Scalp Microbiome and Sebum Composition in Japanese Male Individuals with and without Androgenetic Alopecia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subject Recruitment
2.2. Sample Collection
2.3. Analysis of the Skin Microbiome
2.4. Analysis of Scalp Sebum
2.4.1. Free Fatty Acid Analysis
2.4.2. Glyceride Analysis
2.4.3. Free Cholesterol and Squalene Analysis
2.4.4. Wax Ester Analysis
2.5. Statistics
3. Results
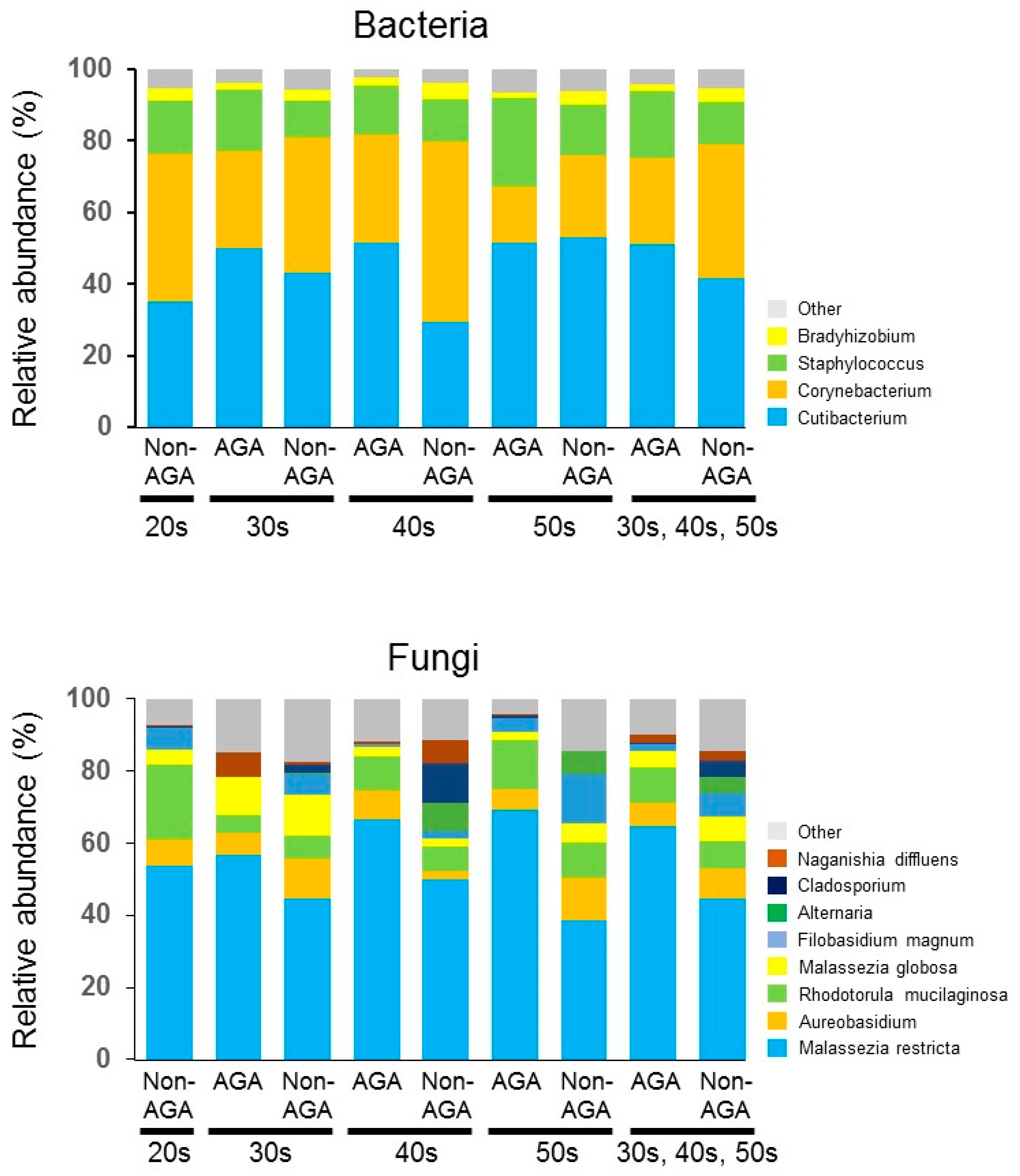
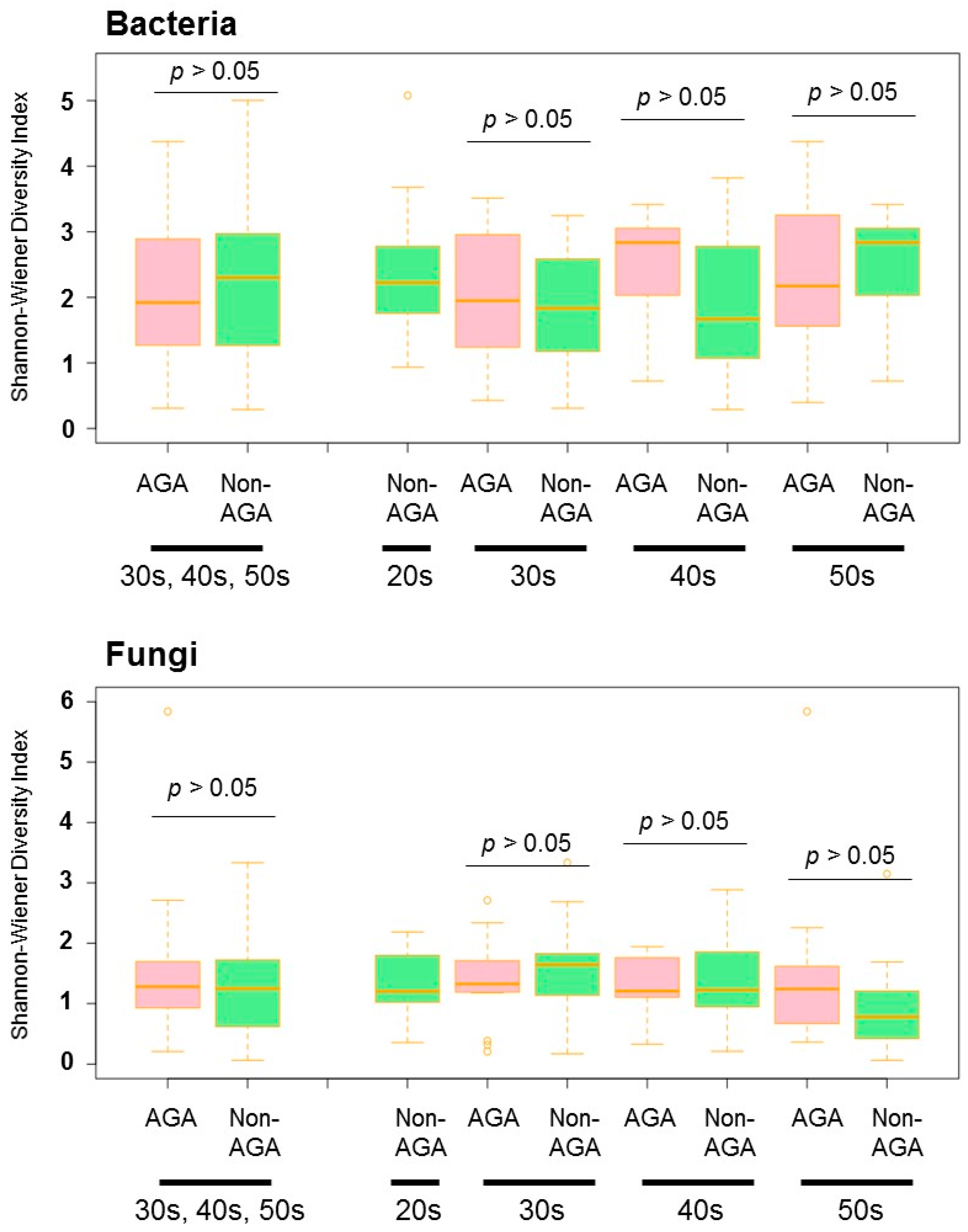
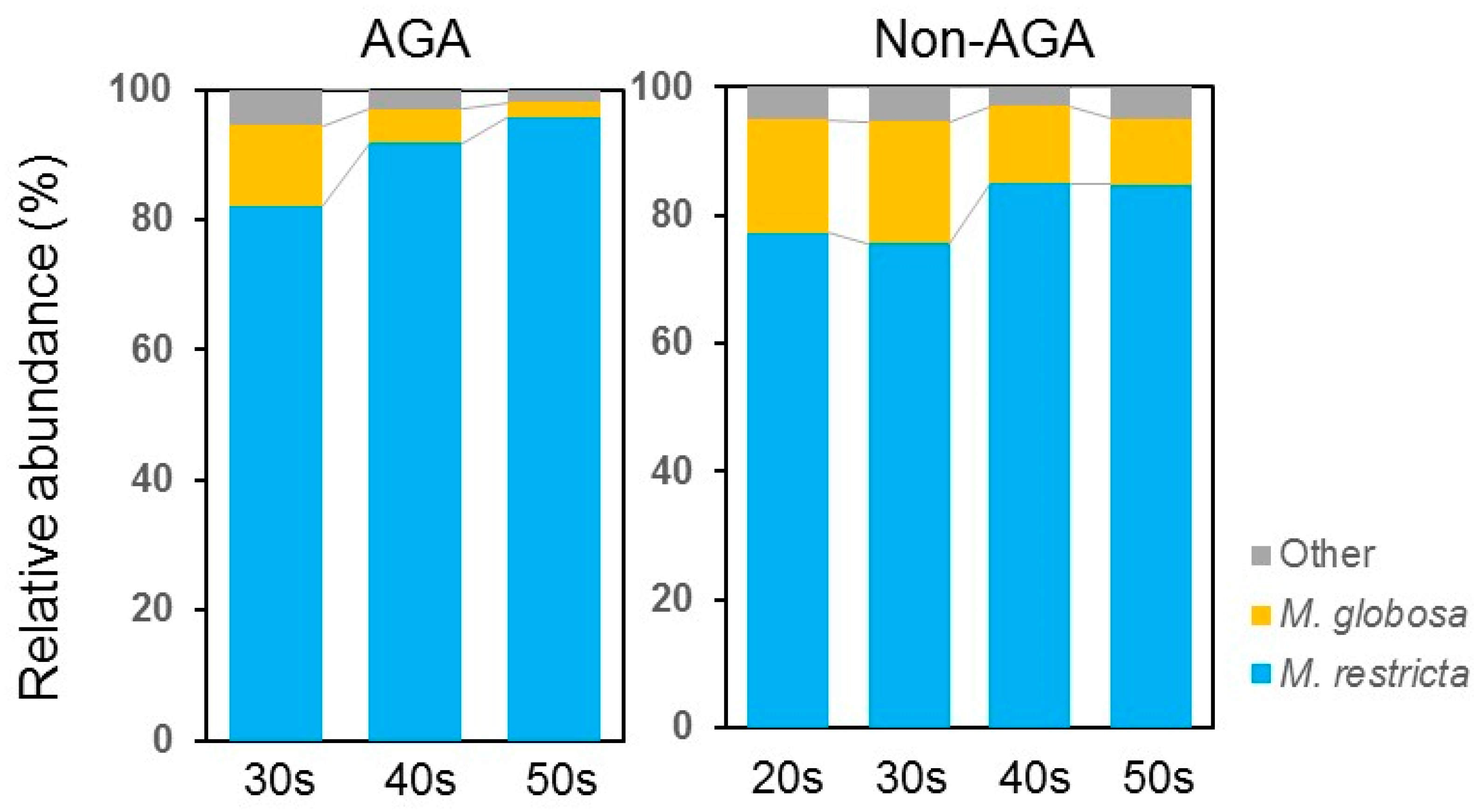
3.1. Skin Microbiome Analysis
3.2. Sebum Composition
3.3. Free Fatty Acids
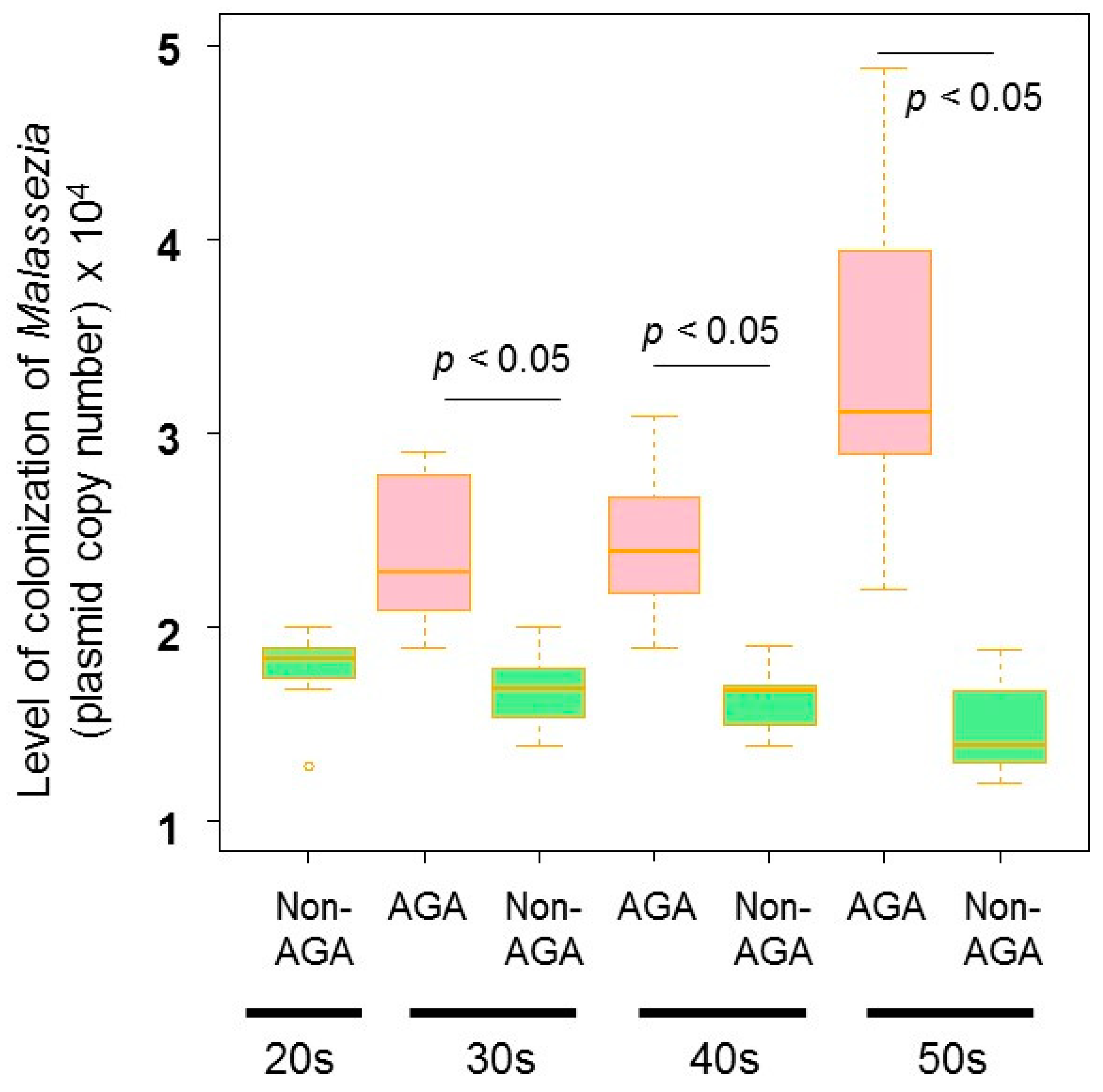
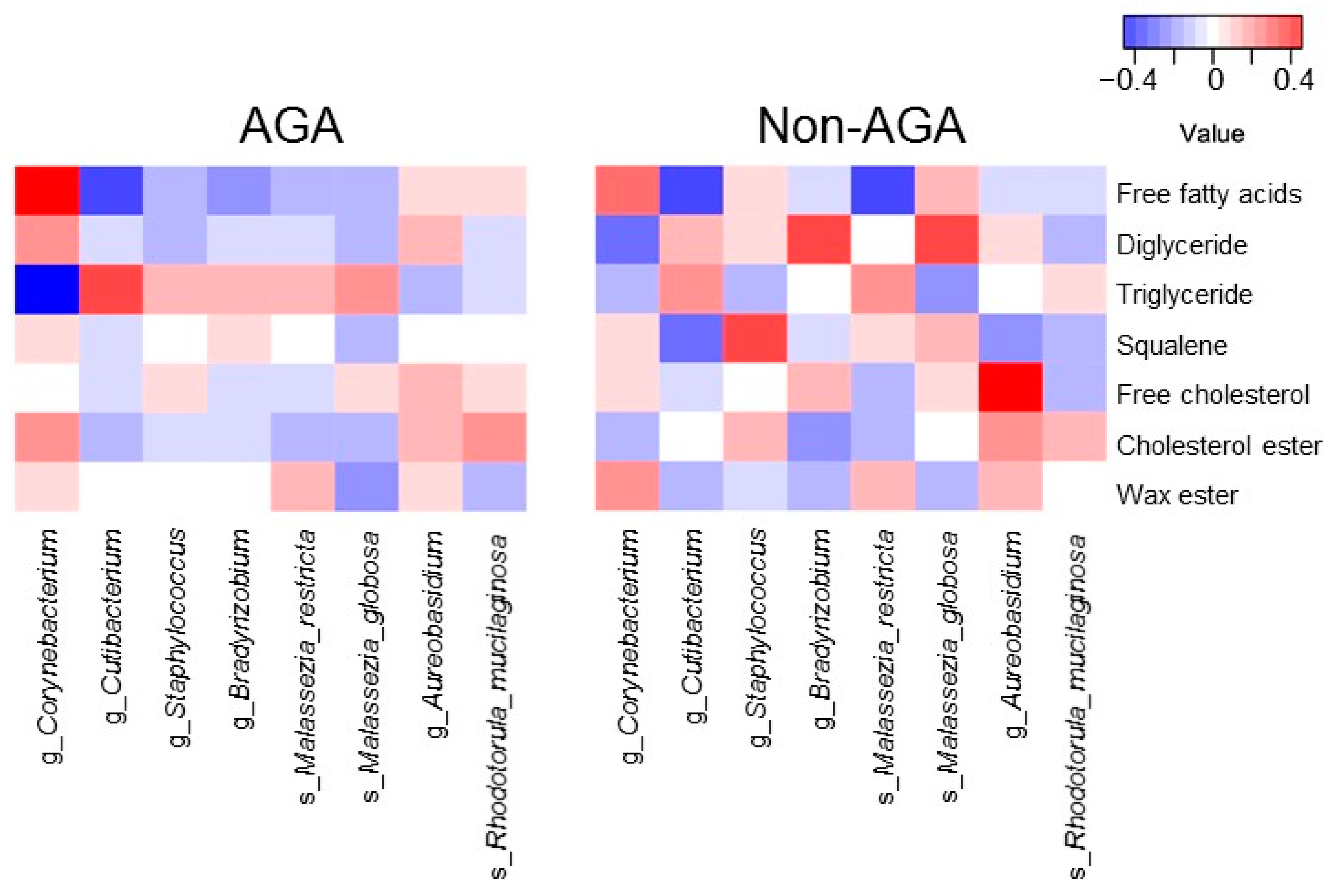
3.4. Relationship between the Scalp Microbiome and Sebum Composition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drake, L.A.; Dinehart, S.M.; Farmer, E.R.; Goltz, R.W.; Graham, G.F.; Hordinsky, M.K.; Lewis, C.W.; Pariser, D.M.; Webster, S.B.; Whitaker, D.C.; et al. Guidelines of care for androgenetic alopecia. J. Am. Acad. Dermatol. 1996, 35, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Olsen, E.A. Current and novel methods for assessing efficacy of hair growth promoters in pattern hair loss. J. Am. Acad. Dermatol. 2003, 48, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Olsen, E.A.; Messenger, A.G.; Shapiro, J.; Bergfeld, W.F.; Hordinsky, M.K.; Roberts, J.L.; Stough, D.; Washenik, K.; Whiting, D.A. Evaluation and treatment of male and female pattern hair loss. J. Am. Acad. Dermatol. 2005, 52, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Takashima, I.; Iju, M.; Sudo, M. Alopecia androgenetica—Its incidence in Japanese and associated conditions. In Hair Research; Orfanos, C.E., Montagna, W., Stüttgen, G., Eds.; Springer: Berlin, Germany, 1981; pp. 287–293. ISBN 978-3-642-81652-9. [Google Scholar]
- Severi, G.; Sinclair, R.; Hopper, J.L.; English, D.R.; McCredie, M.R.E.; Boyle, P.; Giles, G.G. Androgenetic alopecia in men aged 40–69 years: Prevalence and risk factors. Br. J. Dermatol. 2003, 149, 1207–1213. [Google Scholar] [CrossRef]
- Yap, C.X.; Sidorenko, J.; Wu, Y.; Kemper, K.E.; Yang, J.; Wray, N.R.; Robinson, M.R.; Visscher, P.M. Dissection of gnetic variation and evidence for pleiotropy in male pattern baldness. Nat. Commun. 2018, 9, 5407. [Google Scholar] [CrossRef]
- Nyholt, D.R.; Gillespie, N.A.; Heath, A.C.; Martin, N.G. Genetic basis of male pattern baldness. J. Investig. Dermatol. 2003, 121, 1561–1564. [Google Scholar] [CrossRef]
- Rexbye, H.; Petersen, I.; Iachina, M.; Mortensen, J.; McGue, M.; Vaupel, J.W.; Christensen, K. Hair loss among elderly men: Etiology and impact on perceived age. J. Gerontol. A Biol. Sci. Med. Sci. 2005, 60, 1077–1082. [Google Scholar] [CrossRef] [Green Version]
- Gatherwright, J.; Liu, M.T.; Amirlak, B.; Gliniak, C.; Totonchi, A.; Guyuron, B. The contribution of endogenous and exogenous factors to male alopecia: A study of identical twins. Plast. Reconstr. Surg. 2013, 131, 794e–801e. [Google Scholar] [CrossRef]
- Downing, D.T.; Strauss, J.S.; Pochi, P.E. Variability in the chemical composition of human skin surface lipids. J. Investig. Dermatol. 1969, 53, 322–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobsen, E.; Billings, J.K.; Frantz, R.A.; Kinney, C.K.; Stewart, M.E.; Downing, D.T. Age-related changes in sebaceous wax ester secretion rates in men and women. J. Investig. Dermatol. 1985, 85, 483–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimshaw, S.G.; Smith, A.M.; Arnold, D.S.; Xu, E.; Hoptroff, M.; Murphy, B. The diversity and abundance of fungi and bacteria on the healthy and dandruff affected human scalp. PLoS ONE 2019, 18, e0225796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, R.; Mittal, P.; Clavaud, C.; Dhakan, D.B.; Hegde, P.; Veeranagaiah, M.M.; Saha, S.; Souverain, L.; Roy, N.; Breton, L.; et al. Comparison of healthy and dandruff scalp microbiome reveals the role of commensals in scalp health. Front. Cell. Infect. Microbiol. 2018, 4, 346. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Guichard, A.; Cheng, Y.; Li, J.; Qin, O.; Wang, X.; Liu, W.; Tan, Y. Sensitive scalp is associated with excessive sebum and perturbed microbiome. J. Cosmet. Dermatol. 2019, 18, 922–928. [Google Scholar] [CrossRef]
- Ho, B.S.; Ho, E.X.P.; Chu, C.W.; Ramasamy, S.; Bigliardi-Qi, M.; de Sessions, P.F.; Bigliardi, P.L. Microbiome in the hair follicle of androgenetic alopecia patients. PLoS ONE 2019, 14, e0216330. [Google Scholar] [CrossRef]
- Huang, J.; Ran, Y.; Pradhan, S.; Yan, W.; Dai, Y. Investigation on microecology of hair root fungi in androgenetic alopecia patients. Mycopathologia 2019, 184, 505–515. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, Z.; Yuan, C.; Liu, X.; Yang, F.; Wang, T.; Wang, J.; Manabe, K.; Qin, O.; Wang, X.; et al. Dandruff is associated with the conjoined interactions between host and microorganisms. Sci. Rep. 2016, 6, 24877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.K.; Batra, R.; Bluhm, R.; Boekhout, T.; Dawson, T.L., Jr. Skin diseases associated with Malassezia species. J. Am. Acad. Dermatol. 2004, 51, 785–798. [Google Scholar] [CrossRef]
- Xu, J.; Saunders, C.W.; Hu, P.; Grant, R.A.; Boekhout, T.; Kuramae, E.E.; Kronstad, J.W.; Deangelis, Y.M.; Reeder, N.L.; JohnStone, K.R.; et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. USA 2007, 104, 18730–18735. [Google Scholar] [CrossRef] [Green Version]
- Iinuma, K.; Sato, T.; Akimoto, N.; Noguchi, N.; Sasatsu, M.; Nishijima, S.; Kurokawa, I.; Ito, A. Involvement of Propionibacterium acnes in the augmentation of lipogenesis in hamster sebaceous glands in vivo and in vitro. J. Investig. Dermatol. 2009, 129, 2113–2119. [Google Scholar] [CrossRef] [Green Version]
- Ogata, T. Development of patterned alopecia. Sogo Rinsho 1953, 2, 101–106. [Google Scholar]
- Sugita, T.; Suto, H.; Unno, T.; Tsuboi, R.; Ogawa, H.; Shinoda, T.; Nishikawa, A. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. J. Clin. Microbiol. 2001, 39, 3486–3490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugita, T.; Yamazaki, T.; Cho, O.; Furukawa, S.; Mukai, C. The skin mycobiome of an astronaut during a 1-year stay on the International Space Station. Med. Mycol. 2021, 59, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Sugita, T.; Tajima, M.; Tsubuku, H.; Tsuboi, R.; Nishikawa, A. Quantitative analysis of cutaneous Malassezia in atopic dermatitis patients using real-time PCR. Microbiol. Immunol. 2006, 50, 549–552. [Google Scholar] [CrossRef]
- Ramasamy, S.; Barnard, E.; Dawson, T.L., Jr.; Li, H. The role of the skin microbiota in acne pathophysiology. Br. J. Dermatol. 2019, 181, 691–699. [Google Scholar] [CrossRef]
- Quan, C.; Chen, X.Y.; Li, X.; Xue, F.; Chen, L.H.; Liu, N.; Wang, B.; Wang, L.Q.; Wang, X.P.; Yang, H.; et al. Psoriatic lesions are characterized by higher bacterial load and imbalance between Cutibacterium and Corynebacterium. J. Am. Acad. Dermatol. 2020, 82, 955–961. [Google Scholar] [CrossRef]
- Rozas, M.; Hart de Ruijter, A.; Fabrega, M.J.; Zorgani, A.; Guell, M.; Paetzold, B.; Brillet, F. From dysbiosis to healthy skin: Major contributions of Cutibacterium acnes to skin homeostasis. Microorganisms 2021, 9, 628. [Google Scholar] [CrossRef]
- Pinto, D.; Sorbellini, E.; Marzani, B.; Rucco, M.; Giuliani, G.; Rinaldi, F. Scalp bacterial shift in alopecia areata. PLoS ONE 2019, 14, e0215206. [Google Scholar] [CrossRef] [Green Version]
- Claesen, J.; Spagnolo, J.B.; Ramos, S.F.; Kurita, K.L.; Byrd, A.L.; Aksenov, A.A.; Melnik, A.V.; Wong, W.R.; Wang, S.; Hernandez, R.D.; et al. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl. Med. 2020, 12, eaay5445. [Google Scholar] [CrossRef]
- Kang, D.; Shi, B.; Erfe, M.C.; Craft, N.; Li, H. Vitamin B12 modulates the transcriptome of the skin microbiota in acne pathogenesis. Sci. Transl. Med. 2015, 7, 293ra103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trüeb, R.M. Is androgenetic alopecia a photoaggravated dermatosis? Dermatology (Basel) 2003, 207, 343–348. (In Basel) [Google Scholar] [CrossRef] [PubMed]
- Barnard, E.; Johnson, T.; Ngo, T.; Arora, U.; Leuterio, G.; McDowell, A.; Li, H. Porphyrin production and regulation in cutaneous propionibacteria. mSphere 2020, 5, e00793-19. [Google Scholar] [CrossRef] [Green Version]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Sugita, T.; Suzuki, M.; Goto, S.; Nishikawa, A.; Hiruma, M.; Yamazaki, T.; Makimura, K. Quantitative analysis of the cutaneous Malassezia microbiota in 770 healthy Japanese by age and gender using a real-time PCR assay. Med. Mycol. 2010, 48, 229–233. [Google Scholar] [CrossRef] [Green Version]
- Tajima, M.; Sugita, T.; Nishikawa, A.; Tsuboi, R. Molecular analysis of Malassezia microflora in seborrheic dermatitis patients: Comparison with other diseases and healthy subjects. J. Investig. Dermatol. 2008, 128, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Lwin, S.M.; Kimber, I.; McFadden, J.P. Acne, quorum sensing and danger. Clin. Exp. Dermatol. 2014, 39, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Ro, B.I.; Dawson, T.L., Jr. The role of sebaceous gland activity and scalp microfloral metabolism in the etiology of seborrheic dermatitis and dandruff. J. Investig. Dermatol. Symp. Proc. 2005, 10, 194–197. [Google Scholar] [CrossRef] [Green Version]
- Mahé, Y.F.; Michelet, J.F.; Billoni, N.; Jarrousse, F.; Buan, B.; Commo, S.; Saint-Léger, D.; Bernard, B.A. Androgenetic alopecia and microinflammation. Int. J. Dermatol. 2000, 39, 576–584. [Google Scholar] [CrossRef]
- Maibach, H.I.; Feldmann, R.; Payne, B.; Hutshell, T. Scalp and forehead sebum production in male patterned alopecia 1. International Symposium, Rapallo, July 1967: Proceedings. In Biopathology of Pattern Alopecia; Baccaredda-Boy, A., Moretti, G., Frey, J.R., Eds.; Karger: Basel, Switzerland; New York, NY, USA, 1968; pp. 171–176. [Google Scholar] [CrossRef]
- Nelson, S.C.; Yau, Y.C.; Richardson, S.E.; Matlow, A.G. Improved detection of Malassezia species in lipid-supplemented Peds Plus blood culture bottles. J. Clin. Microbiol. 1995, 33, 1005–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.; Hu, X.; He, Y.; Huang, D. Lipidomics demonstrates the association of sex hormones with sebum. J. Cosmet. Dermatol. 2021, 20, 2015–2019. [Google Scholar] [CrossRef] [PubMed]
- Pochi, P.E.; Downing, D.T.; Strauss, J.S. Sebaceous gland response in man to prolonged total caloric deprivation. J. Investig. Dermatol. 1970, 55, 303–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, R.N.; Braue, A.; Varigos, G.A.; Mann, N.J. The effect of a low glycemic load diet on acne vulgaris and the fatty acid composition of skin surface triglycerides. J. Dermatol. Sci. 2008, 50, 41–52. [Google Scholar] [CrossRef]
- Kaminaga, H.; Shinomiya, T. Stress and skin: The change of physiological properties of overcrowding stress in guinea pigs. Jpn. J. Dermatol. 1997, 107, 615–622. [Google Scholar] [CrossRef]
- Sato, T.; Akimoto, N.; Takahashi, A.; Ito, A. Triptolide suppresses ultraviolet B-enhanced sebum production by inhibiting the biosynthesis of triacylglycerol in hamster sebaceous glands in vivo and in vitro. Exp. Ther. Med. 2017, 14, 361–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Component | AGA Group | Non-AGA Group | p-Value |
|---|---|---|---|
| Triglyceride | 54.4 ± 10.5 | 46.6 ± 10.7 | 0.0001 |
| Diglyceride | 14.3 ± 2.3 | 15.2 ± 2.7 | 0.0614 |
| Free fatty acid | 13.6 ± 7.5 | 18.7 ± 7.2 | 0.0002 |
| Squalene | 10.7 ± 2.0 | 12.1 ± 2.2 | 0.0004 |
| Cholesterol ester | 5.6 ± 2.2 | 5.8 ± 1.9 | 0.6741 |
| Free cholesterol | 0.8 ± 0.3 | 1.0 ± 0.5 | 0.0135 |
| Wax ester | 0.6 ± 0.5 | 0.5 ± 0.5 | 0.6810 |
| Type of Free Fatty Acids | AGA Group | Non-AGA Group | p-Value |
|---|---|---|---|
| C16:0 | 33.10 ± 5.15 | 31.35 ± 3.29 | 0.0276 |
| (C16:1_1) | 12.53 ± 3.08 | 12.29 ± 2.43 | 0.6380 |
| C14:0 | 11.55 ± 1.89 | 11.11 ± 1.24 | 0.1381 |
| (C18:1) | 11.22 ± 2.40 | 11.13 ± 2.13 | 0.8455 |
| C15:0 | 8.16 ± 1.72 | 8.36 ± 1.40 | 0.4889 |
| C18:0 | 3.70 ± 1.39 | 3.48 ± 0.59 | 0.2562 |
| C17:0 | 2.84 ± 0.67 | 2.93 ± 0.58 | 0.4427 |
| (Branched C16:0_3) | 1.79 ± 0.91 | 2.23 ± 0.93 | 0.0108 |
| (Branched C14:0) | 1.47 ± 0.85 | 1.87 ± 0.82 | 0.0113 |
| (Branched C15:0_3) | 1.31 ± 0.60 | 1.26 ± 0.38 | 0.5798 |
| (Branched C16:1) | 1.14 ± 0.74 | 1.40 ± 0.61 | 0.0414 |
| (C17:1_2) | 1.11 ± 0.44 | 1.19 ± 0.32 | 0.2561 |
| (C16:1_2) | 1.06 ± 0.46 | 1.11 ± 0.35 | 0.5138 |
| (C14:1) | 0.96 ± 0.48 | 0.99 ± 0.38 | 0.7238 |
| C24:0 | 0.82 ± 0.45 | 0.80 ± 0.31 | 0.7859 |
| C12:0 | 0.79 ± 0.47 | 0.78 ± 0.30 | 0.8865 |
| (C17:1_3) | 0.79 ± 0.40 | 0.84 ± 0.28 | 0.4200 |
| (C15:1) | 0.68 ± 0.34 | 0.70 ± 0.30 | 0.8021 |
| C18:2n6t | 0.61 ± 0.35 | 0.69 ± 0.29 | 0.1469 |
| (Branched C18:1) | 0.56 ± 0.52 | 0.75 ± 0.49 | 0.0425 |
| (Branched C17:0_1) | 0.52 ± 0.36 | 0.63 ± 0.27 | 0.0542 |
| (Branched C17:0_2) | 0.39 ± 0.30 | 0.43 ± 0.24 | 0.4769 |
| C13:0 | 0.39 ± 0.32 | 0.50 ± 0.25 | 0.0333 |
| (C17:1_1) | 0.29 ± 0.28 | 0.32 ± 0.24 | 0.5850 |
| (C18:2) | 0.28 ± 0.26 | 0.33 ± 0.24 | 0.2331 |
| C20:0 | 0.26 ± 0.27 | 0.35 ± 0.23 | 0.0674 |
| C22:0 | 0.23 ± 0.26 | 0.31 ± 0.19 | 0.0483 |
| (C20:2) | 0.20 ± 0.24 | 0.23 ± 0.22 | 0.4610 |
| (Branched C15:0_1) | 0.19 ± 0.23 | 0.27 ± 0.22 | 0.0421 |
| C18:2n6c | 0.18 ± 0.23 | 0.31 ± 0.29 | 0.0097 |
| (Branched C15:0_2) | 0.16 ± 0.30 | 0.16 ± 0.21 | 0.9575 |
| C18:1n9c | 0.13 ± 0.25 | 0.07 ± 0.17 | 0.1856 |
| Others_3 | 0.12 ± 0.19 | 0.18 ± 0.19 | 0.1365 |
| Others_4 | 0.11 ± 0.20 | 0.18 ± 0.23 | 0.0964 |
| (Branched C16:0_2) | 0.09 ± 0.16 | 0.12 ± 0.17 | 0.3721 |
| (C26:0) | 0.07 ± 0.15 | 0.09 ± 0.14 | 0.5248 |
| (Branched C25:0) | 0.04 ± 0.13 | 0.04 ± 0.10 | 0.9482 |
| (C19:0) | 0.03 ± 0.08 | 0.07 ± 0.12 | 0.0629 |
| (Branched C13:0_2) | 0.03 ± 0.08 | 0.03 ± 0.07 | 0.9727 |
| Others_1 | 0.03 ± 0.07 | 0.04 ± 0.09 | 0.2804 |
| (Branched C16:0_1) | 0.02 ± 0.05 | 0.02 ± 0.06 | 0.6507 |
| (C25:0) | 0.01 ± 0.05 | 0.03 ± 0.07 | 0.1634 |
| (Branched C13:0_1) | 0.01 ± 0.05 | 0.02 ± 0.07 | 0.4160 |
| C23:0 | 0.01 ± 0.03 | 0.01 ± 0.03 | 0.7156 |
| (Branched C15:1) | 0.01 ± 0.03 | 0.00 ± 0.00 | 0.0624 |
| Others_2 | 0.01 ± 0.03 | 0.00 ± 0.00 | 0.1292 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, K.; Inoue, M.; Cho, O.; Mizutani, R.; Shimizu, Y.; Nagahama, T.; Sugita, T. Scalp Microbiome and Sebum Composition in Japanese Male Individuals with and without Androgenetic Alopecia. Microorganisms 2021, 9, 2132. https://doi.org/10.3390/microorganisms9102132
Suzuki K, Inoue M, Cho O, Mizutani R, Shimizu Y, Nagahama T, Sugita T. Scalp Microbiome and Sebum Composition in Japanese Male Individuals with and without Androgenetic Alopecia. Microorganisms. 2021; 9(10):2132. https://doi.org/10.3390/microorganisms9102132
Chicago/Turabian StyleSuzuki, Kazuhiro, Mizuna Inoue, Otomi Cho, Rumiko Mizutani, Yuri Shimizu, Tohru Nagahama, and Takashi Sugita. 2021. "Scalp Microbiome and Sebum Composition in Japanese Male Individuals with and without Androgenetic Alopecia" Microorganisms 9, no. 10: 2132. https://doi.org/10.3390/microorganisms9102132
APA StyleSuzuki, K., Inoue, M., Cho, O., Mizutani, R., Shimizu, Y., Nagahama, T., & Sugita, T. (2021). Scalp Microbiome and Sebum Composition in Japanese Male Individuals with and without Androgenetic Alopecia. Microorganisms, 9(10), 2132. https://doi.org/10.3390/microorganisms9102132





