. Protective effects of bifidobacteria on intestines in newborn rats with necrotizing enterocolitis and its regulation on TLR2 and TLR. Genet. Mol. Res. 2015, 14, 11505–11514. [Google Scholar] [CrossRef]
- Wu, W.; Wang, Y.; Zou, J.; Long, F.; Yan, H.; Zeng, L.; Chen, Y. Bifidobacterium adolescentis protects against necrotizing enterocolitis and upregulates TOLLIP and SIGIRR in premature neonatal rats. BMC Pediatr. 2017, 17, 1. [Google Scholar] [CrossRef]
- Khailova, L.; Dvorak, K.; Arganbright, K.M.; Halpern, M.D.; Kinouchi, T.; Yajima, M.; Dvorak, B. Bifidobacterium bifidum improves intestinal integrity in a rat model of necrotizing enterocolitis. Am. J. Physiol. Liver Physiol. 2009, 297, G940–G949. [Google Scholar] [CrossRef]
- Satoh, T.; Izumi, H.; Iwabuchi, N.; Odamaki, T.; Namba, K.; Abe, F.; Xiao, J. Bifidobacterium breve prevents necrotising enterocolitis by suppressing inflammatory responses in a preterm rat model. Benef. Microbes 2016, 7, 75–82. [Google Scholar] [CrossRef]
- Underwood, M.A.; Arriola, J.; Gerber, C.W.; Kaveti, A.; Kalanetra, K.M.; Kananurak, A.; Bevins, C.L.; Mills, D.A.; Dvorak, B. Bifidobacterium longum subsp. infantis in experimental necrotizing enterocolitis: Alterations in inflammation, innate immune response, and the microbiota. Pediatr. Res. 2014, 76, 326–333. [Google Scholar] [CrossRef]
- Underwood, M.A.; Kananurak, A.; Coursodon, C.F.; Adkins-Reick, C.K.; Chu, H.; Bennett, S.H.; Wehkamp, J.; Castillo, P.A.; Leonard, B.; Tancredi, D.; et al. Bifidobacterium bifidum in a rat model of necrotizing enterocolitis: Antimicrobial peptide and protein responses. Pediatr. Res. 2012, 71, 546–551. [Google Scholar] [CrossRef]
- Wu, S.-F.; Chiu, H.-Y.; Chen, A.-C.; Lin, H.-Y.; Lin, H.-C.; Caplan, M. Efficacy of Different Probiotic Combinations on Death and Necrotizing Enterocolitis in a Premature Rat Model. J. Pediatr. Gastroenterol. Nutr. 2013, 57, 23–28. [Google Scholar] [CrossRef]
- Geerlings, S.Y.; Kostopoulos, I.; De Vos, W.M.; Belzer, C. Akkermansia muciniphila in the Human Gastrointestinal Tract: When, Where, and How? Microorganisms 2018, 6, 75. [Google Scholar] [CrossRef] [PubMed]
- Yasui, H.; Kiyoshima, J.; Ushijima, H. Passive Protection against Rotavirus-Induced Diarrhea of Mouse Pups Born to and Nursed by Dams Fed Bifidobacterium breve YIT. J. Infect. Dis. 1995, 172, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Duffy, L.C.; Griffiths, E.; Dryja, D.; Leavens, A.; Rossman, J.; Rich, G.; Riepenhoff-Talty, M.; Locniskar, M. Immune Responses in Rhesus Rotavirus-Challenged Balb/c Mice Treated with Bifidobacteria and Prebiotic Supplements. Pediatr. Res. 2002, 51, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Lee, D.K.; Ha, N.J.; Shin, H.S. Antiviral effects of Lactobacillus ruminis SPM0211 and Bifidobacterium longum SPM1205 and SPM1206 on rotavirus-infected Caco-2 cells and a neonatal mouse model. J. Microbiol. 2015, 53, 796–803. [Google Scholar] [CrossRef]
- Izumi, H.; Minegishi, M.; Sato, Y.; Shimizu, T.; Sekine, K.; Takase, M. Bifidobacterium breve alters immune function and ameliorates DSS-induced inflammation in weanling rats. Pediatr. Res. 2015, 78, 407–416. [Google Scholar] [CrossRef]
- Wagner, R.D.; Pierson, C.; Warner, T.; Dohnalek, M.; Farmer, J.; Roberts, L.; Hilty, M.; Balish, E. Biotherapeutic effects of probiotic bacteria on candidiasis in immunodeficient mice. Infect. Immun. 1997, 65, 4165–4172. [Google Scholar] [CrossRef]
- Weng, M.; Ganguli, K.; Zhu, W.; Shi, H.N.; Walker, W.A. Conditioned medium from Bifidobacteria infantis protects against Cronobacter sakazakii-induced intestinal inflammation in newborn mice. Am. J. Physiol. Liver Physiol. 2014, 306, G779–G787. [Google Scholar] [CrossRef]
- Wong, C.B.; Iwabuchi, N.; Xiao, J.-Z. Exploring the Science behind Bifidobacterium breve M-16V in Infant Health. Nutrients 2019, 11, 1724. [Google Scholar] [CrossRef]
- Desbonnet, L.; Clarke, G.; Traplin, A.; O’Sullivan, O.; Crispie, F.; Moloney, R.D.; Cotter, P.; Dinan, T.; Cryan, J. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain Behav. Immun. 2015, 48, 165–173. [Google Scholar] [CrossRef]
- Weizman, Z.; Alsheikh, A. Safety and tolerance of a probiotic formula in early infancy comparing two probiotic agents: A pilot study. J. Am. Coll. Nutr. 2006, 25, 415–419. [Google Scholar] [CrossRef]
- Smilowitz, J.T.; Moya, J.; Breck, M.A.; Cook, C.; Fineberg, A.; Angkustsiri, K.; Underwood, M.A. Safety and tolerability of Bifidobacterium longum subspecies infantis EVC001 supplementation in healthy term breastfed infants: A phase I clinical trial. BMC Pediatr. 2017, 17, 133. [Google Scholar] [CrossRef]
- Weizman, Z.; Asli, G.; Alsheikh, A. Effect of a Probiotic Infant Formula on Infections in Child Care Centers: Comparison of Two Probiotic Agents. Pediatrics 2005, 115, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Grandy, G.; Medina, M.; Soria, R.; Terán, C.G.; Araya, M. Probiotics in the treatment of acute rotavirus diarrhoea. A randomized, double-blind, controlled trial using two different probiotic preparations in Bolivian children. BMC Infect. Dis. 2010, 10, 253. [Google Scholar] [CrossRef]
- Vandenplas, Y.; de Hert, S.G. PROBIOTICAL-study group Randomised clinical trial: The synbiotic food supplement Probiotical vs. placebo for acute gastroenteritis in children. Aliment. Pharmacol. Ther. 2011, 34, 862–867. [Google Scholar] [CrossRef]
- Escribano, J.; Ferré, N.; Gispert-Llaurado, M.; Luque, V.; Rubio-Torrents, C.; Zaragoza-Jordana, M.; Polanco, I.; Codoñer, F.M.; Chenoll, E.; Morera, M.; et al. Bifidobacterium longum subsp infantis CECT7210-supplemented formula reduces diarrhea in healthy infants: A randomized controlled trial. Pediatr. Res. 2018, 83, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Chouraqui, J.-P.; Van Egroo, L.-D.; Fichot, M.-C. Acidified Milk Formula Supplemented with Bifidobacterium lactis: Impact on Infant Diarrhea in Residential Care Settings. J. Pediatr. Gastroenterol. Nutr. 2004, 38, 288–292. [Google Scholar] [CrossRef]
- Ziegler, E.E.; Jeter, J.M.; Drulis, J.M.; Nelson, S.E.; Haschke, F.; Steenhout, P.; Brown, C.; Maire, J.-C.; Hager, C. Formula with reduced content of improved, partially hydrolyzed protein and probiotics: Infant growth and health. Monatsschrift Kinderheilkd. 2003, 151, S65–S71. [Google Scholar] [CrossRef]
- Urban, M.; Bolton, K.; Mokhachane, M.; Mphahlele, R.; Bomela, H.; Monaheng, L.; Beckh-Arnold, E.; Cooper, P. Growth of infants born to HIV-infected women when fed a biologically acidified starter formula with and without probiotics. S. Afr. J. Clin. Nutr. 2008, 21, 28–32. [Google Scholar] [CrossRef]
- Velaphi, S.C.; Cooper, P.A.; Bolton, K.D.; Mokhachane, M.; Mphahlele, R.M.; Beckh-Arnold, E.; Monaheng, L.; Haschke-Becher, E. Growth and metabolism of infants born to women infected with human immunodeficiency virus and fed acidified whey-adapted starter formulas. Nutrition 2008, 24, 203–211. [Google Scholar] [CrossRef]
- Corthésy, B. Role of secretory IgA in infection and maintenance of homeostasis. Autoimmun. Rev. 2013, 12, 661–665. [Google Scholar] [CrossRef]
- Bakker-Zierikzee, A.M.; Tol, E.A.F.; Kroes, H.; Alles, M.S.; Kok, F.J.; Bindels, J.G. Faecal SIgA secretion in infants fed on pre- or probiotic infant formula. Pediatr. Allergy Immunol. 2006, 17, 134–140. [Google Scholar] [CrossRef]
- Holscher, H.; Czerkies, L.A.; Cekola, P.; Litov, R.; Benbow, M.; Santema, S.; Alexander, D.D.; Perez, V.; Sun, S.; Saavedra, J.M.; et al. Bifidobacterium lactisBb12 Enhances Intestinal Antibody Response in Formula-Fed Infants. J. Parenter. Enter. Nutr. 2012, 36, 106S–117S. [Google Scholar] [CrossRef]
- Taniuchi, S.; Hattori, K.; Yamamoto, A.; Sasai, M.; Hatano, Y.; Kojima, T.; Kobayashi, Y.; Iwamoto, H.; Yaeshima, T. Administration of bifidobacterium to infants with atopic dermatitis: Changes in fecal microflora and clinical symptoms. J. Appl. Res. 2005, 5, 387–396. Available online: https://www.cochranelibrary.com/central/doi/10.1002/central/CN-00569597/full (accessed on 22 November 2021).
- Matsuda, F.; Chowdhury, M.; Saha, A.; Asahara, T.; Nomoto, K.; Tarique, A.; Ahmed, T.; Nishibuchi, M.; Cravioto, A.; Qadri, F. Evaluation of a probiotics, Bifidobacterium breve BBG-01, for enhancement of immunogenicity of an oral inactivated cholera vaccine and safety: A randomized, double-blind, placebo-controlled trial in Bangladeshi children under 5 years of age. Vaccine 2011, 29, 1855–1858. [Google Scholar] [CrossRef] [PubMed]
- Mohan, R.; Koebnick, C.; Schildt, J.; Schmidt, S.; Mueller, M.; Possner, M.; Radke, M.; Blaut, M. Effects of Bifidobacterium lactis Bb12 Supplementation on Intestinal Microbiota of Preterm Infants: A Double-Blind, Placebo-Controlled, Randomized Study. J. Clin. Microbiol. 2006, 44, 4025–4031. [Google Scholar] [CrossRef] [PubMed]
- Alcon-Giner, C.; Dalby, M.J.; Caim, S.; Ketskemety, J.; Shaw, A.; Sim, K.; Lawson, M.A.; Kiu, R.; LeClaire, C.; Chalklen, L.; et al. Microbiota Supplementation with Bifidobacterium and Lactobacillus Modifies the Preterm Infant Gut Microbiota and Metabolome: An Observational Study. Cell Rep. Med. 2020, 1, 100077. [Google Scholar] [CrossRef]
- Kitajima, H.; Sumida, Y.; Tanaka, R.; Yuki, N.; Takayama, H.; Fujimura, M. Early administration of Bifidobacterium breve to preterm infants: Randomised controlled trial. Arch. Dis. Child.-Fetal Neonatal Ed. 1997, 76, F101–F107. [Google Scholar] [CrossRef]
- Stratiki, Z.; Costalos, C.; Sevastiadou, S.; Kastanidou, O.; Skouroliakou, M.; Giakoumatou, A.; Petrohilou, V. The effect of a bifidobacter supplemented bovine milk on intestinal permeability of preterm infants. Early Hum. Dev. 2007, 83, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Mihatsch, W.; Vossbeck, S.; Eikmanns, B.; Hoegel, J.; Pohlandt, F. Effect of Bifidobacterium lactis on the Incidence of Nosocomial Infections in Very-Low-Birth-Weight Infants: A Randomized Controlled Trial. Neonatology 2010, 98, 156–163. [Google Scholar] [CrossRef]
- Dilli, D.; Aydin, B.; Fettah, N.D.; Özyazıcı, E.; Beken, S.; Zenciroğlu, A.; Okumuş, N.; Özyurt, B.M.; Ipek, M.; Akdağ, A.; et al. The ProPre-Save Study: Effects of Probiotics and Prebiotics Alone or Combined on Necrotizing Enterocolitis in Very Low Birth Weight Infants. J. Pediatr. 2015, 166, 545–551.e1. [Google Scholar] [CrossRef]
- Patole, S.K.; Rao, S.C.; Keil, A.D.; Nathan, E.A.; Doherty, D.A.; Simmer, K. Benefits of Bifidobacterium breve M-16V Supplementation in Preterm Neonates—A Retrospective Cohort Study. PLoS ONE 2016, 11, e0150775. [Google Scholar] [CrossRef]
- Fujii, T.; Ohtsuka, Y.; Lee, T.; Kudo, T.; Shoji, H.; Sato, H.; Nagata, S.; Shimizu, T.; Yamashiro, Y. Bifidobacterium breve en-hances transforming growth factor β1 signaling by regulating smad7 expression in preterm infants. J. Pediatric Gas-Troenterol. Nutr. 2006, 43, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Costeloe, K.; Hardy, P.; Juszczak, E.; Wilks, M.; Millar, M.R. Bifidobacterium breve BBG-001 in very preterm infants: A randomised controlled phase 3 trial. Lancet 2015, 387, 649–660. [Google Scholar] [CrossRef]
- Li, Y.; Shimizu, T.; Hosaka, A.; Kaneko, N.; Ohtsuka, Y.; Yamashiro, Y. Effects of bifidobacterium breve supplementation on intestinal flora of low birth weight infants. Pediatr. Int. 2004, 46, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Satoh, Y.; Shinohara, K.; Umezaki, H.; Shoji, H.; Satoh, H.; Ohtsuka, Y.; Shiga, S.; Nagata, S.; Shimizu, T.; Yamashiro, Y. Bifidobacteria pre-vents nec and infection in preterm infants. Int. J. Probiotics Prebiotics 2007, 2, 149–154. [Google Scholar]
- Hikaru, U.; Koichi, S.; Yayoi, S.; Hiromichi, S.; Hiroaki, S.; Yoshikazu, O.; Seigo, S.; Satoru, N.; Toshiaki, S.; Yuichiro, Y. Bifidobacteria prevents preterm infants from developing infection and sepsis. Int. J. Probiotics Prebiot-Ics 2010, 5, 33–36. [Google Scholar]
- Wang, C.; Shoji, H.; Sato, H.; Nagata, S.; Ohtsuka, Y.; Shimizu, T.; Yamashiro, Y. Effects of Oral Administration of Bifidobacterium breve on Fecal Lactic Acid and Short-chain Fatty Acids in Low Birth Weight Infants. J. Pediatr. Gastroenterol. Nutr. 2007, 44, 252–257. [Google Scholar] [CrossRef]
- Athalye-Jape, G.; Minaee, N.; Nathan, E.; Simmer, K.; Patole, S. Outcomes in preterm small versus appropriate for gestation infants after Bifidobacterium breve M-16 V supplementation. J. Matern. Neonatal Med. 2018, 33, 2209–2215. [Google Scholar] [CrossRef]
- Lin, H.-C.; Hsu, C.-H.; Chen, H.-L.; Chung, M.-Y.; Hsu, J.-F.; Lien, R.-I.; Tsao, L.-Y.; Chen, C.-H.; Su, B.-H. Oral Probiotics Prevent Necrotizing Enterocolitis in Very Low Birth Weight Preterm Infants: A Multicenter, Randomized, Controlled Trial. Pediatrics 2008, 122, 693–700. [Google Scholar] [CrossRef]
- Braga, T.D.; Da Silva, G.A.P.; De Lira, P.I.C.; Lima, M.D.C. Efficacy of Bifidobacterium breve and Lactobacillus casei oral supplementation on necrotizing enterocolitis in very-low-birth-weight preterm infants: A double-blind, randomized, controlled trial. Am. J. Clin. Nutr. 2010, 93, 81–86. [Google Scholar] [CrossRef]
- Ishizeki, S.; Sugita, M.; Takata, M.; Yaeshima, T. Effect of administration of bifidobacteria on intestinal microbiota in low-birth-weight infants and transition of administered bifidobacteria: A comparison between one-species and three-species administration. Anaerobe 2013, 23, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Patole, S.; Keil, A.D.; Chang, A.; Nathan, E.; Doherty, D.; Simmer, K.; Esvaran, M.; Conway, P. Effect of Bifidobacterium breve M-16V Supplementation on Fecal Bifidobacteria in Preterm Neonates—A Randomised Double Blind Placebo Controlled Trial. PLoS ONE 2014, 9, e89511. [Google Scholar] [CrossRef] [PubMed]
- Totsu, S.; Yamasaki, C.; Terahara, M.; Uchiyama, A.; Kusuda, S. Probiotics Study Group in Japan Bifidobacterium and enteral feeding in preterm infants: Cluster-randomized trial. Pediatr. Int. 2014, 56, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.L.; Bokulich, N.A.; Kalanetra, K.M.; Mirmiran, M.; Elumalai, J.; Haapanen, L.; Schegg, T.; Rutledge, J.C.; Raff, G.; Mills, D.A.; et al. Probiotic administration in congenital heart disease: A pilot study. J. Perinatol. 2013, 33, 691–697. [Google Scholar] [CrossRef] [PubMed]
| Probiotic Strain | Dose | Target | Animal Model (n)/Start at Postnatal Age/Treatment | Clinical Outcome Results | Refs. |
|---|---|---|---|---|---|
| B. breve M16V | 1 × 109 CFU/mL drinking water | Caesarean section | NIH Swiss mice (n = 6–14)/1 d through nursing dams/daily to 21 d | Restored early life deficit in the Bifidobacterium spp. abundance. Restored neonatal recognition abilities and maternal attachment deficits | [102] |
| B. breve UCC2003 | 2 × 106 CFU | Intestinal barrier | C57BL/6J mice (n = 10)/2 w by oral gavage/3 consecutive days | Changes in neonatal intestinal epithelial cells transcriptome genes/pathways involved in epithelial barrier function | [159] |
| B. breve M-16V | ≈4 × 107 CFU | Mucosal immunity | Lewis rats (n = 8)/6 d by oral gavage/daily to 18 d | Improved development of mucosal immunity in early life. Enhanced intestinal IgA synthesis | [160] |
| B. breve M-16V | 5 × 108 CFU | Mucosal immunity | F344/Du rats (n = 9–14)/1 d or 21 d by oral gavage/daily 2 w | Reduced expression of inflammatory molecules during the new-born period. Promoted tolerance by unregulated expression of CD3 during the weaning period | [161] |
| B. breve M-16V | 5 × 107 CFU + 20 mg oligosaccharides | Mucosal immunity | C57BL6 mice (n = 7–14)/6 d or 14 d by oral gavage/daily 1 w | Differentially expressed genes related to metabolism and immune responses. Enhanced gut immune and endocrine development in suckling mice | [162,163] |
| B. longum AH1206, B. breve AH1205 | 2 × 109 CFU | Mucosal immunity | BALB/c mice (n = 8–10)/1 d by oral gavage/daily 6 w BALB/c and Swiss Webster GF mice (n = 8–10)/6 w by oral gavage/daily 4 w | B. longum augmented T reg cell in neonatal and adult mice B. breve enhanced the Peyer’s Patch and the splenic T reg cells when administered from birth | [164] |
| B. infantis n/a | n/a | Oral tolerance | BALB/c mice (n = 5)/Neonatal or adolescent by oral gavage/n/a Oral challenge with OVA | Bifidobacteria administered in neonates, but not at a later age, restored the susceptibility of Th2 responses to oral tolerance induction | [165] |
| B. infantis n/a | n/a | Oral tolerance | BALB/c mice (n = 5)/Dams by oral gavage/n/a | Restored oral tolerance at similar levels of SPF mice | [166] |
| B. bifidum TMC3115 | 1 × 109 CFUfreeze-dried | Allergy | BALB/c mice (n = 54)/1 d by oral gavage/3 w Intraperitoneal OVA challenge | Minor increases in serum IgE levels induced by OVA-challenge in adult stage and significantly higher TNF-α and IL-10 levels | [167] |
| B. lactis BB-12 | 1 × 109 CFU | Allergy | BALB/c mice (n = 6–9)/1 d by oral gavage/every second day to 8 w of life Aerosolized OVA challenge | Suppression of all aspects of the asthmatic phenotype: airway reactivity, antigen-specific immunoglobulin E production and pulmonary eosinophilia | [168] |
| B. breve M16V | Diet supplemented with 1 × 106 CFU | Allergy | BALB/c mice (n = 6–21)/Pregnant and nursing dams by diet/2 w Exposition to air pollutant and aerosolized | Maternal supplementation with bifidobacteria prevents their offspring from allergic airway inflammation accelerated by the prenatal exposure to an air pollutant aerosol | [169] |
| B. longum subsp. longum CCM7952 | 2 × 108 CFU | Allergy | BALB/c mice (n = 8–12)/mono-associated GF dams by oral gavage/1 dose Subcutaneously sensitization with pollen allergen | Neonatal mother-to-offspring mono-colonization with bifidobacteria significantly reduced the development of allergen-specific immune responses | [170] |
| B. breve M16V | Diet with FOS and 2 × 109 CFU/g | Allergy | C3H/HeOuJ mice (n = 6–8)/3 w by diet/9 d Intradermal whey-challenge | Partial non-responsiveness to whey protein in mice orally exposed to β-lactoglobulin-derived peptides | [171] |
| B. breve M16V | Diet with FOS/GOS and 2 × 109 CFU/g | Allergy | C3H/HeOuJ mice (n = 6)/3 w by diet/7 w Intradermal whey-challenge | Reduction of the allergic effector response in a murine model of IgE-mediated hypersensitivity | [172] |
| B. pseudolongum | 2 × 107 cells | Allergy | BALB/c mice (n = 6)/5 w by oral gavage/2 w DNFB-induced contact hypersensitivity model | Reduction in the initial phase of the disease | [173] |
| B. bifidum TMC3115 | 1 × 109 CFU + antibiotics | Allergy | BALB/c mice (n = 18)/1 d by oral gavage/21 d | Significantly mitigated altered composition of the intestinal microbiota, serum total IgE levels, and the morphology and function of the intestinal epithelium | [174] |
| B. dentium ATCC27678, B. infantis ATCC15697, B. breve ATCC15698, B. bifidum ATCC29521 | 1.1 × 109 CFU | Neurodevelopment | GF Swiss Webster mice (n = 17)/1 d by oral gavage/every other day to 21 d, when weekly 21 d–6 w | Infant-type Bifidobacterium species mimics colonization with a complex microbiota. Restored aspects of normal anxiety-like behavior in a strongly sex-dependent manner. Improved recognition memory | [148,175] |
| B. pseudocatenulatum CECT7765 | 1 × 108 CFU | Chronic stress | C57Bl/6J mice (n = 18)/2 d by oral gavage/3 w Dairy maternal separation | Attenuation of some aspects of the excessive stress response of the HPA axis, particularly corticosterone production at baseline and in response to acute stress in adulthood | [176] |
| B. infantis | 1 × 109 CFU | Acute stress | BALB/c mice (n = 18–24)/mono-associated GF dams by oral gavage/1 dose | Reversed the exaggerated HPA stress response by GF mice | [177] |
| B. infantis (Chr Hansen) | 1 × 109 CFU/freeze-dried | NEC | SD rats (n = 24)/1 d by oral gavage/3 d | Reduction in the incidence of NEC | [178] |
| B. infantis BB-02 | 3 × 106 CFU | NEC | C57BL/6 mice (n = 4–27)/1 d by oral gavage/3 d | Attenuation of the increase in intestinal permeability and decrease of the incidence of NEC | [179] |
| Bifidobacterium sp. | 1× 1010 CFU/mL in microcapsules | NEC | SPF SD rats (n = 15)/1 d by oral gavage/3 d | Reduced NEC and intestinal damage severity. | [180] |
| B. adolescentis | 1 × 108 CFU | NEC | SD rats (n = 15)/1 d by oral gavage/3 d | Prevention of NEC and significantly decreased the rate of NEC-like intestinal injury. | [181] |
| B. bifidum OLB6378 | 5 × 106 CFU | NEC | SD rats (n = 30)/1 d by oral gavage/4 d | Decreased the incidence of NEC and normalized the expression and localization of tight junction and adherents junction proteins in the ileum | [182] |
| B. breve M-16V | 6 × 107 CFU | NEC | SD rats (n = 17)/1 d by oral gavage/4 d | Suppressed the increased expression of molecules related to inflammation and barrier function that resulted from NEC induction | [183] |
| B. longum subsp. infantis ATCC15697 | 5 × 106 CFU | NEC | SD rats (n = 19)/1 d by oral gavage/4 d | Significantly reduced associated inflammation and incidence of NEC | [184] |
| B. bifidum OLB6378 | 5 × 106 CFU | NEC | SD rats (n = 30)/1 d by oral gavage/4 d | Attenuation of induction of antimicrobial peptides and NEC incidence | [185] |
| B. bifidum PM-A0218 and B. longum PM-A0101 | 1 × 108 CFU | NEC | SD rats (n = 12)/1 d by oral gavage/3 d | Lower mortality | [186] |
| Bifidobacterium sp. | 1 × 108 CFU/daily | NEC | SD rats (n = 20)/1 d by oral gavage/3 d | Decreased incidence and reduced the severity of NEC. Inhibition of proinflammatory cytokine secretion and improvement of intestinal barrier integrity. | [187] |
| B. breve YIT4064 | 0.05% of diet, heat-killed | RV-induced diarrhea | BALB/c mice (n = 39)/dams before and after delivery by diet/9 w /pups 5 d-old challenged with RV | Pups born and nursed by dams fed with bifidobacteria were more strongly protected against RV-induced diarrhea. | [188] |
| B. bifidum ATCC15696 and B. infantis ATCC15697 | 0.75 × 108 CFU/mL and 0.75 × 108 CFU/mL | RV-induced diarrhea | BALB/c mice (n = 35)/1 d by oral gavage/7 w: 1 dose/weekly Pups 5 d-old challenged with RV | Significantly delayed and early resolution of diarrhea | [189] |
| B. longum SPM1206 and B. longum SPM1205 | Sonicated extract of 2 × 108 CFU | RV-induced diarrhea | BALB/c (n = n/a)/12 d by oral gavage/3 d Previous RV infection | Inhibited rotavirus gene expression and replication with significant increase of IFN-α and IFN-β gene expression | [190] |
| B. breve M16V | 2.5 × 109 CFU + starch | Colitis | F344 rats (n = 6–12)/21 d by oral gavage/3 w DSS-colitis | Bifidobacteria modulates normal systemic T-cell immune functions. Under inflammatory conditions ameliorates DSS-induced colitis in weanling rats. | [191] |
| B. animalis | 1 × 107 CFU | Pathogen inhibition | C57BL/6 athymic bg/bg-nu/nu and euthymic bg/bg-nu/+ mice (n = 4–27)/Mono-associated dams by oral and anal inoculum/1 dose Infection with Candida albicans | Reduced incidence and severity | [192] |
| B. infantis ATCC15697 | culture supernatant | Pathogen inhibition | C57BL/6 mice (n = 23–26)/1 d by oral gavage/8 d Infection with Cronobacter sakazakii | Protection against C. sakazakii-induced intestinal inflammation | [193] |
| Probiotic Strain | Dose | Aim | Study Design/Study Population | Clinical Outcome Results | Refs. |
|---|---|---|---|---|---|
| B. longum subsp. infantis CECT7210 (B. infantis IM1) | 1 × 107 CFU/g formula | To reduce diarrhea incidence in healthy term infants | RDBC (12 w)/n = 190 (age < 3 m) | Reduction diarrhea episodes, well tolerance and lower constipation prevalence | [201] |
| B. animalis subsp. lactis BB-12 | 1.5 × 108 CFU/L milk formula supplemented | To prevent acute diarrhea | RCT (52 w)/n = 90 (age < 8 m) | Reduced risk of diarrhea by a factor of 1.9 (range, 1.33–2.6) | [202] |
| B. animalis subsp. lactis | 3.6 × 107 CFU/g formula | To determined growth and stool characteristics | Double-blind study/ (up to 4-m age) n = 88 (age 6–10 d) | Protection against diarrheal illness | [203] |
| B. animalis subsp. lactis | 1 × 107–108 CFU/g formula | To study the growth of HIV-exposed uninfected infants | RDBPC (4 m)/n = 131 (age 37–42 w) | Well growth, increased head growth and a trend towards increased weight gain | [204] |
| B. lactis CNCM I-3446 | 67 kcal/100 mL formula Ad libitum | To study the growth of HIV-exposed uninfected infants | RDBPC (6 m)/n = 132 (age 37–42 w) | Growth and metabolism in HIV-negative infants fed were not affected | [205] |
| B. animalis subsp. lactis BB-12 | 6 × 109 CFU/100 mL formula | To study the effects on the sIgA levels | RDBC (32 w)/n = 57 (age 0–32 w) | Trend towards higher fecal sIgA levels—statistically significant at the age of 16 weeks | [207] |
| B. animalis subsp. lactis BB-12 | 1 × 106 CFU/g formula | To study the effects on intestinal immunity and inflammation | Prospective RDBC (6 w)/n = 172 (age 6 w) | Increment of fecal sIgA | [208] |
| B. breve M-16V | 5 × 109 CFU/g cow’s milk | To elucidate the effect on the intestinal microbiota of infants with cow’s milk hypersensitivity and atopic dermatitis | RCT (12 w)/n = 17 (age 2–5 y) | Improvement in the allergic symptoms and increment of bifidobacteria in feces | [209] |
| B. breve strain Yakult (BBG-01) | 4 × 109 CFU/g | To enhances the immunogenicity of oral cholera vaccine | RDBPC (4 w)/n = 128 (age 2–5 y) | Well tolerance. Post vaccinal immunostimulatory effect was not evident | [210] |
| Probiotic Strain | Dose | Aim | Study Design/STUDY Population | Clinical Outcome Results | Refs. |
|---|---|---|---|---|---|
| B. lactis BB-12 | 2.9 × 109 CFU/g formula | To reduce the potentially harmful bacteria | RDBPC (1 m)/n = 69 (GA < 37 w) | Bifidobacteria increment. Enterobacteria and clostridia reduction. | [211] |
| B. bifidum and L. acidophilus | 2.0 × 109 CFU/twice day | To explore the gut microbiota composition and fecal metabolome | Observational study (99 d)/n = 101 (GA < 34 w) | Predominance of Bifidobacterium and a lower abundance of pathobionts. Higher fecal acetate and lactate | [212] |
| B. breve YIT4010 | 5.0 × 109 CFU/day | To investigate the colonization with B. breve | RCT (4 m)/n = 91 (GA 25–28 w) | B. breve colonization. Fewer abnormal abdominal signs. Better weight gain. | [213] |
| B. lactis BB-12 | 2.0 × 107 CFU/g of dry milk | To determine the effect on intestinal permeability, somatic growth, tolerance, rates of sepsis and NEC | Prospective randomized case–control study (30 d)/n = 41 (GA 27–37 w) | Reduced intestinal permeability and increased head growth | [214] |
| B. lactis BB-12 | 1.2 × 1010 CFU/kg/day | To reduce the incidence of nosocomial infections | RCT (6 w)/n = 183 (GA 23–26 and 27–29 w) | The incidence was not reduced, and no adverse effect was observed | [215] |
| B. lactis | 5.0 × 109 CFU/day | To prevent NEC | Prospective RCT (8 w)/n = 400 (GA 28.8 ± 1.9 w) | Significant reduction in the incidence of NEC | [216] |
| B. breve M-16V | 3.0 × 109 CFU/day | To reduce the incidence of NEC | Retrospective cohort study (over the course of 2 y)/n = 1755 (GA < 34 w) | Decrease incidence of NEC ≥ Stage II or all-cause mortality | [217] |
| B. breve M-16V | 1.0 × 109 cells/0.5 mL/twice a day | To examine the effect on the immunologic system in relation to TGF-β | RCT (59 ± 29.3 d)/n = 19 (GA 31.3 ± 3.16 w) | Up-regulation of TGF-β1 signaling and attenuation of inflammatory and allergic reactions | [218] |
| B. breve BBG-001 | 8.2–9.2 × 1010 CFU | To reduce NEC, LOS, and death in preterm infants | Multicenter blinded randomized controlled phase 3 study (36 w)/n = 1315 (GA 23–30 w) | There was no evidence of benefit for this intervention in this population | [219] |
| B. breve | 1.6 × 108 CFU/twice a day | To evaluate positive effect on gut microbiota | RCT (7 w)/n = 30 (GA 27.8–37.6 w) | Promotion on Bifidobacterium colonization and normal gut microbiota development | [220] |
| B. breve M-16V | 1.10 × 109 CFU/day | To prevent NEC and infection | Control study (4 y)/n = 338 (GA 27–36 w) | NEC and infection were prevented | [221] |
| B. breve M-16V | 1 × 109 CFU/g | To prevent infection and sepsis | RCT (91.8 ± 54.1 d)/n = 108 (GA 28.1 ± 2.8 w) | Development of infections and sepsis were significantly lower | [222] |
| B. breve M-16V | 1.6 × 108 CFU twice daily until discharge | To determine the effects on fecal lactic acid and SCFAs | RCT (4 w)/n = 66 (GA 23–36 w) | Butyric acid was reduced | [223] |
| B. breve M-16V | 3 × 109 CFU/day | To compare clinical outcomes between preterm SGA vs. AGA infants after probiotic administration | Retrospective cohort study (3 y)/n = 1380 (GA < 34 w) | NEC, LOS, and all-cause mortality rates were similar in preterm SGA vs AGA infants | [224] |
| B. bifidum and L. acidophilus | 1 × 109 CFU/125 mg/kg twice daily | To prevent NEC | Prospective, blinded, randomized, multicenter controlled(6 w)/n = 580 (GA < 34 w and <1500 g) | Incidence reduction of death or NEC | [225] |
| B. breve and L. casei | 3.5 × 107 to 3.5 × 109 CFU | To prevent the occurrence of NEC stage ≥ 2 by criteria of Bell | RDBC (30 d)/n = 231 (GA 29.5 ± 2.5 w) | Reduced the occurrence of NEC (Bell’s stage ≥ 2) | [226] |
| B. breve M-16V | 5 × 108 CFU/day | To investigate the effects on the intestinal microbiota | RCT (6 w)/n = 46 (GA 29.9 ± 2.3 w) | Promotion of bifidobacteria colonization and the formation of a healthy microbiota | [227] |
| B. breve M-16V | 3 × 109 CFU/day | To increase fecal B. breve counts without adverse effects | RDBPC (6 w)/n = 159 (GA < 33 w) | Increased the proportion of neonates with detectable B. breve | [228] |
| B. bifidum OLB6378 | 2.5 × 109 viable cells/500 mg/twice a day | To evaluate the benefit on enteral feeding | RCT study (21 d)/n = 585 (GA 28.6 ± 2.9 w) | Acceleration of the establishment of enteral feeding after birth | [229] |
| B. longum subsp. infantis ATCC 15697 | 4.2 × 109 CFU/two times daily | To investigate the impact on the fecal microbiota and plasma cytokines in neonates with congenital heart disease | RCT (for 8 w)/n = 16 (GA 38.4 ± 1.2 w) | There was no significant alteration in the gut microbiota and plasma interleukin (IL)10, interferon (IFN) ɣ and IL1β levels were higher | [230] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saturio, S.; Nogacka, A.M.; Alvarado-Jasso, G.M.; Salazar, N.; de los Reyes-Gavilán, C.G.; Gueimonde, M.; Arboleya, S. Role of Bifidobacteria on Infant Health. Microorganisms 2021, 9, 2415. https://doi.org/10.3390/microorganisms9122415
Saturio S, Nogacka AM, Alvarado-Jasso GM, Salazar N, de los Reyes-Gavilán CG, Gueimonde M, Arboleya S. Role of Bifidobacteria on Infant Health. Microorganisms. 2021; 9(12):2415. https://doi.org/10.3390/microorganisms9122415
Chicago/Turabian StyleSaturio, Silvia, Alicja M. Nogacka, Guadalupe M. Alvarado-Jasso, Nuria Salazar, Clara G. de los Reyes-Gavilán, Miguel Gueimonde, and Silvia Arboleya. 2021. "Role of Bifidobacteria on Infant Health" Microorganisms 9, no. 12: 2415. https://doi.org/10.3390/microorganisms9122415
APA StyleSaturio, S., Nogacka, A. M., Alvarado-Jasso, G. M., Salazar, N., de los Reyes-Gavilán, C. G., Gueimonde, M., & Arboleya, S. (2021). Role of Bifidobacteria on Infant Health. Microorganisms, 9(12), 2415. https://doi.org/10.3390/microorganisms9122415






