Phlebotomus papatasi Antimicrobial Peptides in Larvae and Females and a Gut-Specific Defensin Upregulated by Leishmania major Infection
Abstract
1. Introduction
2. Materials and Methods
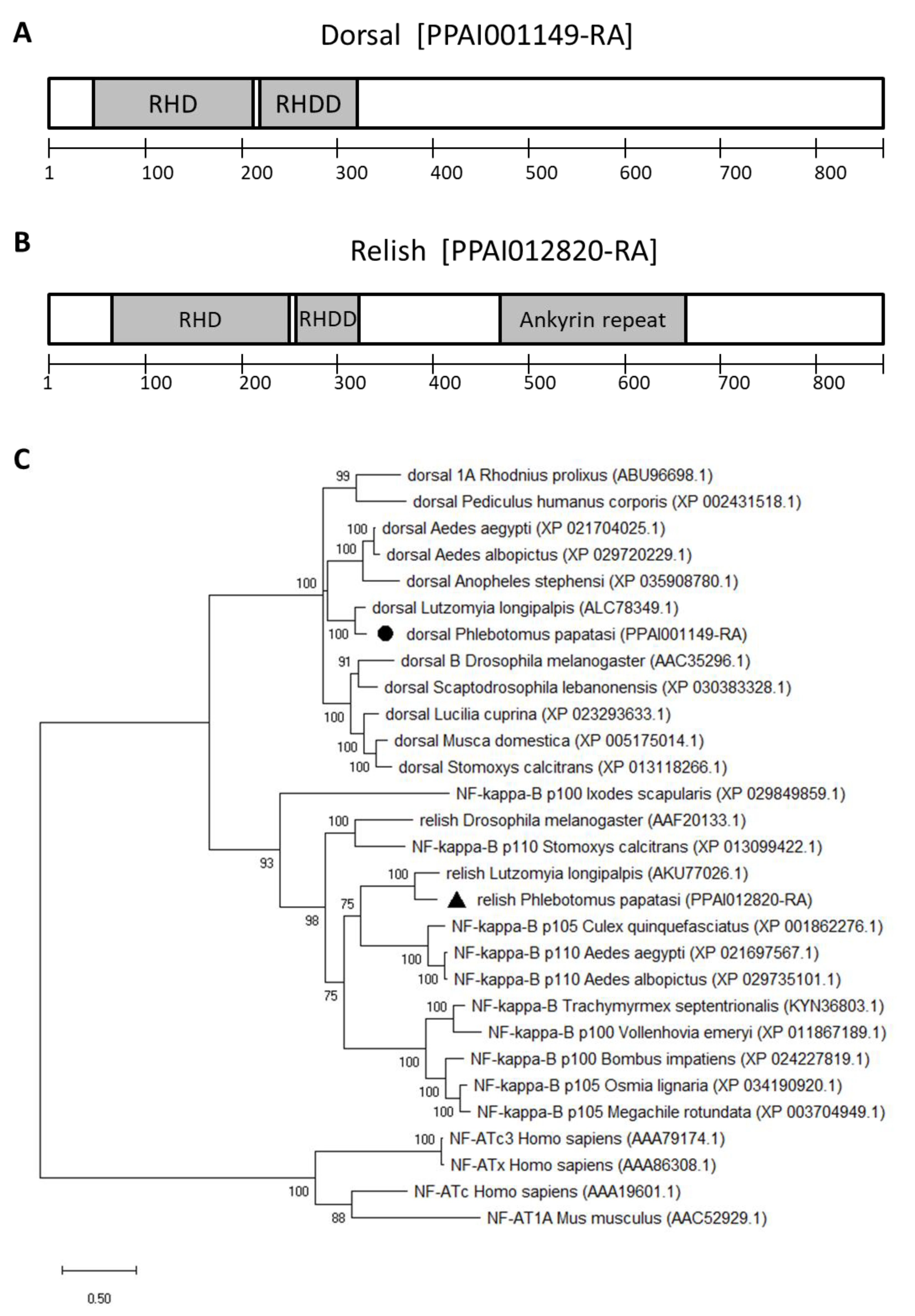
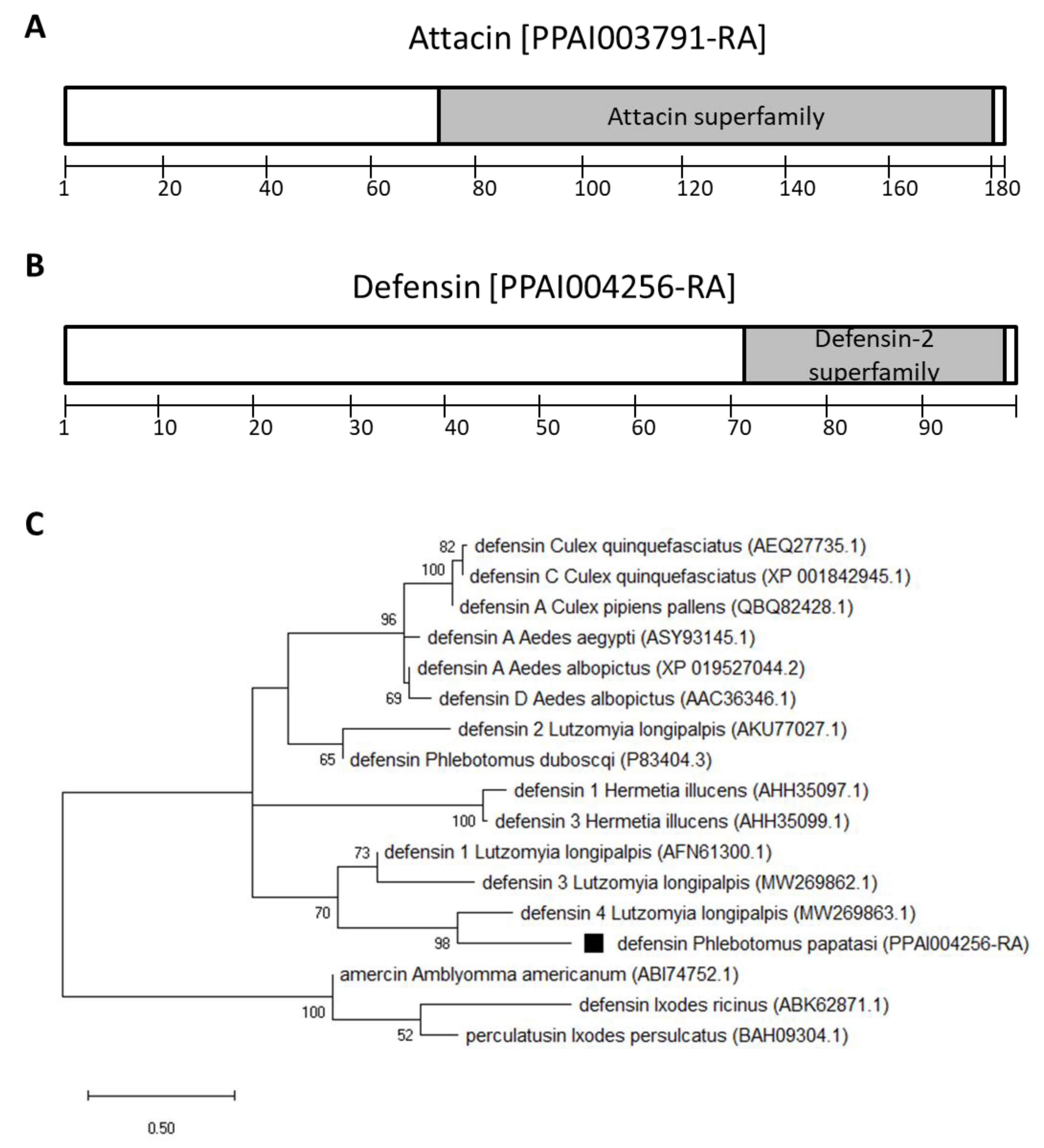
2.1. P. papatasi Immunity Genes
2.2. L. major Culture
2.3. P. papatasi Colony Rearing
2.4. Larvae Experimental Feeding
2.5. Depletion of Sand Fly Gut Bacteria
2.6. Leishmania Experimental Infection
2.7. RNA Extraction, cDNA Synthesis, and PCR
2.8. Relative Gene Expression by qPCR
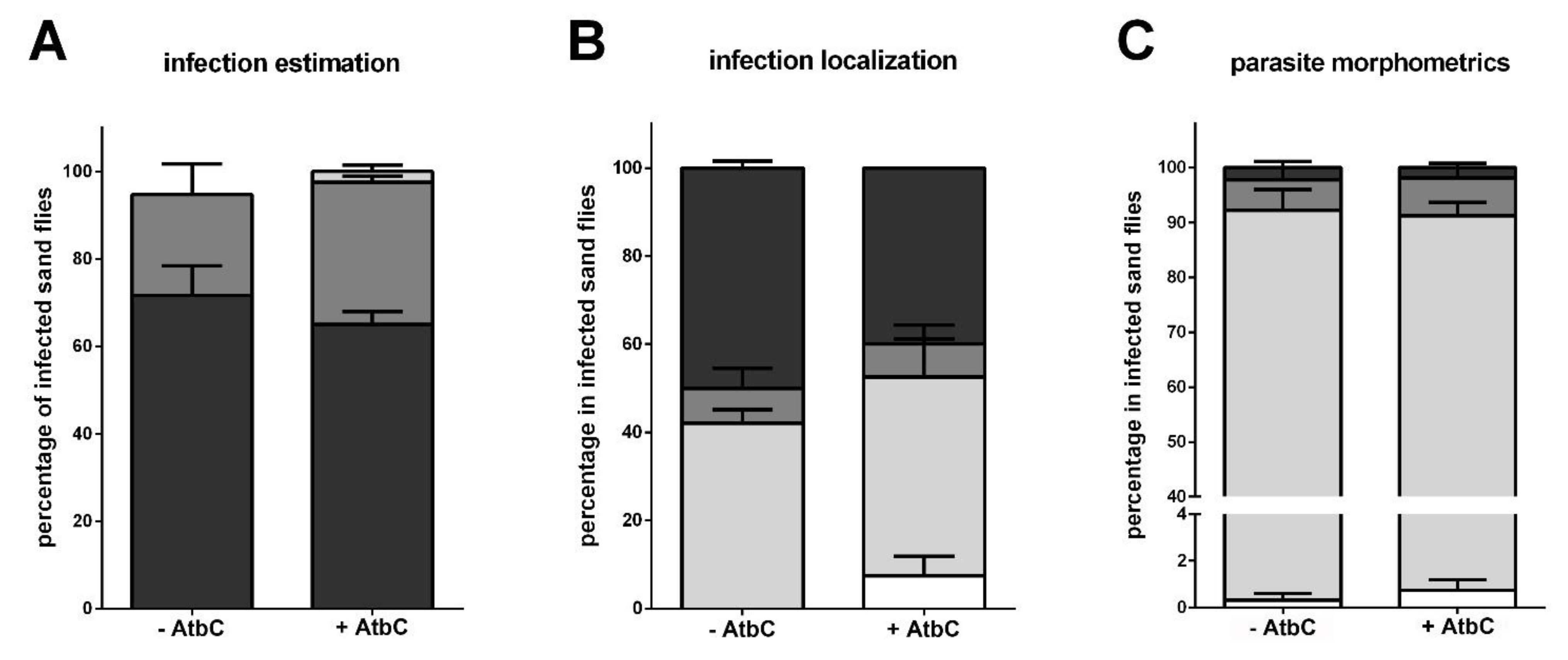
2.9. Leishmania Infection Estimation and Morphometrics
| Reference | Gene | Sequence |
|---|---|---|
| Nadkarni et al. 2002 [35] | Bacteria 16S rRNA | 5′ TCCTACGGGAGGCAGCAGT 3′ |
| 5′ GGACTACCAGGGTATCTAATCCTGTT 3′ | ||
| Di-Blasi et al. 2015 [36] | Leishmania actin | 5′ GTCGTCGATAAAGCCGAAGGTGGTT 3′ |
| 5′ TTGGGCCAGACTCGTCGTACTCGCT 3′ | ||
| (PPAI004850) | P. papatasi actin | 5′ GCACATCCCTGGAGAAATCCTAT 3′ |
| 5′ GGAAAGATGGCTGGAAGAGAGAT 3′ | ||
| (PPAI003791) | P. papatasi attacin | 5′ GCCATTTCTGCTGCGTACTC 3′ |
| 5′ GAGGCACCAAGTACACGACA 3′ | ||
| (PPAI004256) | P. papatasi defensin | 5′ GCCCGGTTAAAGACGATGTAAAG 3′ |
| 5′ AGTTGGTCCAAGGATATCGCAAG 3′ | ||
| (PPAI001149) | P. papatasi dorsal | 5′ GCTGCAAATCCTGCAAAGA 3′ |
| 5′ CCCAAGGAGGTCACAGGTTA 3′ | ||
| Louradour et al. 2019 [17] | P. papatasi relish | 5′ ATCCATCCTTTATGCAACCG 3′ |
| 5′ GCCTTTGAGTCGCAGTATCC 3′ | ||
| (PPAI008202) | P. papatasi ribosomal protein L8 | 5′ GACATGGATACCTCAAGGGAGTC 3′ |
| 5′ TTGCGGATCTTATAGCGATAGGG 3′ | ||
| VectorBase gene identification shown in parenthesis. | ||
2.10. Statistical Analysis
3. Results
3.1. Transcription Factors and Antimicrobial Peptide Genes
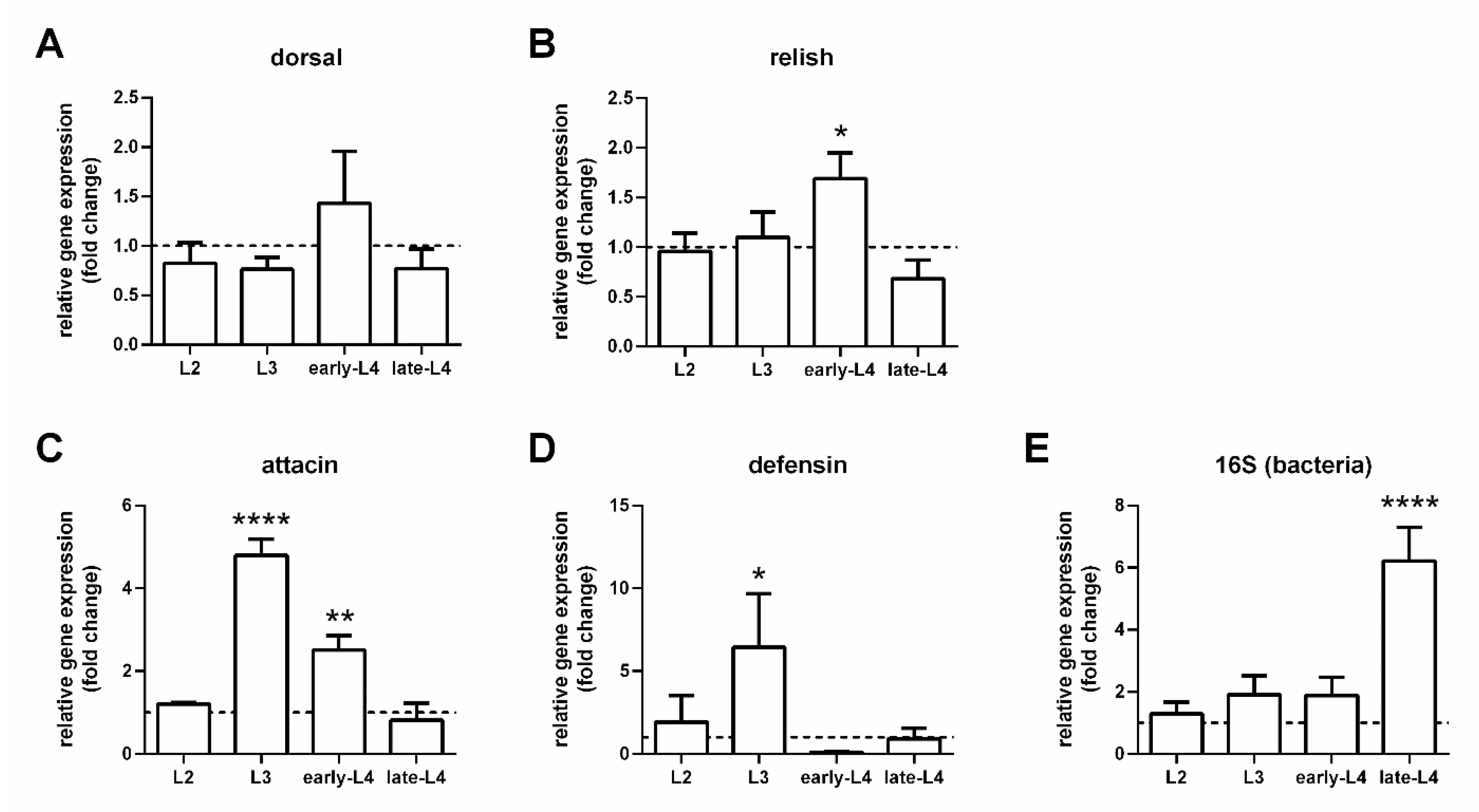
3.2. Expression of Immunity Genes in Larval Guts
3.3. Expression of Immunity Genes in Infected Females with Depleted Gut Bacteria
3.4. Immunity Genes and Infection Progression in Infected Females with Recovered Gut Bacteria
3.5. Defensin Gut-Specific Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maroli, M.; Feliciangeli, M.D.; Bichaud, L.; Charrel, R.N.; Gradoni, L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med. Vet. Entomol. 2013, 27, 123–147. [Google Scholar] [CrossRef] [PubMed]
- WHO. Leishmaniasis. Available online: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 15 September 2021).
- Hamarsheh, O. Distribution of Leishmania major zymodemes in relation to populations of Phlebotomus papatasi sand flies. Parasit. Vectors 2011, 4, 9. [Google Scholar] [CrossRef]
- Dostalova, A.; Volf, P. Leishmania development in sand flies: Parasite-vector interactions overview. Parasit. Vectors 2012, 5, 276. [Google Scholar] [CrossRef]
- Bates, P.A.; Depaquit, J.; Galati, E.A.; Kamhawi, S.; Maroli, M.; McDowell, M.A.; Picado, A.; Ready, P.D.; Salomon, O.D.; Shaw, J.J.; et al. Recent advances in phlebotomine sand fly research related to leishmaniasis control. Parasit. Vectors 2015, 8, 131. [Google Scholar] [CrossRef]
- Schlein, Y.; Polacheck, I.; Yuval, B. Mycoses, bacterial infections and antibacterial activity in sandflies (Psychodidae) and their possible role in the transmission of leishmaniasis. Parasitology 1985, 90 Pt 1, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.J.; el Kordy, E.; Shehata, M.; Lane, R.P. The prevalence of a microbiota in the digestive tract of Phlebotomus papatasi. Ann. Trop. Med. Parasitol. 1996, 90, 669–673. [Google Scholar] [CrossRef]
- Guernaoui, S.; Garcia, D.; Gazanion, E.; Ouhdouch, Y.; Boumezzough, A.; Pesson, B.; Fontenille, D.; Sereno, D. Bacterial flora as indicated by PCR-temperature gradient gel electrophoresis (TGGE) of 16S rDNA gene fragments from isolated guts of phlebotomine sand flies (Diptera: Psychodidae). J. Vector Ecol. 2011, 36, S144–S147. [Google Scholar] [CrossRef]
- Akhoundi, M.; Bakhtiari, R.; Guillard, T.; Baghaei, A.; Tolouei, R.; Sereno, D.; Toubas, D.; Depaquit, J.; Abyaneh, M.R. Diversity of the bacterial and fungal microflora from the midgut and cuticle of phlebotomine sand flies collected in North-Western Iran. PLoS ONE 2012, 7, e50259. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, J.; Braig, H.R.; Rowton, E.D.; Ghosh, K. Naturally occurring culturable aerobic gut flora of adult Phlebotomus papatasi, vector of Leishmania major in the Old World. PLoS ONE 2012, 7, e35748. [Google Scholar] [CrossRef] [PubMed]
- Maleki-Ravasan, N.; Oshaghi, M.A.; Afshar, D.; Arandian, M.H.; Hajikhani, S.; Akhavan, A.A.; Yakhchali, B.; Shirazi, M.H.; Rassi, Y.; Jafari, R.; et al. Aerobic bacterial flora of biotic and abiotic compartments of a hyperendemic Zoonotic Cutaneous Leishmaniasis (ZCL) focus. Parasit. Vectors 2015, 8, 63. [Google Scholar] [CrossRef]
- Karakuş, M.; Karabey, B.; Orçun Kalkan, Ş.; Özdemir, G.; Oğuz, G.; Erişöz Kasap, Ö.; Alten, B.; Töz, S.; Özbel, Y. Midgut Bacterial Diversity of Wild Populations of Phlebotomus (P.) papatasi, the Vector of Zoonotic Cutaneous Leishmaniasis (ZCL) in Turkey. Sci. Rep. 2017, 7, 14812. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, C.; Karas, P.A.; Vasileiadis, S.; Ligda, P.; Saratsis, A.; Sotiraki, S.; Karpouzas, D.G. Host Species Determines the Composition of the Prokaryotic Microbiota in Phlebotomus Sandflies. Pathogens 2020, 9, 428. [Google Scholar] [CrossRef] [PubMed]
- Telleria, E.L.; Martins-Da-Silva, A.; Tempone, A.J.; Traub-Cseko, Y.M. Leishmania, microbiota and sand fly immunity. Parasitology 2018, 145, 1336–1353. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef] [PubMed]
- Telleria, E.L.; Sant’Anna, M.R.V.; Ortigão-Farias, J.R.; Pitaluga, A.N.; Dillon, V.M.; Bates, P.A.; Traub-Csekö, Y.M.; Dillon, R.J. Caspar-like gene depletion reduces leishmania infection in sand fly host Lutzomyia longipalpis. J. Biol. Chem. 2012, 287, 12985–12993. [Google Scholar] [CrossRef]
- Louradour, I.; Ghosh, K.; Inbar, E.; Sacks, D.L. CRISPR/Cas9 mutagenesis in Phlebotomus papatasi: The immune deficiency pathway impacts vector competence for Leishmania major. mBio 2019, 10, e01941-19. [Google Scholar] [CrossRef]
- Boulanger, N.; Lowenberger, C.; Volf, P.; Ursic, R.; Sigutova, L.; Sabatier, L.; Svobodova, M.; Beverley, S.M.; Spath, G.; Brun, R.; et al. Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania major. Infect. Immun. 2004, 72, 7140–7146. [Google Scholar] [CrossRef]
- Telleria, E.L.; Tinoco-Nunes, B.; Leštinová, T.; de Avellar, L.M.; Tempone, A.J.; Pitaluga, A.N.; Volf, P.; Traub-Csekö, Y.M. Lutzomyia longipalpis Antimicrobial Peptides: Differential Expression during Development and Potential Involvement in Vector Interaction with Microbiota and Leishmania. Microorganisms 2021, 9, 1271. [Google Scholar] [CrossRef]
- Blank, V.; Kourilsky, P.; Israël, A. NF-κB and related proteins: Rel/dorsal homologies meet ankyrin-like repeats. Trends Biochem. Sci. 1992, 17, 135–140. [Google Scholar] [CrossRef]
- Tinoco-Nunes, B.; Telleria, E.L.; Da Silva-Neves, M.; Marques, C.; Azevedo-Brito, D.A.; Pitaluga, A.N.; Traub-Csekö, Y.M. The sandfly Lutzomyia longipalpis LL5 embryonic cell line has active Toll and Imd pathways and shows immune responses to bacteria, yeast and Leishmania. Parasit. Vectors 2016, 9, 222. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A General Empirical Model of Protein Evolution Derived from Multiple Protein Families Using a Maximum-Likelihood Approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Volf, P.; Volfova, V. Establishment and maintenance of sand fly colonies. J. Vector Ecol. 2011, 36, S1–S9. [Google Scholar] [CrossRef]
- Lawyer, P.; Killick-Kendrick, M.; Rowland, T.; Rowton, E.; Volf, P. Laboratory colonization and mass rearing of phlebotomine sand flies (Diptera, Psychodidae). Parasite 2017, 24, 42. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.H.; Bahr, S.M.; Serafim, T.D.; Ajami, N.J.; Petrosino, J.F.; Meneses, C.; Kirby, J.R.; Valenzuela, J.G.; Kamhawi, S.; Wilson, M.E. The Gut Microbiome of the Vector Lutzomyia longipalpis Is Essential for Survival of Leishmania infantum. mBio 2017, 8, e01121-16. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Benkova, I.; Volf, P. Effect of temperature on metabolism of Phlebotomus papatasi (Diptera: Psychodidae). J. Med. Entomol. 2007, 44, 150–154. [Google Scholar] [CrossRef]
- Myskova, J.; Votypka, J.; Volf, P. Leishmania in Sand Flies: Comparison of Quantitative Polymerase Chain Reaction with Other Techniques to Determine the Intensity of Infection. J. Med. Entomol. 2008, 45, 133–138. [Google Scholar] [CrossRef][Green Version]
- Sadlova, J.; Price, H.P.; Smith, B.A.; Votypka, J.; Volf, P.; Smith, D.F. The stage-regulated HASPB and SHERP proteins are essential for differentiation of the protozoan parasite Leishmania major in its sand fly vector, Phlebotomus papatasi. Cell. Microbiol. 2010, 12, 1765–1779. [Google Scholar] [CrossRef]
- Rasband, W. ImageJ Website. Available online: https://imagej.nih.gov/ij/ (accessed on 10 August 2019).
- Walters, L.L.; Chaplin, G.L.; Modi, G.B.; Tesh, R.B. Ultrastructural biology of Leishmania (Viannia) panamensis (=Leishmania braziliensis panamensis) in Lutzomyia gomezi (Diptera: Psychodidae): A natural host-parasite association. Am. J. Trop. Med. Hyg. 1989, 40, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Nadkarni, M.A.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 148, 257–266. [Google Scholar] [CrossRef]
- Di-Blasi, T.; Lobo, A.R.; Nascimento, L.M.; Córdova-Rojas, J.L.; Pestana, K.; Marín-Villa, M.; Tempone, A.J.; Telleria, E.L.; Ramalho-Ortigão, M.; McMahon-Pratt, D.; et al. The flagellar protein FLAG1/SMP1 is a candidate for leishmania-sand fly interaction. Vector-Borne Zoonotic Dis. 2015, 15, 202–209. [Google Scholar] [CrossRef]
- Graef, I.A.; Gastier, J.M.; Francke, U.; Crabtree, G.R. Evolutionary relationships among rel domains indicate functional diversification by recombination. Proc. Natl. Acad. Sci. USA 2001, 98, 5740–5745. [Google Scholar] [CrossRef]
- Moorthy, A.K.; Huang, D.B.; Wang, V.Y.F.; Vu, D.; Ghosh, G. X-ray Structure of a NF-κB p50/RelB/DNA Complex Reveals Assembly of Multiple Dimers on Tandem κB Sites. J. Mol. Biol. 2007, 373, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Dushay, M.S.; Åsling, B.; Hultmark, D. Origins of immunity: Relish, a compound rel-like gene in the antibacterial defense of Drosophila. Proc. Natl. Acad. Sci. USA 1996, 93, 10343–10347. [Google Scholar] [CrossRef]
- Huguet, C.; Crepieux, P.; Laudet, V. Rel/NF-kB transcription factors and IkB inhibitors: Evolution from a unique common ancestor. Oncogene 1997, 15, 2965–2974. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gilmore, T.D.; Wolenski, F.S. NF-κB: Where did it come from and why? Immunol. Rev. 2012, 246, 14–35. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Liang, H.; Hou, Y. Functional analysis of a NF-κB transcription factor in the immune defense of Oriental fruit fly, Bactrocera dorsalis Hendel (Diptera: Tephritidae). Bull. Entomol. Res. 2017, 107, 251–260. [Google Scholar] [CrossRef]
- Mapalo, M.A.; Arakawa, K.; Baker, C.M.; Persson, D.K.; Mirano-Bascos, D.; Giribet, G. The Unique Antimicrobial Recognition and Signaling Pathways in Tardigrades with a Comparison Across Ecdysozoa. G3 Genes Genomes Genet. 2020, 10, 1137–1148. [Google Scholar] [CrossRef]
- Hedengren, M.; Borge, K.; Hultmark, D. Expression and evolution of the Drosophila attacin/diptericin gene family. Biochem. Biophys. Res. Commun. 2000, 279, 574–581. [Google Scholar] [CrossRef]
- Buonocore, F.; Fausto, A.M.; Pelle, G.D.; Roncevic, T.; Gerdol, M.; Picchietti, S. Attacins: A Promising Class of Insect Antimicrobial Peptides. Antibiotics 2021, 10, 212. [Google Scholar] [CrossRef] [PubMed]
- Bulet, P.; Stöcklin, R.; Menin, L. Anti-microbial peptides: From invertebrates to vertebrates. Immunol. Rev. 2004, 198, 169–184. [Google Scholar] [CrossRef]
- Wei, L.; Mu, L.; Wang, Y.; Bian, H.; Li, J.; Lu, Y.; Han, Y.; Liu, T.; Lv, J.; Feng, C.; et al. Purification and characterization of a novel defensin from the salivary glands of the black fly, Simulium bannaense. Parasit. Vectors 2015, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Freitak, D.; Wheat, C.W.; Heckel, D.G.; Vogel, H. Immune system responses and fitness costs associated with consumption of bacteria in larvae of Trichoplusia ni. BMC Biol. 2007, 5, 56. [Google Scholar] [CrossRef]
- Heerman, M.; Weng, J.L.; Hurwitz, I.; Durvasula, R.; Ramalho-Ortigao, M. Bacterial Infection and Immune Responses in Lutzomyia longipalpis Sand Fly Larvae Midgut. PLoS Negl. Trop. Dis. 2015, 9, e0003923. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. The microbial dimension in insect nutritional ecology. Funct. Ecol. 2009, 23, 38–47. [Google Scholar] [CrossRef]
- De Melo Ximenes, M.d.F.F.; Maciel, J.C.; Jerônimo, S.M.B. Characteristics of the Biological Cycle of Lutzomyia evandroi Costa Lima & Antunes, 1936 (Diptera: Psychodidae) under Experimental Conditions. Mem. Inst. Oswaldo Cruz 2001, 96, 883–886. [Google Scholar] [CrossRef]
- Feliciangeli, M.D. Natural breeding places of phlebotomine sandflies. Med. Vet. Entomol. 2004, 18, 71–80. [Google Scholar] [CrossRef]
- Marayati, B.F.; Schal, C.; Ponnusamy, L.; Apperson, C.S.; Rowland, T.E.; Wasserberg, G. Attraction and oviposition preferences of Phlebotomus papatasi (Diptera: Psychodidae), vector of Old-World cutaneous leishmaniasis, to larval rearing media. Parasit. Vectors 2015, 8, 663. [Google Scholar] [CrossRef] [PubMed]
- Adler, S.; Theodor, O. Transmission of Disease Agents by Phlebotomine Sand Flies. Annu. Rev. Entomol. 1957, 2, 203–226. [Google Scholar] [CrossRef]
- Volf, P.; Kiewegova, A.; Nemec, A. Bacterial colonisation in the gut of Phlebotomus duboseqi (Diptera: Psychodidae): Transtadial passage and the role of female diet. Folia Parasitol. 2002, 49, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Dey, R.; Joshi, A.B.; Oliveira, F.; Pereira, L.; Guimaraes-Costa, A.B.; Serafim, T.D.; de Castro, W.; Coutinho-Abreu, I.V.; Bhattacharya, P.; Townsend, S.; et al. Gut Microbes Egested during Bites of Infected Sand Flies Augment Severity of Leishmaniasis via Inflammasome-Derived IL-1β. Cell Host Microbe 2018, 23, 134–143.e6. [Google Scholar] [CrossRef] [PubMed]
- Sacks, D.L. Leishmania-sand fly interactions controlling species-specific vector competence. Cell. Microbiol. 2001, 3, 189–196. [Google Scholar] [CrossRef]
- Pruzinova, K.; Sadlova, J.; Seblova, V.; Homola, M.; Votypka, J.; Volf, P. Comparison of Bloodmeal Digestion and the Peritrophic Matrix in Four Sand Fly Species Differing in Susceptibility to Leishmania donovani. PLoS ONE 2015, 10, e0128203. [Google Scholar] [CrossRef]
- Coutinho-Abreu, I.V.; Sharma, N.K.; Robles-Murguia, M.; Ramalho-Ortigao, M. Targeting the midgut secreted PpChit1 reduces Leishmania major development in its natural vector, the sand fly Phlebotomus papatasi. PLoS Negl. Trop. Dis. 2010, 4, e901. [Google Scholar] [CrossRef] [PubMed]
- Atayde, V.D.; Aslan, H.; Townsend, S.; Hassani, K.; Kamhawi, S.; Olivier, M. Exosome Secretion by the Parasitic Protozoan Leishmania within the Sand Fly Midgut. Cell Rep. 2015, 13, 957–967. [Google Scholar] [CrossRef]
- Corrales, R.M.; Sereno, D.; Mathieu-Daude, F. Deciphering the Leishmania exoproteome: What we know and what we can learn. FEMS Immunol. Med. Microbiol. 2010, 58, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Forrest, D.M.; Batista, M.; Marchini, F.K.; Tempone, A.J.; Traub-Csekö, Y.M. Proteomic analysis of exosomes derived from procyclic and metacyclic-like cultured Leishmania infantum chagasi. J. Proteom. 2020, 227, 103902. [Google Scholar] [CrossRef]
- Kingsolver, M.B.; Hardy, R.W. Making connections in insect innate immunity. Proc. Natl. Acad. Sci. USA 2012, 109, 18639–18640. [Google Scholar] [CrossRef]
- Huang, Z.; Kingsolver, M.B.; Avadhanula, V.; Hardy, R.W. An antiviral role for antimicrobial peptides during the arthropod response to alphavirus replication. J. Virol. 2013, 87, 4272–4280. [Google Scholar] [CrossRef]
- Telleria, E.L.; Sant’Anna, M.R.V.; Alkurbi, M.O.; Pitaluga, A.N.; Dillon, R.J.; Traub-Csekö, Y.M. Bacterial feeding, Leishmania infection and distinct infection routes induce differential defensin expression in Lutzomyia longipalpis. Parasit. Vectors 2013, 6, 12. [Google Scholar] [CrossRef]
- Sloan, M.A.; Sadlova, J.; Lestinova, T.; Sanders, M.J.; Cotton, J.A.; Volf, P.; Ligoxygakis, P. The Phlebotomus papatasi systemic transcriptional response to trypanosomatid-contaminated blood does not differ from the non-infected blood meal. Parasit. Vectors 2021, 14, 15. [Google Scholar] [CrossRef]
- Garver, L.S.; Dong, Y.; Dimopoulos, G. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog. 2009, 5, e1000335. [Google Scholar] [CrossRef] [PubMed]
- Garver, L.S.; Bahia, A.C.; Das, S.; Souza-Neto, J.A.; Shiao, J.; Dong, Y.; Dimopoulos, G. Anopheles Imd pathway factors and effectors in infection intensity-dependent anti-Plasmodium action. PLoS Pathog. 2012, 8, e1002737. [Google Scholar] [CrossRef]
- Mitri, C.; Jacques, J.-C.; Thiery, I.; Riehle, M.M.; Xu, J.; Bischoff, E.; Morlais, I.; Nsango, S.E.; Vernick, K.D.; Bourgouin, C. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 2009, 5, e1000576. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Aguilar, R.; Xi, Z.; Warr, E.; Mongin, E.; Dimopoulos, G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006, 2, e52. [Google Scholar] [CrossRef] [PubMed]
- Kamhawi, S. Phlebotomine sand flies and Leishmania parasites: Friends or foes? Trends Parasitol. 2006, 22, 439–445. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Sant’Anna, M.R.; Diaz-Albiter, H.; Aguiar-Martins, K.; Al Salem, W.S.; Cavalcante, R.R.; Dillon, V.M.; Bates, P.A.; Genta, F.A.; Dillon, R.J. Colonisation resistance in the sand fly gut: Leishmania protects Lutzomyia longipalpis from bacterial infection. Parasit. Vectors 2014, 7, 329. [Google Scholar] [CrossRef]
- Campolina, T.B.; Villegas, L.E.M.; Monteiro, C.C.; Pimenta, P.F.P.; Secundino, N.F.C. Tripartite interactions: Leishmania, microbiota and Lutzomyia longipalpis. PLoS Negl. Trop. Dis. 2020, 14, e0008666. [Google Scholar] [CrossRef]
- Sheehan, G.; Farrell, G.; Kavanagh, K. Immune priming: The secret weapon of the insect world. Virulence 2020, 11, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Manniello, M.D.; Moretta, A.; Salvia, R.; Scieuzo, C.; Lucchetti, D.; Vogel, H.; Sgambato, A.; Falabella, P. Insect antimicrobial peptides: Potential weapons to counteract the antibiotic resistance. Cell. Mol. Life Sci. 2021 789 2021, 78, 4259–4282. [Google Scholar] [CrossRef]
- Andoh, M.; Ueno, T.; Kawasaki, K. Tissue-dependent induction of antimicrobial peptide genes after body wall injury in house fly (Musca domestica) larvae. Drug Discov. Ther. 2018, 12, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Broderick, N.A.; Poidevin, M.; Pradervand, S.; Lemaitre, B. Drosophila Intestinal Response to Bacterial Infection: Activation of Host Defense and Stem Cell Proliferation. Cell Host Microbe 2009, 5, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, R.V.D.M.; Dias, D.B.S.; Bretãs, J.A.C.; Mazzoni, C.J.; Souza, N.A.; Albano, R.M.; Wagner, G.; Davila, A.M.R.; Peixoto, A.A. The Transcriptome of Lutzomyia longipalpis (Diptera: Psychodidae) Male Reproductive Organs. PLoS ONE 2012, 7, e34495. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jochim, R.C.; Teixeira, C.R.; Laughinghouse, A.; Mu, J.; Oliveira, F.; Gomes, R.B.; Elnaiem, D.E.; Valenzuela, J.G. The midgut transcriptome of Lutzomyia longipalpis: Comparative analysis of cDNA libraries from sugar-fed, blood-fed, post digested and Leishmania infantum chagasi-infected sand flies. BMC Genom. 2008, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Do Nascimento, V.V.; Mello, É.D.O.; Carvalho, L.P.; De Melo, E.J.T.; Carvalho, A.D.O.; Fernandes, K.V.S.; Gomes, V.M. PvD1 defensin, a plant antimicrobial peptide with inhibitory activity against Leishmania amazonensis. Biosci. Rep. 2015, 35, e00248. [Google Scholar] [CrossRef]



| Sand Fly Groups | AtbC in Sucrose Meal before Blood Meal | AtbC in Blood Meal | Leishmania in Blood Meal | AtbC in Sucrose Meal after Blood Meal |
|---|---|---|---|---|
| Control group | + | + | - | + |
| Experimental group 1 (EG1) | + | + | + | + |
| Experimental group 2 (EG2) | + | − | + | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kykalová, B.; Tichá, L.; Volf, P.; Loza Telleria, E. Phlebotomus papatasi Antimicrobial Peptides in Larvae and Females and a Gut-Specific Defensin Upregulated by Leishmania major Infection. Microorganisms 2021, 9, 2307. https://doi.org/10.3390/microorganisms9112307
Kykalová B, Tichá L, Volf P, Loza Telleria E. Phlebotomus papatasi Antimicrobial Peptides in Larvae and Females and a Gut-Specific Defensin Upregulated by Leishmania major Infection. Microorganisms. 2021; 9(11):2307. https://doi.org/10.3390/microorganisms9112307
Chicago/Turabian StyleKykalová, Barbora, Lucie Tichá, Petr Volf, and Erich Loza Telleria. 2021. "Phlebotomus papatasi Antimicrobial Peptides in Larvae and Females and a Gut-Specific Defensin Upregulated by Leishmania major Infection" Microorganisms 9, no. 11: 2307. https://doi.org/10.3390/microorganisms9112307
APA StyleKykalová, B., Tichá, L., Volf, P., & Loza Telleria, E. (2021). Phlebotomus papatasi Antimicrobial Peptides in Larvae and Females and a Gut-Specific Defensin Upregulated by Leishmania major Infection. Microorganisms, 9(11), 2307. https://doi.org/10.3390/microorganisms9112307







