Evaluation of Tetracycline Resistance and Determination of the Tentative Microbiological Cutoff Values in Lactic Acid Bacterial Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Cultural Conditions
2.2. Antibiotic Susceptibility Testing
2.3. Identification of Tetracycline Resistance Genes
2.4. Statistical Analysis and Determination of Tentative Microbiological Cutoff Values (TMCOFFs)
2.5. Sample Collection and RT-PCR
3. Results
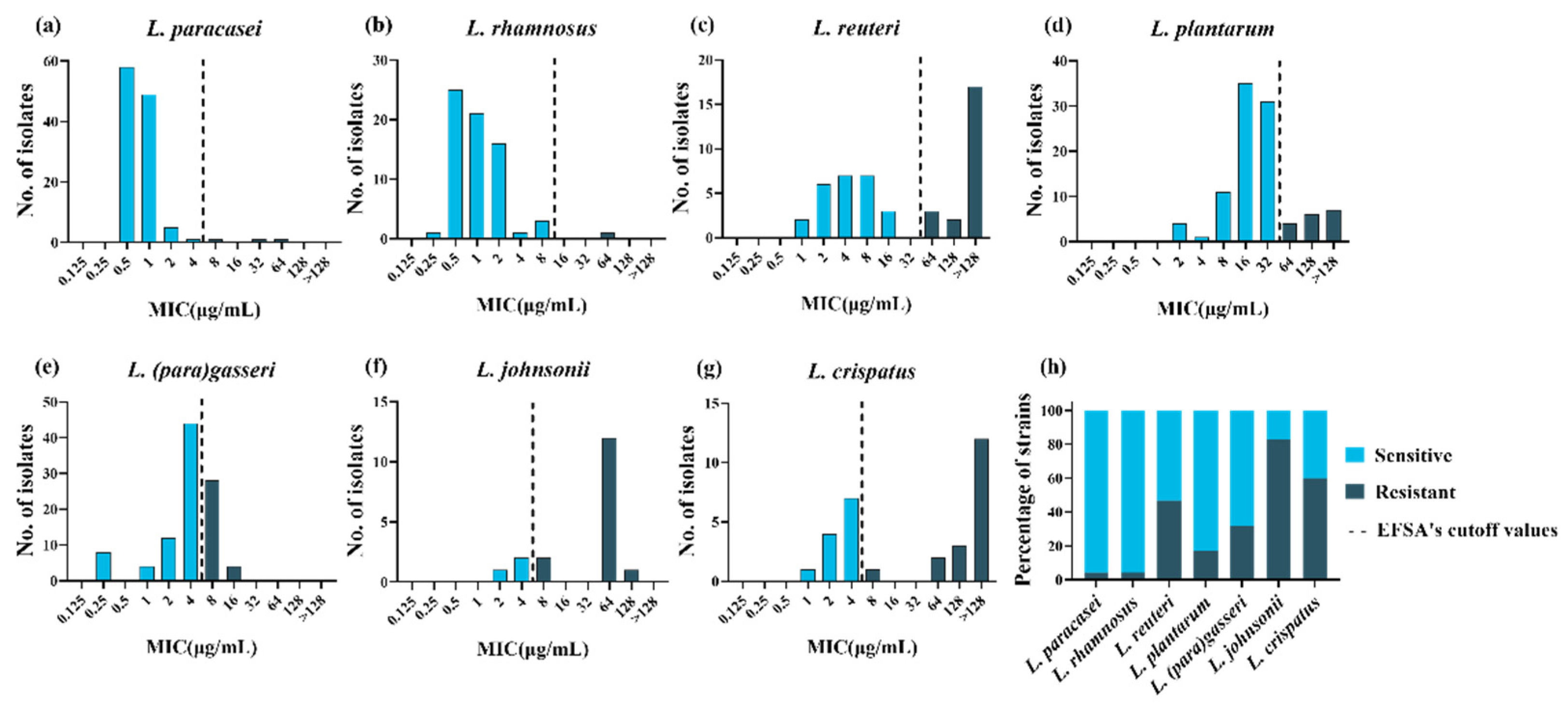
3.1. Determination of the MICs and Identification of the Resistance Phenotype
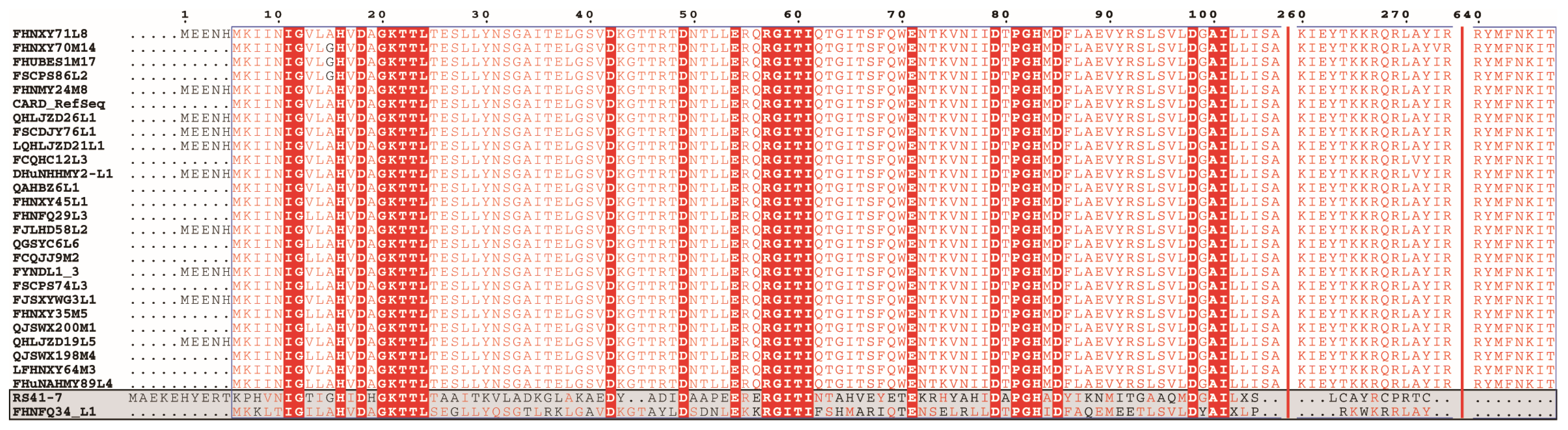
3.2. Identification of ARGs and Their Correlation with Phenotype
3.3. Definition of New Susceptibility–Resistance Cutoff Values
3.4. Prevalence and Distribution of Tetracycline Resistance Genes in LAB
3.5. Expression of Tetracycline Resistance Gene Based on RT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jamal, M.; Shareef, M.; Sajid, S. Lincomycin and tetracycline resistance in poultry. Review. Matrix Sci. Pharma. 2017, 1, 33–38. [Google Scholar] [CrossRef]
- Rudra, P.; Hurst-Hess, K.; Lappierre, P.; Ghosh, P. High levels of intrinsic tetracycline resistance in Mycobacterium abscessus are conferred by a tetracycline-modifying monooxygenase. Antimicrob. Agents Chemothe. 2018, 62, e00119-18. [Google Scholar] [CrossRef] [Green Version]
- Tao, R.; Ying, G.-G.; Su, H.-C.; Zhou, H.-W.; Sidhu, J.P. Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl rivers in South China. Environ. Pollut. 2010, 158, 2101–2109. [Google Scholar] [CrossRef]
- Guarddon, M.; Miranda, J.M.; Rodríguez, J.A.; Vázquez, B.I.; Cepeda, A.; Franco, C.M. Real-time polymerase chain reaction for the quantitative detection of tetA and tetB bacterial tetracycline resistance genes in food. Int. J. Food Microbiol. 2011, 146, 284–289. [Google Scholar] [CrossRef]
- Arredondo, A.; Àlvarez, G.; Nart, J.; Mor, C.; Blanc, V.; León, R. Detection and expression analysis of tet(B) in Streptococcus oralis. J. Oral Microbiol. 2019, 11, 1643204. [Google Scholar] [CrossRef] [Green Version]
- Speer, B.S.; Shoemaker, N.B.; Salyers, A.A. Bacterial resistance to tetracycline: Mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 1992, 5, 387–399. [Google Scholar] [CrossRef]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Phillips, I.; Casewell, M.; Cox, T.; De Groot, B.; Friis, C.; Jones, R.; Nightingale, C.; Preston, R.; Waddell, J. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J. Antimicrob. Chemother. 2004, 53, 28–52. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-n.; Han, Y.; Zhou, Z.-j. Lactic acid bacteria in traditional fermented Chinese foods. Food Res. Int. 2011, 44, 643–651. [Google Scholar] [CrossRef]
- Damania, P.; Patel, R.; Shaw, R.; Kataria, R.P.; Wadia, A. Development of antimicrobial packaging materials for food preservation using bacteriocin from Lactobacillus casei. Microbiol. Res. 2016, 7, 19–22. [Google Scholar] [CrossRef] [Green Version]
- Leuschner, R.G.; Robinson, T.P.; Hugas, M.; Cocconcelli, P.S.; Richard-Forget, F.; Klein, G.; Licht, T.R.; Nguyen-The, C.; Querol, A.; Richardson, M. Qualified presumption of safety (QPS): A generic risk assessment approach for biological agents notified to the European Food Safety Authority (EFSA). Trends Food Sci. Technol. 2010, 21, 425–435. [Google Scholar] [CrossRef]
- Shazali, N.; Foo, H.L.; Loh, T.C.; Choe, D.W.; Rahim, R.A. Prevalence of antibiotic resistance in lactic acid bacteria isolated from the faeces of broiler chicken in Malaysia. Gut Pathog. 2014, 6, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Çataloluk, O.; Gogebakan, B. Presence of drug resistance in intestinal lactobacilli of dairy and human origin in Turkey. FEMS Microbiol. Lett. 2004, 236, 7–12. [Google Scholar] [CrossRef]
- Abriouel, H.; Muñoz, M.d.C.C.; Lerma, L.L.; Montoro, B.P.; Bockelmann, W.; Pichner, R.; Kabisch, J.; Cho, G.-S.; Franz, C.M.; Galvez, A.; et al. New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Res. Int. 2015, 78, 465–481. [Google Scholar] [CrossRef]
- Broaders, E.; Gahan, C.G.; Marchesi, J.R. Mobile genetic elements of the human gastrointestinal tract: Potential for spread of antibiotic resistance genes. Gut microbes. 2013, 4, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Sadiq, F.A.; Han, X.; Zhao, J.; Zhang, H.; Ross, R.P.; Lu, W.; Chen, W. Identification, characterization, and phylogenetic analysis of eight new inducible prophages in Lactobacillus. Virus Res. 2020, 286, 198003. [Google Scholar] [CrossRef]
- Das, D.J.; Shankar, A.; Johnson, J.B.; Thomas, S. Critical insights into antibiotic resistance transferability in probiotic Lactobacillus. Nutrition. 2020, 69, 110567. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastro. Hepat. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Hazards, E.P.o.B. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA J. 2013, 11, 3449. [Google Scholar] [CrossRef] [Green Version]
- Additives, E.P.o.; Feed, P.o.S.u.i.A. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Kushiro, A.; Chervaux, C.; Cools-Portier, S.; Perony, A.; Legrain-Raspaud, S.; Obis, D.; Onoue, M.; van de Moer, A. Antimicrobial susceptibility testing of lactic acid bacteria and bifidobacteria by broth microdilution method and Etest. Int. J. Food Microbiol. 2009, 132, 54–58. [Google Scholar] [CrossRef]
- McLain, J.E.; Cytryn, E.; Durso, L.M.; Young, S. Culture-based methods for detection of antibiotic resistance in agroecosystems: Advantages, challenges, and gaps in knowledge. J. Environ. Qual. 2016, 45, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Kahlmeter, G.; Giske, C.G.; Kirn, T.J.; Sharp, S.E. Point-counterpoint: Differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute recommendations for reporting antimicrobial susceptibility results. J. Clin. Microbiol. 2019, 57, e01129-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boolchandani, M.; D’Souza, A.W.; Dantas, G. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019, 20, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Satola, S.W.; Read, T.D. Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 2019, 57, e01405-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Organization for Standardization (ISO). Milk and Milk Products—Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB); ISO 10932:2010 (IDF 223:2010); ISO: Geneva, Switzerland, 2010; Available online: https://www.iso.org/standard/46434.html (accessed on 12 September 2019).
- Additives, E.P.o.; Feed, P.o.S.u.i.A.; Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; Bastos, M.d.L.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [CrossRef]
- Yu, M.; Zhao, Y. Comparative resistomic analyses of Lysobacter species with high intrinsic multidrug resistance. J. Glob. Antimicrob. Resist. 2019, 19, 320–327. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 13, 1194–1202. [Google Scholar] [CrossRef]
- Turnidge, J.; Kahlmeter, G.; Kronvall, G. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin. Microbiol. Infect. 2006, 12, 418–425. [Google Scholar] [CrossRef]
- Kronvall, G.r. Normalized resistance interpretation as a tool for establishing epidemiological MIC susceptibility breakpoints. J. Clin. Microbiol. 2010, 48, 4445–4452. [Google Scholar] [CrossRef] [Green Version]
- Cafarchia, C.; Iatta, R.; Immediato, D.; Puttilli, M.R.; Otranto, D. Azole susceptibility of Malassezia pachydermatis and Malassezia furfur and tentative epidemiological cut-off values. Med. Mycol. 2015, 53, 743–748. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.-j.; Singh, R.P.; Lan, X.; Zhang, C.-s.; Sheng, D.-h.; Li, Y.-q. Whole transcriptome analysis and gene deletion to understand the chloramphenicol resistance mechanism and develop a screening method for homologous recombination in Myxococcus xanthus. Microb. Cell Factories 2019, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kelm-Nelson, C.A.; Stevenson, S.A.; Ciucci, M.R. Data in support of qPCR primer design and verification in a Pink1−/− rat model of Parkinson disease. Data Brief 2016, 8, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.J.; Tian, A.J.; Lv, S.M.; Du, A.D.; Chen, B.L.; Zhang, S.X. Detection of resistance and resistance genes of swine Escherichia coli in Guizhou to tetracyclines. China Anim. Husb. Vet. Med. 2018, 45, 1367–1373. [Google Scholar] [CrossRef]
- Uchida, M.; Amakasu, H.; Satoh, Y.; Murata, M. Combinations of lactic acid bacteria and yeast suitable for preparation of marine silage. Fish. Sci. 2004, 70, 507–517. [Google Scholar] [CrossRef]
- Wang, Q.; Xiong, Y.; Zhang, S.; Sui, Y.; Yu, C.; Liu, P.; Li, H.; Guo, W.; Gao, Y.; Przepiorski, A. The Dynamics of Metabolic Characterization in iPSC-Derived Kidney Organoid Differentiation via a Comparative Omics Approach. Front. Genet. 2021, 12, 132. [Google Scholar] [CrossRef]
- Hua, S.; Kittler, R.; White, K.P. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009, 137, 1259–1271. [Google Scholar] [CrossRef] [Green Version]
- Flórez, A.B.; Egervärn, M.; Danielsen, M.; Tosi, L.; Morelli, L.; Lindgren, S.; Mayo, B. Susceptibility of Lactobacillus plantarum strains to six antibiotics and definition of new susceptibility–resistance cutoff values. Microb. Drug Resist. 2006, 12, 252–256. [Google Scholar] [CrossRef]
- Cui, C.-Y.; He, Q.; Jia, Q.-L.; Li, C.; Chen, C.; Wu, X.-T.; Zhang, X.-J.; Lin, Z.-Y.; Zheng, Z.-J.; Liao, X.-P. Evolutionary Trajectory of the Tet (X) Family: Critical Residue Changes towards High-Level Tigecycline Resistance. Msystems 2021, 6, e00050-21. [Google Scholar] [CrossRef]
- Shao, Y.; Zhang, W.; Guo, H.; Pan, L.; Zhang, H.; Sun, T. Comparative studies on antibiotic resistance in Lactobacillus casei and Lactobacillus plantarum. Food Control. 2015, 50, 250–258. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef] [Green Version]
- Authority, E.F.S. Opinion of the Scientific Panel on additives and products or substances used in animal feed (FEEDAP) on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human or veterinary importance. EFSA J. 2005, 3, 223. [Google Scholar] [CrossRef]
- Danielsen, M.; Wind, A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 2003, 82, 1–11. [Google Scholar] [CrossRef]
- Matzrafi, M.; Shaar-Moshe, L.; Rubin, B.; Peleg, Z. Unraveling the transcriptional basis of temperature-dependent Pinoxaden resistance in Brachypodium hybridum. Front. Plant Sci. 2017, 8, 1064. [Google Scholar] [CrossRef] [Green Version]
- Andersson, D.I. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 2003, 6, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Comunian, R.; Daga, E.; Dupré, I.; Paba, A.; Devirgiliis, C.; Piccioni, V.; Perozzi, G.; Zonenschain, D.; Rebecchi, A.; Morelli, L.; et al. Susceptibility to tetracycline and erythromycin of Lactobacillus paracasei strains isolated from traditional Italian fermented foods. Int. J. Food Microbiol. 2010, 138, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Egervärn, M.; Roos, S.; Lindmark, H. Identification and characterization of antibiotic resistance genes in Lactobacillus reuteri and Lactobacillus plantarum. J. Appl. Microbiol. 2009, 107, 1658–1668. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; D’HAENE, K.; Danielsen, M.; Maettoe, J.; Egervaern, M.; Vandamme, P. Phenotypic and molecular assessment of antimicrobial resistance in Lactobacillus paracasei strains of food origin. J. Food Prot. 2008, 71, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D. The tetracycline resistome. Cell. Mol. Life Sci. 2010, 67, 419–431. [Google Scholar] [CrossRef]
- Zhou, X.; Yang, B.; Stanton, C.; Ross, P.; Zhao, J.; Zhang, H.; Chen, W. Comparative analysis of Lactobacillus gasseri from Chinese subjects reveals a new species-level taxa. BMC Genom. 2020, 21, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, L.; Ross, P.; Zhao, J.; Zhang, H.; Chen, W. Comparative genomics of Lactobacillus crispatus from the gut and vagina reveals genetic diversity and lifestyle adaptation. Genes 2020, 11, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Yang, B.; Chen, Y.; Stanton, C.; Ross, R.P.; Zhao, J.; Zhang, H.; Chen, W. Comparative genomic analyses of Lactobacillus rhamnosus isolated from Chinese subjects. Food Biosci. 2020, 36, 100659. [Google Scholar] [CrossRef]
- Pei, Z.; Sadiq, F.A.; Han, X.; Zhao, J.; Zhang, H.; Ross, R.P.; Lu, W.; Chen, W. Comprehensive Scanning of Prophages in Lactobacillus: Distribution, Diversity, Antibiotic Resistance Genes, and Linkages with CRISPR-Cas Systems. Msystems 2021, 6, e01211-20. [Google Scholar] [CrossRef] [PubMed]

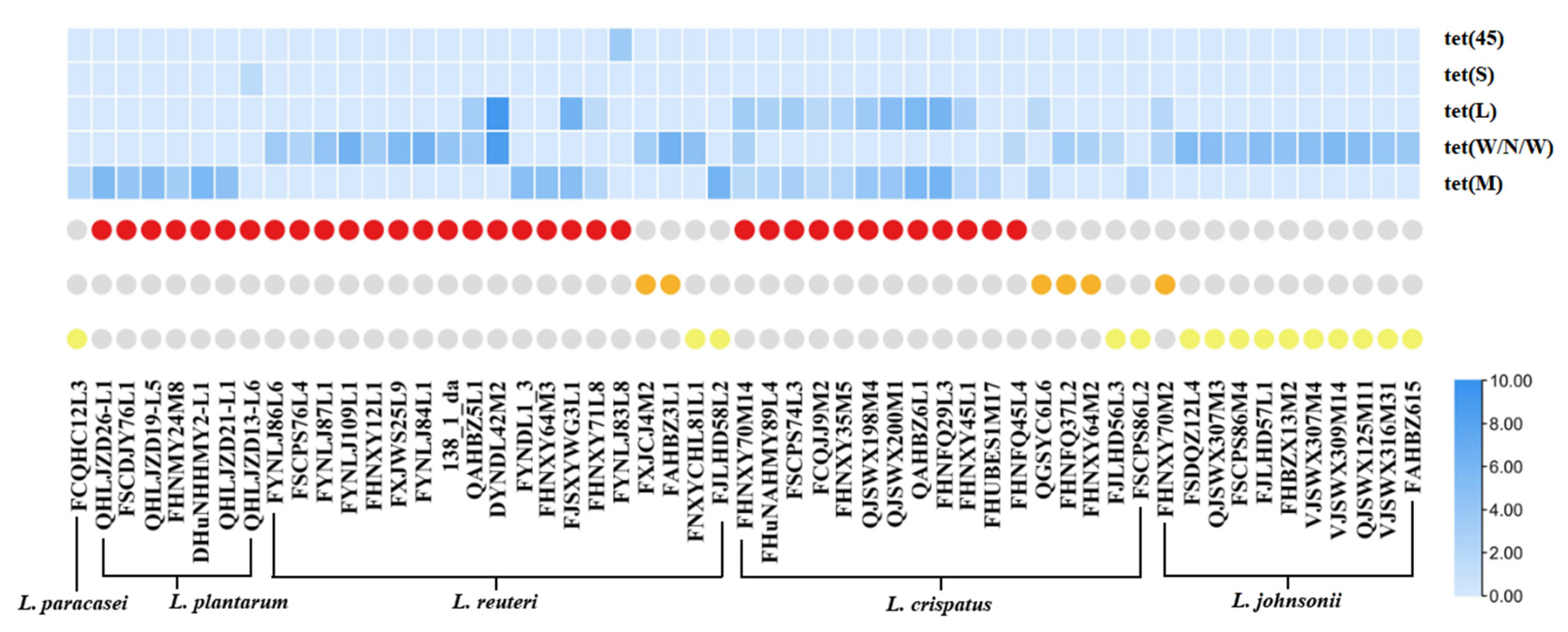
| Tetracycline Resistance Genes | Number of Tetracycline-Resistant Strains |
|---|---|
| tet(M) | L. paracasei (1), L. plantarum (6), L. reuteri (3), L.crispatus (2) |
| tet(W/N/W) | L. reuteri (11), L.johnsonii (11), L.crispatus (4) |
| tet(S) | L. plantarum (1) |
| tet(45) | L. reuteri (1) |
| tet(M) and tet(L) | L. reuteri (2), L.crispatus (10) |
| tet(W/N/W) and tet(L) | L. reuteri (2), L.johnsonii (1) |
| tet(M), tet(W/N/W), and tet(L) | L.crispatus (1) |
| No tetracycline resistance genes | L. paracasei (2), L. rhamnosus (1), L. plantarum (10), L. reuteri (3), L.johnsonii (3), L.crispatus (1), L. (para)gasseri (32) |
| TMCOFFs Obtained Using the Indicated Method (%) a | |||||
|---|---|---|---|---|---|
| Species | EFSA Cut Off | Method of Turnidge et al. b | Method of Kronvall | Eyeball Method | Median for the Method |
| L. (para)gasseri | 4 (68%) | 16 (100%) | 256 (100%) | 16 (100%) | 16 (100%) |
| L. johnsonii | 4 (17%) | 32 (38%) | 16 (38%) | 16 (38%) | 16 (38%) |
| L. crispatus | 4 (40%) | 8 (50%) | 16 (50%) | 16 (50%) | 16 (50%) |
| L. plantarum | 32 (83%) | 64 (87%) | 64 (87%) | 64 (87%) | 64 (87%) |
| Species | Total Strain Number | TETR | tet(M) | tet(W/N/W) | tet(L) | tet(S) | tet(45) |
|---|---|---|---|---|---|---|---|
| L. paracasei | 116 | 3 | 1 | 0 | 0 | 0 | 0 |
| L. rhamnosus | 68 | 1 | 0 | 0 | 0 | 0 | 0 |
| L. plantarum | 99 | 17 | 6 | 0 | 0 | 1 | 0 |
| L. reuteri | 47 | 22 | 5 | 13 | 4 | 0 | 1 |
| L. johnsonii | 18 | 15 | 0 | 12 | 1 | 0 | 0 |
| L. crispatus | 30 | 18 | 13 | 5 | 11 | 0 | 0 |
| L.(para)gasseri | 100 | 32 | 0 | 0 | 0 | 0 | 0 |
| Total | 478 | 108 | 25 | 30 | 16 | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Q.; Pei, Z.; Fang, Z.; Wang, H.; Zhu, J.; Lee, Y.-k.; Zhang, H.; Zhao, J.; Lu, W.; Chen, W. Evaluation of Tetracycline Resistance and Determination of the Tentative Microbiological Cutoff Values in Lactic Acid Bacterial Species. Microorganisms 2021, 9, 2128. https://doi.org/10.3390/microorganisms9102128
Ma Q, Pei Z, Fang Z, Wang H, Zhu J, Lee Y-k, Zhang H, Zhao J, Lu W, Chen W. Evaluation of Tetracycline Resistance and Determination of the Tentative Microbiological Cutoff Values in Lactic Acid Bacterial Species. Microorganisms. 2021; 9(10):2128. https://doi.org/10.3390/microorganisms9102128
Chicago/Turabian StyleMa, Qingqing, Zhangming Pei, Zhifeng Fang, Hongchao Wang, Jinlin Zhu, Yuan-kun Lee, Hao Zhang, Jianxin Zhao, Wenwei Lu, and Wei Chen. 2021. "Evaluation of Tetracycline Resistance and Determination of the Tentative Microbiological Cutoff Values in Lactic Acid Bacterial Species" Microorganisms 9, no. 10: 2128. https://doi.org/10.3390/microorganisms9102128
APA StyleMa, Q., Pei, Z., Fang, Z., Wang, H., Zhu, J., Lee, Y.-k., Zhang, H., Zhao, J., Lu, W., & Chen, W. (2021). Evaluation of Tetracycline Resistance and Determination of the Tentative Microbiological Cutoff Values in Lactic Acid Bacterial Species. Microorganisms, 9(10), 2128. https://doi.org/10.3390/microorganisms9102128








