Contrasting Patterns and Drivers of Soil Fungal Communities between Two Ecosystems Divided by the Treeline
Abstract
:1. Introduction
2. Materials and Methods
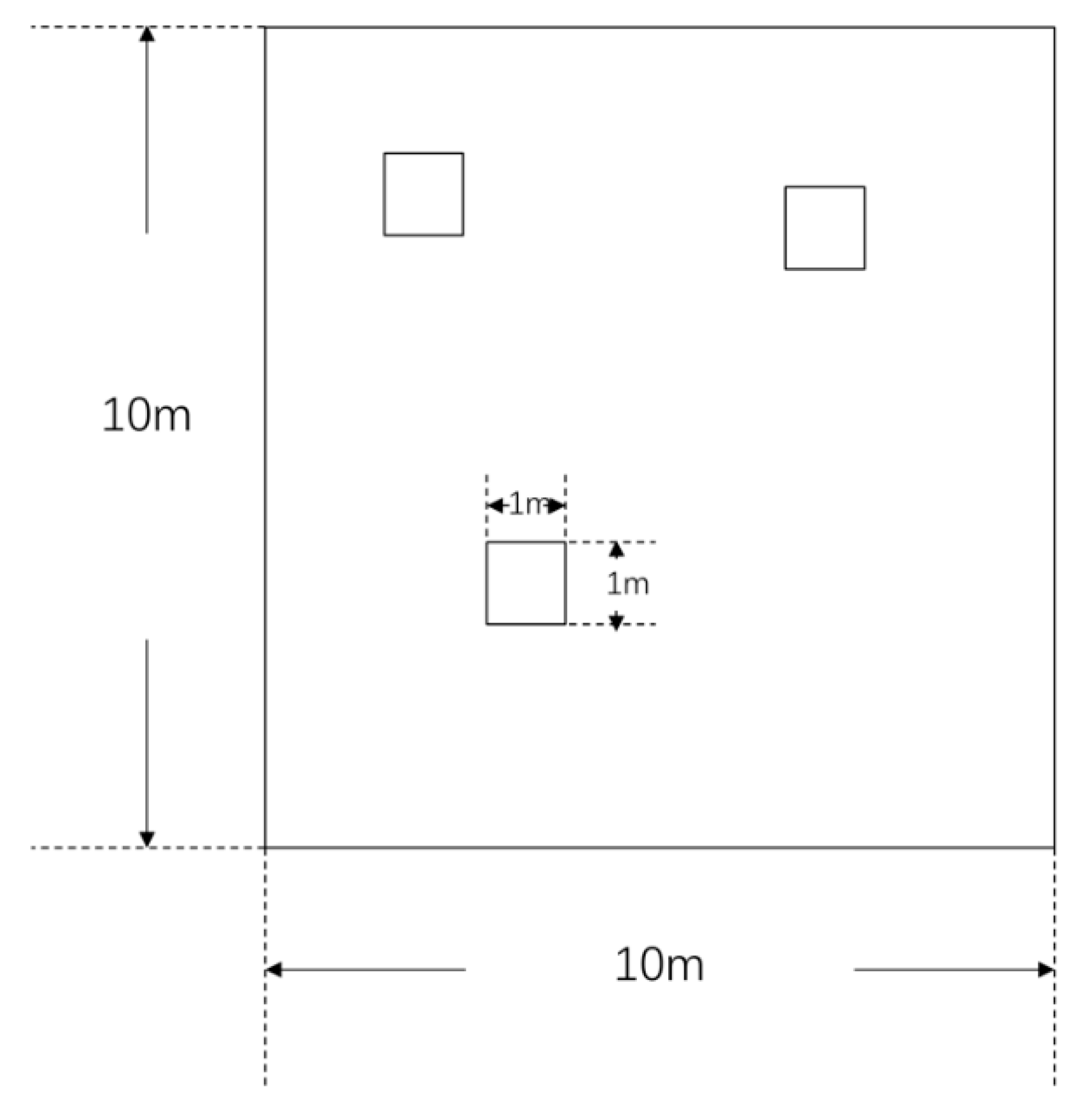
2.1. Study Site and Soil Sampling
2.2. Measurement of Soil Parameters
2.3. DNA Extraction and Sequencing
2.4. Statistical Analysis
3. Results
3.1. Plant Parameters and Soil Properties
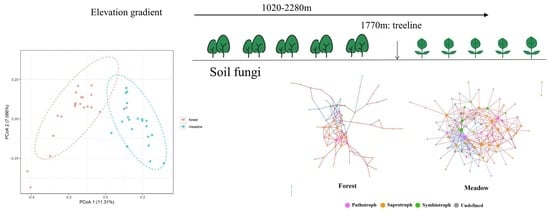
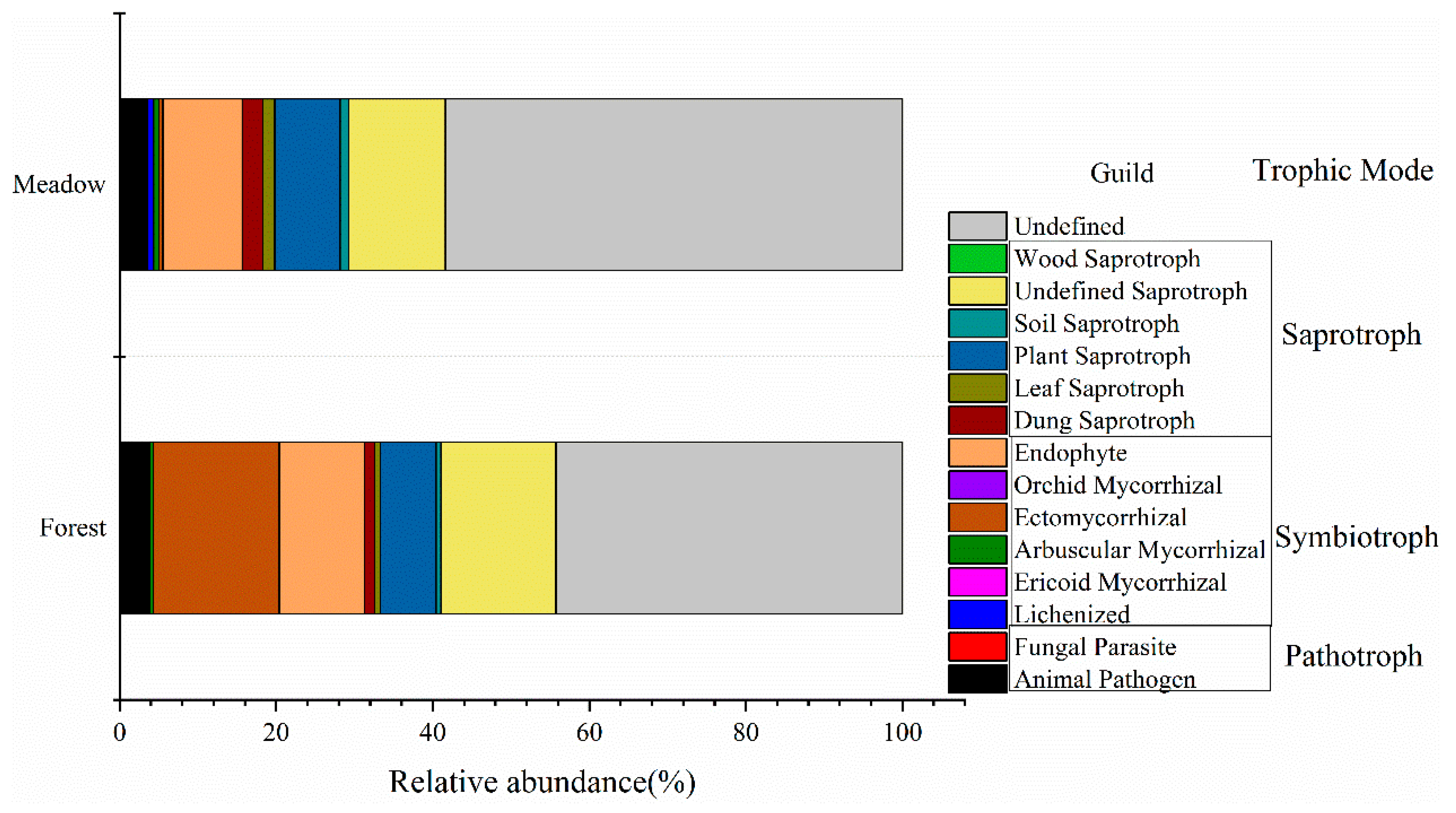
3.2. Fungal Community Diversity and Composition
3.3. Relationship of Biotic and Abiotic Factors with Fungal Community Structure
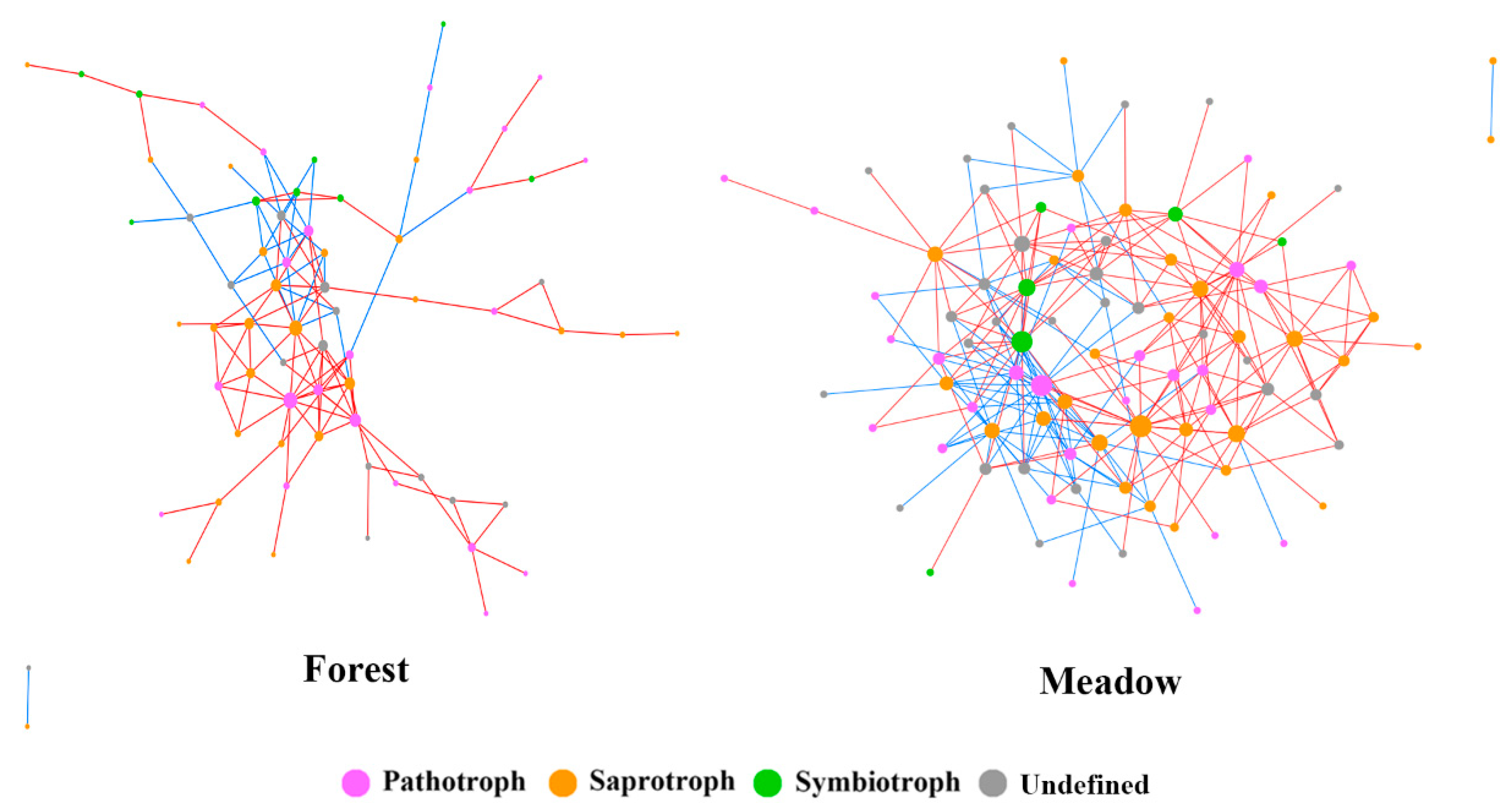
3.4. Fungal Ecological Network Analysis above and below the Treeline
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, P.H.; Bellingham, P.J. Towards integrated ecological research in tropical montane cloud forests. J. Trop. Ecol. 2016, 32, 345–354. [Google Scholar] [CrossRef]
- Malhi, Y.; Silman, M.; Salinas, N.; Bush, M.; Meir, P.; Saatchi, S. Introduction: Elevation gradients in the tropics: Laboratories for ecosystem ecology and global change research. Glob. Chang. Biol. 2010, 16, 3171–3175. [Google Scholar] [CrossRef]
- Korner, C.; Paulsen, J. A world-wide study of high altitude treeline temperatures. J. Biogeogr. 2004, 31, 713–732. [Google Scholar] [CrossRef]
- Mayor, J.R.; Sanders, N.J.; Classen, A.T.; Bardgett, R.D.; Clement, J.C.; Fajardo, A.; Lavorel, S.; Sundqvist, M.K.; Bahn, M.; Chisholm, C.; et al. Elevation alters ecosystem properties across temperate treelines globally. Nature 2017, 542, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Harsch, M.A.; Hulme, P.E.; McGlone, M.S.; Duncan, R.P. Are treelines advancing? A global meta-analysis of treeline response to climate warming. Ecol. Lett. 2009, 12, 1040–1049. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, L.F.; Chen, Y.M.; Zhang, J.; Liu, Y. Litter stoichiometric traits have stronger impact on humification than environment conditions in an alpine treeline ecotone. Plant Soil 2020, 453, 545–560. [Google Scholar] [CrossRef]
- Kammer, A.; Hagedorn, F.; Shevchenko, I.; Leifeld, J.; Guggenberger, G.; Goryacheva, T.; Rigling, A.; Moiseev, P. Treeline shifts in the Ural mountains affect soil organic matter dynamics. Glob. Chang. Biol. 2009, 15, 1570–1583. [Google Scholar] [CrossRef]
- Withington, C.L.; Sanford, R.L. Decomposition rates of buried substrates increase with altitude in the forest-alpine tundra ecotone. Soil Biol. Biochem. 2007, 39, 68–75. [Google Scholar] [CrossRef]
- Li, G.X.; Xu, G.R.; Shen, C.C.; Tang, Y.; Zhang, Y.X.; Ma, K.M. Contrasting elevational diversity patterns for soil bacteria between two ecosystems divided by the treeline. Sci. China-Life Sci. 2016, 59, 1177–1186. [Google Scholar] [CrossRef]
- Clipson, N.; Otte, M.; Landy, E. Fungi in Biogeochemical Cycles: Biogeochemical Roles of Fungi in Marine and Estuarine Habitats; Cambridge University Press: Cambridge, UK, 2006; pp. 436–461. [Google Scholar]
- Yang, Y.; Dou, Y.X.; Huang, Y.M.; An, S.S. Links between Soil Fungal Diversity and Plant and Soil Properties on the Loess Plateau. Front. Microbiol. 2017, 8, 2198. [Google Scholar] [CrossRef]
- Geml, J.; Pastor, N.; Fernandez, L.; Pacheco, S.; Semenova, T.A.; Becerra, A.G.; Wicaksono, C.Y.; Nouhra, E.R. Large-scale fungal diversity assessment in the Andean Yungas forests reveals strong community turnover among forest types along an altitudinal gradient. Mol. Ecol. 2014, 23, 2452–2472. [Google Scholar] [CrossRef]
- Looby, C.I.; Martin, P.H. Diversity and function of soil microbes on montane gradients: The state of knowledge in a changing world. FEMS Microbiol. Ecol. 2020, 96, fiaa122. [Google Scholar] [CrossRef]
- Mukhtar, H.; Lin, C.M.; Wunderlich, R.F.; Cheng, L.C.; Ko, M.C.; Lin, Y.P. Climate and land cover shape the fungal community structure in topsoil. Sci. Total Environ. 2021, 751, 141721. [Google Scholar] [CrossRef]
- Corneo, P.E.; Pellegrini, A.; Cappellin, L.; Gessler, C.; Pertot, I. Moderate Warming in Microcosm Experiment Does Not Affect Microbial Communities in Temperate Vineyard Soils. Microb. Ecol. 2014, 67, 659–670. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.W.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Root, R.B. NICHE EXPLOITATION PATTERN OF BLUE-GRAY GNATCATCHER. Ecol. Monogr. 1967, 37, 317–350. [Google Scholar] [CrossRef]
- Hawkins, C.P.; Macmahon, J.A. GUILDS—THE MULTIPLE MEANINGS OF A CONCEPT. Annu. Rev. Entomol. 1989, 34, 423–451. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Polme, S.; Koljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 125688. [Google Scholar] [CrossRef] [Green Version]
- Xie, F.; Ma, A.Z.; Zhou, H.C.; Liang, Y.; Yin, J.; Ma, K.; Zhuang, X.L.; Zhuang, G.Q. Revealing Fungal Communities in Alpine Wetlands through Species Diversity, Functional Diversity and Ecological Network Diversity. Microorganisms 2020, 8, 632. [Google Scholar] [CrossRef]
- Toju, H.; Kishida, O.; Katayama, N.; Takagi, K. Networks Depicting the Fine-Scale Co-Occurrences of Fungi in Soil Horizons. PLoS ONE 2016, 11, e0165987. [Google Scholar] [CrossRef] [Green Version]
- Luo, F.; Yang, Y.; Zhong, J.; Gao, H.; Khan, L.; Thompson, D.K.; Zhou, J. Constructing gene co-expression networks and predicting functions of unknown genes by random matrix theory. BMC Bioinform. 2007, 8, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.Z.; Deng, Y.; Luo, F.; He, Z.L.; Tu, Q.C.; Zhi, X.Y. Functional Molecular Ecological Networks. mBio 2010, 1, 1592–1601. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.Z.; Deng, Y.; Luo, F.; He, Z.L.; Yang, Y.F. Phylogenetic Molecular Ecological Network of Soil Microbial Communities in Response to Elevated CO2. mBio 2011, 2, e00122-11. [Google Scholar] [CrossRef] [Green Version]
- Cornfield, A.H. Ammonia released on treating soils with N sodium hydroxide as a possible means of predicting the nitrogen-supplying power of soils. Nature 1960, 187, 260–261. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. In PCR Protocols; Academic Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Hoiland, K.; Kjoller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification of fungi-recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, W.H.; Wu, H.L.; Ouyang, S.; Lei, P.F.; Hu, Y.J.; Ge, T.D.; Ye, J.; Kuzyakov, Y. Contrasting patterns and drivers of soil fungal communities in subtropical deciduous and evergreen broadleaved forests. Appl. Microbiol. Biotechnol. 2019, 103, 5421–5433. [Google Scholar] [CrossRef]
- Mitchell, R.J.; Hester, A.J.; Campbell, C.D.; Chapman, S.J.; Cameron, C.M.; Hewison, R.L.; Potts, J.M. Is vegetation composition or soil chemistry the best predictor of the soil microbial community? Plant Soil 2010, 333, 417–430. [Google Scholar] [CrossRef]
- Wang, D.; He, N.P.; Wang, Q.; Lu, Y.L.; Wang, Q.F.; Xu, Z.W.; Zhu, J.X. Effects of Temperature and Moisture on Soil Organic Matter Decomposition Along Elevation Gradients on the Changbai Mountains, Northeast China. Pedosphere 2016, 26, 399–407. [Google Scholar] [CrossRef]
- Shen, C.C.; Gunina, A.; Luo, Y.; Wang, J.J.; He, J.Z.; Kuzyakov, Y.; Hemp, A.; Classen, A.T.; Ge, Y. Contrasting patterns and drivers of soil bacterial and fungal diversity across a mountain gradient. Environ. Microbiol. 2020, 22, 3287–3301. [Google Scholar] [CrossRef]
- Yeh, C.F.; Soininen, J.; Teittinen, A.; Wang, J.J. Elevational patterns and hierarchical determinants of biodiversity across microbial taxonomic scales. Mol. Ecol. 2019, 28, 86–99. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Zhang, Y.; Shi, N.N.; Zheng, Y.; Chen, L.; Wubet, T.; Bruelheide, H.; Both, S.; Buscot, F.; Ding, Q.; et al. Community assembly of ectomycorrhizal fungi along a subtropical secondary forest succession. New Phytol. 2015, 205, 771–785. [Google Scholar] [CrossRef]
- Leopold, D.R. Ericoid fungal diversity: Challenges and opportunities for mycorrhizal research. Fungal Ecol. 2016, 24, 114–123. [Google Scholar] [CrossRef]
- Genre, A.; Lanfranco, L.; Perotto, S.; Bonfante, P. Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 2020, 18, 649–660. [Google Scholar] [CrossRef]
- Boraks, A.; Plunkett, G.M.; Doro, T.M.; Alo, F.; Sam, C.; Tuiwawa, M.; Ticktin, T.; Amend, A.S. Scale-Dependent Influences of Distance and Vegetation on the Composition of Aboveground and Belowground Tropical Fungal Communities. Microb. Ecol. 2021, 81, 874–883. [Google Scholar] [CrossRef]
- Kivlin, S.N.; Winston, G.C.; Goulden, M.L.; Treseder, K.K. Environmental filtering affects soil fungal community composition more than dispersal limitation at regional scales. Fungal Ecol. 2014, 12, 14–25. [Google Scholar] [CrossRef] [Green Version]
- Fuhrman, J.A. Microbial community structure and its functional implications. Nature 2009, 459, 193–199. [Google Scholar] [CrossRef]
- Ding, J.J.; Zhang, Y.G.; Deng, Y.; Cong, J.; Lu, H.; Sun, X.; Yang, C.Y.; Yuan, T.; Van Nostrand, J.D.; Li, D.Q.; et al. Integrated metagenomics and network analysis of soil microbial community of the forest timberline. Sci. Rep. 2015, 5, 7994. [Google Scholar] [CrossRef] [Green Version]
- Stouffer, D.B.; Bascompte, J. Compartmentalization increases food-web persistence. Proc. Natl. Acad. Sci. USA 2011, 108, 3648–3652. [Google Scholar] [CrossRef] [Green Version]
- Montoya, J.M.; Pimm, S.L.; Sole, R.V. Ecological networks and their fragility. Nature 2006, 442, 259–264. [Google Scholar] [CrossRef]
- Berry, D.; Widder, S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014, 5, 219. [Google Scholar] [CrossRef] [Green Version]


| Forest | Meadow | t-Tests (p Value) | |
|---|---|---|---|
| Plant parameters | |||
| Plant richness | 32.19 ± 7.55 | 30.48 ± 5.96 | 0.419 |
| Herb richness | 24.43 ± 7.85 | 30.48 ± 5.96 | 0.008 |
| Litter thickness (cm) | 1.37 ± 0.93 | 1.13 ± 0.66 | 0.345 |
| Microbial parameters | |||
| Observed richness | 493.14 ± 148.45 | 497.48 ± 170.79 | 0.931 |
| Shannon index | 3.8 ± 0.85 | 4.12 ± 0.73 | 0.207 |
| Soil parameters | |||
| Temperature (°C) | 17.97 ± 1.17 | 15.18 ± 1.69 | <0.001 |
| SM (%) | 0.39 ± 0.11 | 0.5 ± 0.15 | 0.011 |
| Bulk density (g/cm3) | 82.45 ± 14.3 | 73.89 ± 8.26 | 0.022 |
| pH | 6.28 ± 0.6 | 6.34 ± 0.26 | 0.679 |
| Clay (%) | 17.14 ± 5.01 | 15.12 ± 4.91 | 0.196 |
| Silt (%) | 60.59 ± 3.1 | 67.15 ± 4.74 | <0.001 |
| Sand (%) | 22.27 ± 5.46 | 17.73 ± 4.87 | 0.007 |
| TN (%) | 0.33 ± 0.06 | 0.51 ± 0.11 | <0.001 |
| TC (%) | 4.89 ± 0.92 | 7.15 ± 1.7 | <0.001 |
| C:N molar ratio | 17.12 ± 1.31 | 16.29 ± 0.68 | 0.014 |
| AP (mg/kg) | 3.92 ± 0.7 | 6.76 ± 2.48 | <0.001 |
| AN (mg/kg) | 370 ± 62.99 | 585.99 ± 136.39 | <0.001 |
| TP (mg/kg) | 657.51 ± 115.61 | 952.39 ± 207.9 | <0.001 |
| Explanatory Distance | R | p |
|---|---|---|
| Geographic distance | 0.1692 | 0.001 |
| Environmental distance | 0.1467 | 0.019 |
| Variables | R | p |
|---|---|---|
| Herb richness | 0.001 | 0.478 |
| Plant richness | −0.0633 | 0.745 |
| Temperature | 0.1487 | 0.025 |
| SM | −0.029 | 0.644 |
| Litter thickness | −0.0951 | 0.864 |
| BD | 0.0876 | 0.174 |
| pH | 0.0815 | 0.227 |
| Clay | 0.0186 | 0.354 |
| Silt | 0.018 | 0.384 |
| Sand | −0.1241 | 0.957 |
| TN | 0.0414 | 0.304 |
| TC | −0.0214 | 0.592 |
| C:N ratio | 0.106 | 0.151 |
| AP | 0.0548 | 0.281 |
| AN | 0.0246 | 0.369 |
| TP | 0.0462 | 0.264 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Li, G.; Zhang, Y.; Ma, K. Contrasting Patterns and Drivers of Soil Fungal Communities between Two Ecosystems Divided by the Treeline. Microorganisms 2021, 9, 2280. https://doi.org/10.3390/microorganisms9112280
Wang X, Li G, Zhang Y, Ma K. Contrasting Patterns and Drivers of Soil Fungal Communities between Two Ecosystems Divided by the Treeline. Microorganisms. 2021; 9(11):2280. https://doi.org/10.3390/microorganisms9112280
Chicago/Turabian StyleWang, Xueying, Guixiang Li, Yuxin Zhang, and Keming Ma. 2021. "Contrasting Patterns and Drivers of Soil Fungal Communities between Two Ecosystems Divided by the Treeline" Microorganisms 9, no. 11: 2280. https://doi.org/10.3390/microorganisms9112280
APA StyleWang, X., Li, G., Zhang, Y., & Ma, K. (2021). Contrasting Patterns and Drivers of Soil Fungal Communities between Two Ecosystems Divided by the Treeline. Microorganisms, 9(11), 2280. https://doi.org/10.3390/microorganisms9112280