Immunoglobulin A, an Active Liaison for Host-Microbiota Homeostasis
Abstract
1. Introduction
2. Immunoglobulins: The Basics
3. IgA: A Unique Structural and Functional Antibody
3.1. IgA Production in Germinal Centers
3.2. IgA Production in Nongerminal Centers
3.3. IgA Subclasses in Humans and Mice
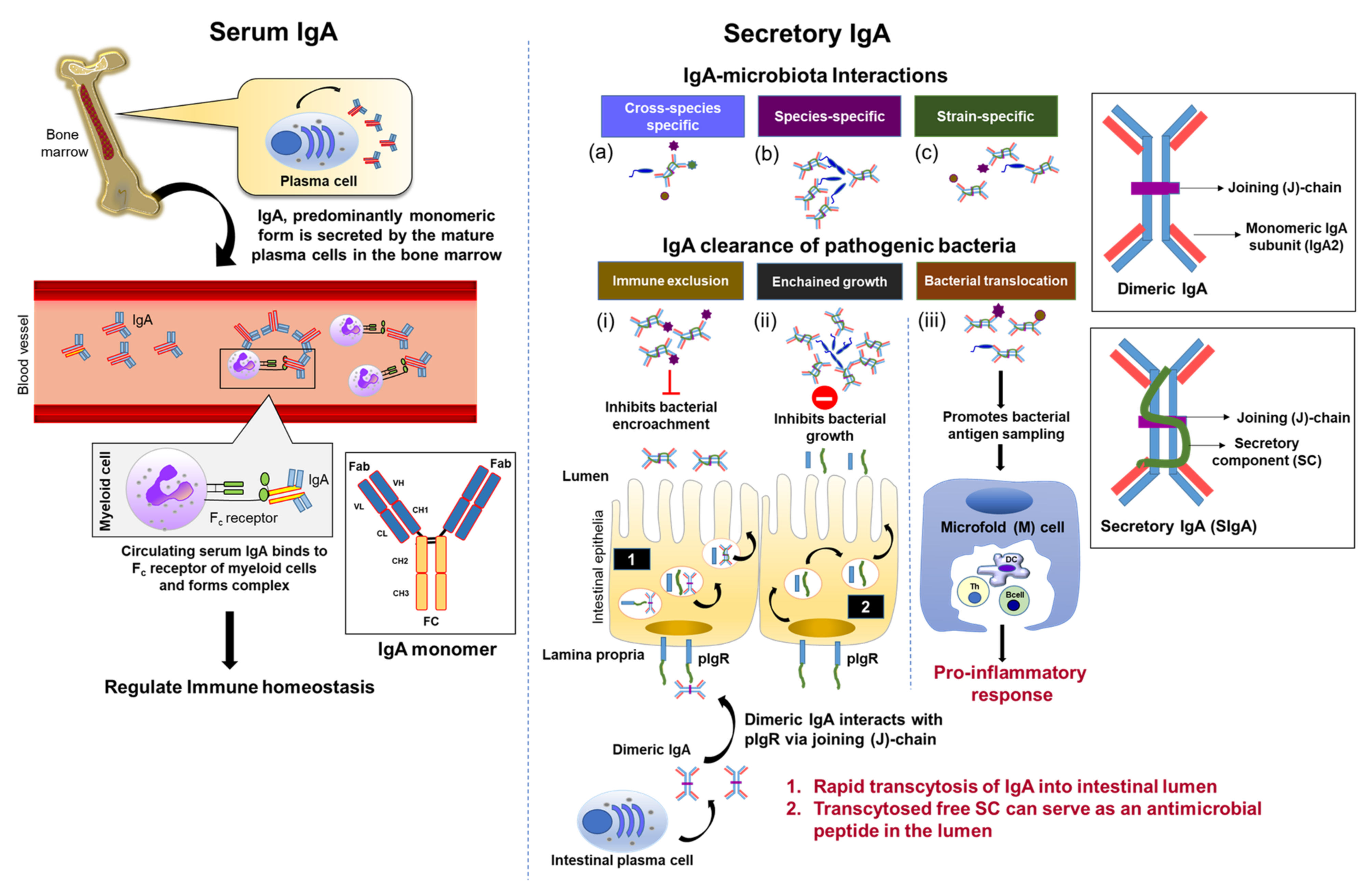
3.4. Systemic vs. Mucosal IgA: A ‘Silent Panic Button’ vs. Robust Interaction
4. SIgA: A Dynamic and Versatile Ally in Host-Microbiota Interactions
4.1. SIgA Is Selectively Reactive against the Gut Microbiota
4.2. SIgA-Mediated Gut Pathogen Clearance and Homeostatic Properties
5. Defects in IgA-Microbiota Axis Lead to Pathological Diseases
5.1. IgA Deficiency in Autoimmunity
5.2. IgA-Microbiota in Necrotizing Enterocolitis
5.3. IgA-Microbiota and Inflammatory Bowel Diseases
5.4. IgA-Microbiota in Colorectal Cancer
5.5. IgA Nephropathy and Vasculitis
5.6. IgA-Microbiota in Salmonella Infection
5.7. IgA-Microbiota in Biliary Infection
| Disease | Involvement of IgA |
|---|---|
| Autoimmune Disease [Section 5.1] | Selective IgA deficiency has a two-hit impact causing pathobiont expansion leading to dysbiosis and spontaneous gut inflammation [122,127] and unmitigated inflammation leading to autoimmune disease development [120]. |
| Necrotizing Enterocolitis (NEC) [Section 5.2] | Dimeric IgA in mother’s milk helps to control the prevalence of Enterobacteriaceae providing a safeguard against NEC [151]. |
| Inflammatory Bowel Diseases (IBD) [Section 5.3] | SIgA aids in preventing IBD pathogenesis by helping to facilitate microbiota stability [172], neutralization of procolitogenic fungi and bacteria via immune exclusion [103,180]. |
| Colorectal Cancer (CRC) [Section 5.4] | IgA antibodies reactive to carcinoembryonic antigen to be cytotoxic to colonic tumor cells [196]. Promotes a proinflammatory tumor microenvironment for oncogenic growth [203]. |
| Nephropathy & Vasculitis [Section 5.5] | IgA aggregates in the glomerulus of the kidney causing inflammation leading to nephropathy [198,199,200]. IgA deposits in the walls of the blood vessel leading to vasculitis [221]. |
| Salmonella Infection [Section 5.6] | Promotes bacterial agglutination to become susceptible to immune exclusion or enchained growth by SigA for clearance [104,249,250]. |
| Biliary Infection [Section 5.7] | SIgA is the predominant antibody in bile and helps to prevent primary and secondary hepatobiliary infection from intestinal or parasitic infection [258,265,266,267,268,269,270]. |
6. Therapeutic Potential of IgA
7. Future Thoughts
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Gutzeit, C.; Magri, G.; Cerutti, A. Intestinal IgA production and its role in host-microbe interaction. Immunol. Rev. 2014, 260, 76–85. [Google Scholar] [CrossRef]
- Weis, A.M.; Round, J.L. Microbiota-antibody interactions that regulate gut homeostasis. Cell Host Microbe. 2021, 29, 334–346. [Google Scholar] [CrossRef]
- Mkaddem, S.B.; Christou, I.; Rossato, E.; Berthelot, L.; Lehuen, A.; Monteiro, R.C. IgA, IgA Receptors, and Their Anti-inflammatory Properties. In Fc Receptors; Daeron, M., Nimmerjahn, F., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 221–235. [Google Scholar]
- Petersen, C.; Bell, R.; Klag, K.A.; Lee, S.H.; Soto, R.; Ghazaryan, A.; Buhrke, K.; Ekiz, H.A.; Ost, K.S.; Boudina, S.; et al. T cell-mediated regulation of the microbiota protects against obesity. Science 2019, 365, 6451. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Gadir, A.; Stephen-Victor, E.; Gerber, G.K.; Noval Rivas, M.; Wang, S.; Harb, H.; Wang, L.; Li, N.; Crestani, E.; Spielman, S.; et al. Microbiota therapy acts via a regulatory T cell MyD88/RORgammat pathway to suppress food allergy. Nat. Med. 2019, 25, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Stanfield, R.L.; Wilson, I.A. Antibody Structure. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Schroeder, H.W., Jr.; Cavacini, L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010, 125, S41–S52. [Google Scholar] [CrossRef] [PubMed]
- Gabrielli, E.; Pericolini, E.; Cenci, E.; Ortelli, F.; Magliani, W.; Ciociola, T.; Bistoni, F.; Conti, S.; Vecchiarelli, A.; Polonelli, L. Antibody complementarity-determining regions (CDRs): A bridge between adaptive and innate immunity. PLoS ONE 2009, 4, e8187. [Google Scholar] [CrossRef]
- Kishikawa, S.; Sato, S.; Kaneto, S.; Uchino, S.; Kohsaka, S.; Nakamura, S.; Kiyono, H. Allograft inflammatory factor 1 is a regulator of transcytosis in M cells. Nat. Commun. 2017, 8, 14509. [Google Scholar] [CrossRef]
- Rios, D.; Wood, M.B.; Li, J.; Chassaing, B.; Gewirtz, A.T.; Williams, I.R. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal. Immunol. 2016, 9, 907–916. [Google Scholar] [CrossRef]
- Neutra, M.R. Current concepts in mucosal immunity. V Role of M cells in transepithelial transport of antigens and pathogens to the mucosal immune system. Am. J. Physiol. 1998, 274, G785–G791. [Google Scholar]
- Lelouard, H.; Fallet, M.; de Bovis, B.; Meresse, S.; Gorvel, J.P. Peyer’s patch dendritic cells sample antigens by extending dendrites through M cell-specific transcellular pores. Gastroenterology 2012, 142, 592–601.e593. [Google Scholar] [CrossRef]
- Lelouard, H.; Henri, S.; De Bovis, B.; Mugnier, B.; Chollat-Namy, A.; Malissen, B.; Meresse, S.; Gorvel, J.P. Pathogenic bacteria and dead cells are internalized by a unique subset of Peyer’s patch dendritic cells that express lysozyme. Gastroenterology 2010, 138, 173–184. [Google Scholar] [CrossRef]
- Reboldi, A.; Arnon, T.I.; Rodda, L.B.; Atakilit, A.; Sheppard, D.; Cyster, J.G. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science 2016, 352, aaf4822. [Google Scholar] [CrossRef]
- Qi, H.; Egen, J.G.; Huang, A.Y.; Germain, R.N. Extrafollicular activation of lymph node B cells by antigen-bearing dendritic cells. Science 2006, 312, 1672–1676. [Google Scholar] [CrossRef]
- Borsutzky, S.; Cazac, B.B.; Roes, J.; Guzman, C.A. TGF-beta receptor signaling is critical for mucosal IgA responses. J. Immunol. 2004, 173, 3305–3309. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, A. The regulation of IgA class switching. Nat. Rev. Immunol. 2008, 8, 421–434. [Google Scholar] [CrossRef]
- Nakamura, Y.; Mimuro, H.; Kunisawa, J.; Furusawa, Y.; Takahashi, D.; Fujimura, Y.; Kaisho, T.; Kiyono, H.; Hase, K. Microfold cell-dependent antigen transport alleviates infectious colitis by inducing antigen-specific cellular immunity. Mucosal. Immunol. 2020, 13, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Gonzalez, R.M.; Niess, J.H.; Zammit, D.J.; Ravindran, R.; Srinivasan, A.; Maxwell, J.R.; Stoklasek, T.; Yadav, R.; Williams, I.R.; Gu, X.; et al. CCR6-mediated dendritic cell activation of pathogen-specific T cells in Peyer’s patches. Immunity 2006, 24, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Lycke, N.Y.; Bemark, M. The regulation of gut mucosal IgA B-cell responses: Recent developments. Mucosal. Immunol. 2017, 10, 1361–1374. [Google Scholar] [CrossRef]
- Hirota, K.; Turner, J.E.; Villa, M.; Duarte, J.H.; Demengeot, J.; Steinmetz, O.M.; Stockinger, B. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 2013, 14, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, M.; Komatsu, N.; Kawamoto, S.; Suzuki, K.; Kanagawa, O.; Honjo, T.; Hori, S.; Fagarasan, S. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science 2009, 323, 1488–1492. [Google Scholar] [CrossRef]
- Martinez-Lopez, M.; Iborra, S.; Conde-Garrosa, R.; Mastrangelo, A.; Danne, C.; Mann, E.R.; Reid, D.M.; Gaboriau-Routhiau, V.; Chaparro, M.; Lorenzo, M.P.; et al. Microbiota Sensing by Mincle-Syk Axis in Dendritic Cells Regulates Interleukin-17 and -22 Production and Promotes Intestinal Barrier Integrity. Immunity 2019, 50, 446–461.e449. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S. T follicular helper cell differentiation, function, and roles in disease. Immunity 2014, 41, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Huus, K.E.; Petersen, C.; Finlay, B.B. Diversity and dynamism of IgA−microbiota interactions. Nature Rev. Immunol. 2021, 21, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Gohda, M.; Kunisawa, J.; Miura, F.; Kagiyama, Y.; Kurashima, Y.; Higuchi, M.; Ishikawa, I.; Ogahara, I.; Kiyono, H. Sphingosine 1-phosphate regulates the egress of IgA plasmablasts from Peyer’s patches for intestinal IgA responses. J. Immunol. 2008, 180, 5335–5343. [Google Scholar] [CrossRef] [PubMed]
- Mora, J.R.; Iwata, M.; Eksteen, B.; Song, S.Y.; Junt, T.; Senman, B.; Otipoby, K.L.; Yokota, A.; Takeuchi, H.; Ricciardi-Castagnoli, P.; et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science 2006, 314, 1157–1160. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, S.; Tran, T.H.; Maruya, M.; Suzuki, K.; Doi, Y.; Tsutsui, Y.; Kato, L.M.; Fagarasan, S. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science 2012, 336, 485–489. [Google Scholar] [CrossRef]
- Perruzza, L.; Strati, F.; Gargari, G.; D’Erchia, A.M.; Fosso, B.; Pesole, G.; Guglielmetti, S.; Grassi, F. Enrichment of intestinal Lactobacillus by enhanced secretory IgA coating alters glucose homeostasis in P2rx7(-/-) mice. Sci. Rep. 2019, 9, 9315. [Google Scholar] [CrossRef]
- Perruzza, L.; Gargari, G.; Proietti, M.; Fosso, B.; D’Erchia, A.M.; Faliti, C.E.; Rezzonico-Jost, T.; Scribano, D.; Mauri, L.; Colombo, D.; et al. T Follicular Helper Cells Promote a Beneficial Gut Ecosystem for Host Metabolic Homeostasis by Sensing Microbiota-Derived Extracellular ATP. Cell Rep. 2017, 18, 2566–2575. [Google Scholar] [CrossRef]
- Melo-Gonzalez, F.; Kammoun, H.; Evren, E.; Dutton, E.E.; Papadopoulou, M.; Bradford, B.M.; Tanes, C.; Fardus-Reid, F.; Swann, J.R.; Bittinger, K.; et al. Antigen-presenting ILC3 regulate T cell-dependent IgA responses to colonic mucosal bacteria. J. Exp. Med. 2019, 216, 728–742. [Google Scholar] [CrossRef]
- McCarron, M.J.; Marie, J.C. TGF-beta prevents T follicular helper cell accumulation and B cell autoreactivity. J. Clin. Investig. 2014, 124, 4375–4386. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Wen, T.; Mingler, M.K.; Caldwell, J.M.; Wang, Y.H.; Chaplin, D.D.; Lee, E.H.; Jang, M.H.; Woo, S.Y.; Seoh, J.Y.; et al. IL-1beta in eosinophil-mediated small intestinal homeostasis and IgA production. Mucosal. Immunol. 2015, 8, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Koscso, B.; Kurapati, S.; Rodrigues, R.R.; Nedjic, J.; Gowda, K.; Shin, C.; Soni, C.; Ashraf, A.Z.; Purushothaman, I.; Palisoc, M.; et al. Gut-resident CX3CR1(hi) macrophages induce tertiary lymphoid structures and IgA response in situ. Sci. Immunol. 2020, 5, eaax0062. [Google Scholar] [CrossRef]
- Litinskiy, M.B.; Nardelli, B.; Hilbert, D.M.; He, B.; Schaffer, A.; Casali, P.; Cerutti, A. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat. Immunol. 2002, 3, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Boone, T.; Delaney, J.; Hawkins, N.; Kelley, M.; Ramakrishnan, M.; McCabe, S.; Qiu, W.R.; Kornuc, M.; Xia, X.Z.; et al. APRIL and TALL-I and receptors BCMA and TACI: System for regulating humoral immunity. Nat. Immunol. 2000, 1, 252–256. [Google Scholar] [CrossRef]
- Castigli, E.; Wilson, S.A.; Scott, S.; Dedeoglu, F.; Xu, S.; Lam, K.P.; Bram, R.J.; Jabara, H.; Geha, R.S. TACI and BAFF-R mediate isotype switching in B cells. J. Exp. Med. 2005, 201, 35–39. [Google Scholar] [CrossRef]
- Castigli, E.; Scott, S.; Dedeoglu, F.; Bryce, P.; Jabara, H.; Bhan, A.K.; Mizoguchi, E.; Geha, R.S. Impaired IgA class switching in APRIL-deficient mice. Proc. Natl. Acad. Sci. USA 2004, 101, 3903–3908. [Google Scholar] [CrossRef]
- Matsuo, K.; Nagakubo, D.; Yamamoto, S.; Shigeta, A.; Tomida, S.; Fujita, M.; Hirata, T.; Tsunoda, I.; Nakayama, T.; Yoshie, O. CCL28-Deficient Mice Have Reduced IgA Antibody-Secreting Cells and an Altered Microbiota in the Colon. J. Immunol. 2018, 200, 800–809. [Google Scholar] [CrossRef]
- Grootjans, J.; Krupka, N.; Hosomi, S.; Matute, J.D.; Hanley, T.; Saveljeva, S.; Gensollen, T.; Heijmans, J.; Li, H.; Limenitakis, J.P.; et al. Epithelial endoplasmic reticulum stress orchestrates a protective IgA response. Science 2019, 363, 993–998. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004, 303, 1662–1665. [Google Scholar] [CrossRef]
- McDole, J.R.; Wheeler, L.W.; McDonald, K.G.; Wang, B.; Konjufca, V.; Knoop, K.A.; Newberry, R.D.; Miller, M.J. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 2012, 483, 345–349. [Google Scholar] [CrossRef]
- Tezuka, H.; Abe, Y.; Iwata, M.; Takeuchi, H.; Ishikawa, H.; Matsushita, M.; Shiohara, T.; Akira, S.; Ohteki, T. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature 2007, 448, 929–933. [Google Scholar] [CrossRef]
- Fritz, J.H.; Rojas, O.L.; Simard, N.; McCarthy, D.D.; Hapfelmeier, S.; Rubino, S.; Robertson, S.J.; Larijani, M.; Gosselin, J.; Ivanov, I.I.; et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature 2011, 481, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Bergqvist, P.; Stensson, A.; Lycke, N.Y.; Bemark, M. T cell-independent IgA class switch recombination is restricted to the GALT and occurs prior to manifest germinal center formation. J. Immunol. 2010, 184, 3545–3553. [Google Scholar] [CrossRef] [PubMed]
- Cao, A.T.; Yao, S.; Gong, B.; Nurieva, R.I.; Elson, C.O.; Cong, Y. Interleukin (IL)-21 promotes intestinal IgA response to microbiota. Mucosal. Immunol. 2015, 8, 1072–1082. [Google Scholar] [CrossRef]
- Bang, Y.-J.; Hu, Z.; Li, Y.; Gattu, S.; Ruhn Kelly, A.; Raj, P.; Herz, J.; Hooper Lora, V. Serum amyloid A delivers retinol to intestinal myeloid cells to promote adaptive immunity. Science 2021, 373, eabf9232. [Google Scholar] [CrossRef] [PubMed]
- Kumar Bharathkar, S.; Parker, B.W.; Malyutin, A.G.; Haloi, N.; Huey-Tubman, K.E.; Tajkhorshid, E.; Stadtmueller, B.M. The structures of secretory and dimeric immunoglobulin A. Elife 2020, 9, e56098. [Google Scholar] [CrossRef]
- Steffen, U.; Koeleman, C.A.; Sokolova, M.V.; Bang, H.; Kleyer, A.; Rech, J.; Unterweger, H.; Schicht, M.; Garreis, F.; Hahn, J.; et al. IgA subclasses have different effector functions associated with distinct glycosylation profiles. Nat. Commun. 2020, 11, 120. [Google Scholar] [CrossRef] [PubMed]
- Woof, J.M.; Russell, M.W. Structure and function relationships in IgA. Mucosal. Immunol. 2011, 4, 590–597. [Google Scholar] [CrossRef]
- Phillips-Quagliata, J.M. Mouse IgA allotypes have major differences in their hinge regions. Immunogenetics 2002, 53, 1033–1038. [Google Scholar] [CrossRef]
- Senior, B.W.; Dunlop, J.I.; Batten, M.R.; Kilian, M.; Woof, J.M. Cleavage of a recombinant human immunoglobulin A2 (IgA2)-IgA1 hybrid antibody by certain bacterial IgA1 proteases. Infect. Immun. 2000, 68, 463–469. [Google Scholar] [CrossRef]
- He, B.; Xu, W.; Santini, P.A.; Polydorides, A.D.; Chiu, A.; Estrella, J.; Shan, M.; Chadburn, A.; Villanacci, V.; Plebani, A.; et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity 2007, 26, 812–826. [Google Scholar] [CrossRef]
- Johansen, F.E.; Braathen, R.; Brandtzaeg, P. Role of J chain in secretory immunoglobulin formation. Scand. J. Immunol. 2000, 52, 240–248. [Google Scholar] [CrossRef]
- Johansen, F.E.; Braathen, R.; Brandtzaeg, P. The J chain is essential for polymeric Ig receptor-mediated epithelial transport of IgA. J. Immunol. 2001, 167, 5185–5192. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Bomsel, M.; Casanova, J.; Vaerman, J.P.; Mostov, K. Stimulation of transcytosis of the polymeric immunoglobulin receptor by dimeric IgA. Proc. Natl. Acad. Sci. USA 1994, 91, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Stadtmueller, B.M.; Huey-Tubman, K.E.; Lopez, C.J.; Yang, Z.; Hubbell, W.L.; Bjorkman, P.J. The structure and dynamics of secretory component and its interactions with polymeric immunoglobulins. Elife 2016, 5, e10640. [Google Scholar] [CrossRef]
- Patel, A.; Jialal, I. Biochemistry, Immunoglobulin A; StatPearls: Treasure Island, FL, USA, 2021. [Google Scholar]
- Xiong, E.; Li, Y.; Min, Q.; Cui, C.; Liu, J.; Hong, R.; Lai, N.; Wang, Y.; Sun, J.; Matsumoto, R.; et al. MZB1 promotes the secretion of J-chain-containing dimeric IgA and is critical for the suppression of gut inflammation. Proc. Natl. Acad. Sci. USA 2019, 116, 13480–13489. [Google Scholar] [CrossRef]
- Crottet, P.; Corthesy, B. Secretory component delays the conversion of secretory IgA into antigen-binding competent F(ab’)2: A possible implication for mucosal defense. J. Immunol. 1998, 161, 5445–5453. [Google Scholar] [PubMed]
- Renegar, K.B.; Jackson, G.D.; Mestecky, J. In vitro comparison of the biologic activities of monoclonal monomeric IgA, polymeric IgA, and secretory IgA. J. Immunol. 1998, 160, 1219–1223. [Google Scholar]
- Phalipon, A.; Cardona, A.; Kraehenbuhl, J.P.; Edelman, L.; Sansonetti, P.J.; Corthesy, B. Secretory component: A new role in secretory IgA-mediated immune exclusion in vivo. Immunity 2002, 17, 107–115. [Google Scholar] [CrossRef]
- Mestas, J.; Hughes, C.C. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef]
- Gibbons, D.L.; Spencer, J. Mouse and human intestinal immunity: Same ballpark, different players; different rules, same score. Mucosal. Immunol. 2011, 4, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.L. Signaling in B cells via Toll-like receptors. Curr. Opin. Immunol. 2005, 17, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, E.B.; Russell, M.W. Dual function of human IgA antibodies: Inhibition of phagocytosis in circulating neutrophils and enhancement of responses in IL-8-stimulated cells. J. Leukoc. Biol. 1995, 57, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Russell, M.W.; Reinholdt, J.; Kilian, M. Anti-inflammatory activity of human IgA antibodies and their Fab alpha fragments: Inhibition of IgG-mediated complement activation. Eur. J. Immunol. 1989, 19, 2243–2249. [Google Scholar] [CrossRef] [PubMed]
- Blank, U.; Launay, P.; Benhamou, M.; Monteiro, R.C. Inhibitory ITAMs as novel regulators of immunity. Immunol. Rev. 2009, 232, 59–71. [Google Scholar] [CrossRef]
- Bakema, J.E.; van Egmond, M. The human immunoglobulin A Fc receptor FcalphaRI: A multifaceted regulator of mucosal immunity. Mucosal. Immunol. 2011, 4, 612–624. [Google Scholar] [CrossRef]
- Pasquier, B.; Launay, P.; Kanamaru, Y.; Moura, I.C.; Pfirsch, S.; Ruffie, C.; Henin, D.; Benhamou, M.; Pretolani, M.; Blank, U.; et al. Identification of FcalphaRI as an inhibitory receptor that controls inflammation: Dual role of FcRgamma ITAM. Immunity 2005, 22, 31–42. [Google Scholar] [PubMed]
- Wolf, H.M.; Fischer, M.B.; Puhringer, H.; Samstag, A.; Vogel, E.; Eibl, M.M. Human serum IgA downregulates the release of inflammatory cytokines (tumor necrosis factor-alpha, interleukin-6) in human monocytes. Blood 1994, 83, 1278–1288. [Google Scholar] [CrossRef]
- Rogier, E.W.; Frantz, A.L.; Bruno, M.E.; Kaetzel, C.S. Secretory IgA is Concentrated in the Outer Layer of Colonic Mucus along with Gut Bacteria. Pathogens 2014, 3, 390–403. [Google Scholar] [CrossRef]
- Kurita, N.; Honda, S.; Shibuya, A. Increased serum IgA in Fcalpha/muR-deficient mice on the (129 x C57BL/6) F1 genetic background. Mol. Immunol. 2015, 63, 367–372. [Google Scholar] [CrossRef]
- Vidarsson, G.; van Der Pol, W.L.; van Den Elsen, J.M.; Vile, H.; Jansen, M.; Duijs, J.; Morton, H.C.; Boel, E.; Daha, M.R.; Corthesy, B.; et al. Activity of human IgG and IgA subclasses in immune defense against Neisseria meningitidis serogroup B. J. Immunol. 2001, 166, 6250–6256. [Google Scholar] [CrossRef]
- Lang, M.L.; Shen, L.; Gao, H.; Cusack, W.F.; Lang, G.A.; Wade, W.F. Fc alpha receptor cross-linking causes translocation of phosphatidylinositol-dependent protein kinase 1 and protein kinase B alpha to MHC class II peptide-loading-like compartments. J. Immunol. 2001, 166, 5585–5593. [Google Scholar] [CrossRef] [PubMed]
- Park, R.K.; Izadi, K.D.; Deo, Y.M.; Durden, D.L. Role of Src in the modulation of multiple adaptor proteins in FcalphaRI oxidant signaling. Blood 1999, 94, 2112–2120. [Google Scholar] [CrossRef]
- Gulle, H.; Samstag, A.; Eibl, M.M.; Wolf, H.M. Physical and functional association of Fc alpha R with protein tyrosine kinase Lyn. Blood 1998, 91, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Van Gool, M.M.J.; van Egmond, M. IgA and FcalphaRI: Versatile Players in Homeostasis, Infection, and Autoimmunity. Immunotargets Ther 2020, 9, 351–372. [Google Scholar] [CrossRef] [PubMed]
- Van der Steen, L.; Tuk, C.W.; Bakema, J.E.; Kooij, G.; Reijerkerk, A.; Vidarsson, G.; Bouma, G.; Kraal, G.; de Vries, H.E.; Beelen, R.H.; et al. Immunoglobulin A: Fc(alpha)RI interactions induce neutrophil migration through release of leukotriene B4. Gastroenterology 2009, 137, 2018–2029. [Google Scholar] [CrossRef] [PubMed]
- Hansen, I.S.; Hoepel, W.; Zaat, S.A.J.; Baeten, D.L.P.; den Dunnen, J. Serum IgA Immune Complexes Promote Proinflammatory Cytokine Production by Human Macrophages, Monocytes, and Kupffer Cells through FcalphaRI-TLR Cross-Talk. J. Immunol. 2017, 199, 4124–4131. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-Bacterial Mutualism in the Human Intestine. Science 2005, 307, 1915. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Janzon, A.; Goodrich, J.K.; Koren, O.; Group, T.S.; Waters, J.L.; Ley, R.E. Interactions between the Gut Microbiome and Mucosal Immunoglobulins A, M, and G in the Developing Infant Gut. mSystems 2019, 4, e00612-19. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef]
- Sutherland, D.B.; Suzuki, K.; Fagarasan, S. Fostering of advanced mutualism with gut microbiota by Immunoglobulin A. Immunol. Rev. 2016, 270, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Tsuruta, T.; Inoue, R.; Nojima, I.; Tsukahara, T.; Hara, H.; Yajima, T. The amount of secreted IgA may not determine the secretory IgA coating ratio of gastrointestinal bacteria. FEMS Immunol. Med. Microbiol. 2009, 56, 185–189. [Google Scholar] [CrossRef]
- Kabbert, J.; Benckert, J.; Rollenske, T.; Hitch, T.C.A.; Clavel, T.; Cerovic, V.; Wardemann, H.; Pabst, O. High microbiota reactivity of adult human intestinal IgA requires somatic mutations. J. Exp. Med. 2020, 217, e20200275. [Google Scholar] [CrossRef] [PubMed]
- Bunker, J.J.; Erickson, S.A.; Flynn, T.M.; Henry, C.; Koval, J.C.; Meisel, M.; Jabri, B.; Antonopoulos, D.A.; Wilson, P.C.; Bendelac, A. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 2017, 358, eaan6619. [Google Scholar] [CrossRef] [PubMed]
- Benckert, J.; Schmolka, N.; Kreschel, C.; Zoller, M.J.; Sturm, A.; Wiedenmann, B.; Wardemann, H. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J. Clin. Investig. 2011, 121, 1946–1955. [Google Scholar] [CrossRef] [PubMed]
- Fransen, F.; Zagato, E.; Mazzini, E.; Fosso, B.; Manzari, C.; El Aidy, S.; Chiavelli, A.; D’Erchia, A.M.; Sethi, M.K.; Pabst, O.; et al. BALB/c and C57BL/6 Mice Differ in Polyreactive IgA Abundance, which Impacts the Generation of Antigen-Specific IgA and Microbiota Diversity. Immunity 2015, 43, 527–540. [Google Scholar] [CrossRef]
- Sterlin, D.; Fadlallah, J.; Adams, O.; Fieschi, C.; Parizot, C.; Dorgham, K.; Rajkumar, A.; Autaa, G.; El-Kafsi, H.; Charuel, J.L.; et al. Human IgA binds a diverse array of commensal bacteria. J. Exp. Med. 2020, 217, e20181635. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, P.; Ding, H.; Canales-Herrerias, P.; Pasricha, P.J.; Sonnenburg, J.L.; Peterson, D.A. Intestinal IgA Regulates Expression of a Fructan Polysaccharide Utilization Locus in Colonizing Gut Commensal Bacteroides thetaiotaomicron. mBio 2019, 10, e02324-19. [Google Scholar] [CrossRef] [PubMed]
- Hapfelmeier, S.; Lawson, M.A.; Slack, E.; Kirundi, J.K.; Stoel, M.; Heikenwalder, M.; Cahenzli, J.; Velykoredko, Y.; Balmer, M.L.; Endt, K.; et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 2010, 328, 1705–1709. [Google Scholar] [CrossRef]
- Grosserichter-Wagener, C.; Radjabzadeh, D.; van der Weide, H.; Smit, K.N.; Kraaij, R.; Hays, J.P.; van Zelm, M.C. Differences in Systemic IgA Reactivity and Circulating Th Subsets in Healthy Volunteers with Specific Microbiota Enterotypes. Front. Immunol. 2019, 10, 341. [Google Scholar] [CrossRef]
- Yang, C.; Mogno, I.; Contijoch, E.J.; Borgerding, J.N.; Aggarwala, V.; Li, Z.; Siu, S.; Grasset, E.K.; Helmus, D.S.; Dubinsky, M.C.; et al. Fecal IgA Levels Are Determined by Strain-Level Differences in Bacteroides ovatus and Are Modifiable by Gut Microbiota Manipulation. Cell Host Microbe. 2020, 27, 467–475.e466. [Google Scholar] [CrossRef]
- Yasui, H.; Nagaoka, N.; Mike, A.; Hayakawa, K.; Ohwaki, M. Detection of Bifidobacterium Strains that Induce Large Quantities of IgA. Microb. Ecol. Health Dis. 1992, 5, 155–162. [Google Scholar]
- Peterson, D.A.; McNulty, N.P.; Guruge, J.L.; Gordon, J.I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2007, 2, 328–339. [Google Scholar] [CrossRef]
- Michetti, P.; Porta, N.; Mahan, M.J.; Slauch, J.M.; Mekalanos, J.J.; Blum, A.L.; Kraehenbuhl, J.P.; Neutra, M.R. Monoclonal immunoglobulin A prevents adherence and invasion of polarized epithelial cell monolayers by Salmonella typhimurium. Gastroenterology 1994, 107, 915–923. [Google Scholar] [CrossRef]
- Yang, Y.; Palm, N.W. Immunoglobulin A and the microbiome. Curr. Opin. Microbiol. 2020, 56, 89–96. [Google Scholar] [CrossRef]
- Hand, T.W.; Reboldi, A. Production and Function of Immunoglobulin A. Ann. Rev. Immunol. 2021, 39, 695–718. [Google Scholar] [CrossRef] [PubMed]
- Mantis, N.J.; Rol, N.; Corthésy, B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal. Immunol. 2011, 4, 603–611. [Google Scholar] [CrossRef]
- Michetti, P.; Mahan, M.J.; Slauch, J.M.; Mekalanos, J.J.; Neutra, M.R. Monoclonal secretory immunoglobulin A protects mice against oral challenge with the invasive pathogen Salmonella typhimurium. Infect. Immunol. 1992, 60, 1786–1792. [Google Scholar] [CrossRef] [PubMed]
- Ost, K.S.; O’Meara, T.R.; Stephens, W.Z.; Chiaro, T.; Zhou, H.; Penman, J.; Bell, R.; Catanzaro, J.R.; Song, D.; Singh, S.; et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature 2021, 596, 114–118. [Google Scholar] [CrossRef]
- Moor, K.; Diard, M.; Sellin, M.E.; Felmy, B.; Wotzka, S.Y.; Toska, A.; Bakkeren, E.; Arnoldini, M.; Bansept, F.; Co, A.D.; et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 2017, 544, 498–502. [Google Scholar] [CrossRef]
- Bansept, F.; Marrec, L.; Bitbol, A.F.; Loverdo, C. Antibody-mediated crosslinking of gut bacteria hinders the spread of antibiotic resistance. Evolution 2019, 73, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Bansept, F.; Schumann-Moor, K.; Diard, M.; Hardt, W.D.; Slack, E.; Loverdo, C. Enchained growth and cluster dislocation: A possible mechanism for microbiota homeostasis. PLoS Comput. Biol. 2019, 15, e1006986. [Google Scholar] [CrossRef]
- Kadaoui, K.A.; Corthesy, B. Secretory IgA mediates bacterial translocation to dendritic cells in mouse Peyer’s patches with restriction to mucosal compartment. J. Immunol. 2007, 179, 7751–7757. [Google Scholar] [CrossRef]
- Rey, J.; Garin, N.; Spertini, F.; Corthesy, B. Targeting of secretory IgA to Peyer’s patch dendritic and T cells after transport by intestinal M cells. J. Immunol. 2004, 172, 3026–3033. [Google Scholar] [CrossRef] [PubMed]
- Bunker, J.J.; Flynn, T.M.; Koval, J.C.; Shaw, D.G.; Meisel, M.; McDonald, B.D.; Ishizuka, I.E.; Dent, A.L.; Wilson, P.C.; Jabri, B.; et al. Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity 2015, 43, 541–553. [Google Scholar] [CrossRef]
- Van der Waaij, L.A.; Limburg, P.C.; Mesander, G.; van der Waaij, D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut 1996, 38, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Vogelzang, A.; Maruya, M.; Miyajima, M.; Murata, M.; Son, A.; Kuwahara, T.; Tsuruyama, T.; Yamada, S.; Matsuura, M.; et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J. Exp. Med. 2018, 215, 2019–2034. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Ladinsky, M.S.; Yu, K.B.; Sanders, J.G.; Yoo, B.B.; Chou, W.C.; Conner, M.E.; Earl, A.M.; Knight, R.; Bjorkman, P.J.; et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018, 360, 795–800. [Google Scholar] [CrossRef]
- Briliute, J.; Urbanowicz, P.A.; Luis, A.S.; Basle, A.; Paterson, N.; Rebello, O.; Hendel, J.; Ndeh, D.A.; Lowe, E.C.; Martens, E.C.; et al. Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci. Nat. Microbiol. 2019, 4, 1571–1581. [Google Scholar] [CrossRef]
- Fehr, K.; Moossavi, S.; Sbihi, H.; Boutin, R.C.T.; Bode, L.; Robertson, B.; Yonemitsu, C.; Field, C.J.; Becker, A.B.; Mandhane, P.J.; et al. Breastmilk Feeding Practices Are Associated with the Co-Occurrence of Bacteria in Mothers’ Milk and the Infant Gut: The CHILD Cohort Study. Cell Host Microbe. 2020, 28, 285–297.e284. [Google Scholar] [CrossRef]
- McLoughlin, K.; Schluter, J.; Rakoff-Nahoum, S.; Smith, A.L.; Foster, K.R. Host Selection of Microbiota via Differential Adhesion. Cell Host Microbe 2016, 19, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, S.; Maruya, M.; Kato, L.M.; Suda, W.; Atarashi, K.; Doi, Y.; Tsutsui, Y.; Qin, H.; Honda, K.; Okada, T.; et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 2014, 41, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Ansaldo, E.; Slayden, L.C.; Ching, K.L.; Koch, M.A.; Wolf, N.K.; Plichta, D.R.; Brown, E.M.; Graham, D.B.; Xavier, R.J.; Moon, J.J.; et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 2019, 364, 1179–1184. [Google Scholar] [CrossRef]
- Bunker, J.J.; Bendelac, A. IgA Responses to Microbiota. Immunity 2018, 49, 211–224. [Google Scholar] [CrossRef]
- Blumberg, R.; Powrie, F. Microbiota, disease, and back to health: A metastable journey. Sci. Transl. Med. 2012, 4, 137rv137. [Google Scholar] [CrossRef]
- Yel, L. Selective IgA deficiency. J. Clin. Immunol. 2010, 30, 10–16. [Google Scholar] [CrossRef]
- Jorgensen, G.H.; Thorsteinsdottir, I.; Gudmundsson, S.; Hammarstrom, L.; Ludviksson, B.R. Familial aggregation of IgAD and autoimmunity. Clin. Immunol 2009, 131, 233–239. [Google Scholar] [CrossRef]
- Jacob, C.M.; Pastorino, A.C.; Fahl, K.; Carneiro-Sampaio, M.; Monteiro, R.C. Autoimmunity in IgA deficiency: Revisiting the role of IgA as a silent housekeeper. J. Clin. Immunol. 2008, 28 (Suppl. 1), S56–S61. [Google Scholar] [CrossRef] [PubMed]
- Shulzhenko, N.; Morgun, A.; Hsiao, W.; Battle, M.; Yao, M.; Gavrilova, O.; Orandle, M.; Mayer, L.; Macpherson, A.J.; McCoy, K.D.; et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med. 2011, 17, 1585–1593. [Google Scholar] [CrossRef]
- Fadlallah, J.; El Kafsi, H.; Sterlin, D.; Juste, C.; Parizot, C.; Dorgham, K.; Autaa, G.; Gouas, D.; Almeida, M.; Lepage, P.; et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med. 2018, 10, eaan1217. [Google Scholar] [CrossRef] [PubMed]
- Sterlin, D.; Fieschi, C.; Malphettes, M.; Larsen, M.; Gorochov, G.; Fadlallah, J. Immune/microbial interface perturbation in human IgA deficiency. Gut Microbes 2019, 10, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, J.R.; Strauss, J.D.; Bielecka, A.; Porto, A.F.; Lobo, F.M.; Urban, A.; Schofield, W.B.; Palm, N.W. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci. Rep. 2019, 9, 13574. [Google Scholar] [CrossRef]
- Fadlallah, J.; Sterlin, D.; Fieschi, C.; Parizot, C.; Dorgham, K.; El Kafsi, H.; Autaa, G.; Ghillani-Dalbin, P.; Juste, C.; Lepage, P.; et al. Synergistic convergence of microbiota-specific systemic IgG and secretory IgA. J. Allergy Clin. Immunol. 2019, 143, 1575–1585.e1574. [Google Scholar] [CrossRef] [PubMed]
- Moll, J.M.; Myers, P.N.; Zhang, C.; Eriksen, C.; Wolf, J.; Appelberg, K.S.; Lindberg, G.; Bahl, M.I.; Zhao, H.; Pan-Hammarstrom, Q.; et al. Gut Microbiota Perturbation in IgA Deficiency Is Influenced by IgA-Autoantibody Status. Gastroenterology 2021, 160, 2423–2434.e2425. [Google Scholar] [CrossRef]
- Nagaishi, T.; Watabe, T.; Kotake, K.; Kumazawa, T.; Aida, T.; Tanaka, K.; Ono, R.; Ishino, F.; Usami, T.; Miura, T.; et al. Immunoglobulin A-specific deficiency induces spontaneous inflammation specifically in the ileum. Gut 2021, in press. [Google Scholar] [CrossRef]
- Hammarstrom, L.; Vorechovsky, I.; Webster, D. Selective IgA deficiency (SIgAD) and common variable immunodeficiency (CVID). Clin. Exp. Immunol. 2000, 120, 225–231. [Google Scholar] [CrossRef]
- Jorgensen, S.F.; Troseid, M.; Kummen, M.; Anmarkrud, J.A.; Michelsen, A.E.; Osnes, L.T.; Holm, K.; Hoivik, M.L.; Rashidi, A.; Dahl, C.P.; et al. Altered gut microbiota profile in common variable immunodeficiency associates with levels of lipopolysaccharide and markers of systemic immune activation. Mucosal. Immunol. 2016, 9, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Rigoni, R.; Fontana, E.; Guglielmetti, S.; Fosso, B.; D’Erchia, A.M.; Maina, V.; Taverniti, V.; Castiello, M.C.; Mantero, S.; Pacchiana, G.; et al. Intestinal microbiota sustains inflammation and autoimmunity induced by hypomorphic RAG defects. J. Exp. Med. 2016, 213, 355–375. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.D.; Khan, M.A.W.; Chatzistamou, I.; Chamseddine, D.; Williams-Kang, K.; Perry, M.; Enos, R.; Murphy, A.; Gomez, G.; Aladhami, A.; et al. Gut Antibody Deficiency in a Mouse Model of CVID Results in Spontaneous Development of a Gluten-Sensitive Enteropathy. Front. Immunol. 2019, 10, 2484. [Google Scholar] [CrossRef]
- Shulzhenko, N.; Dong, X.; Vyshenska, D.; Greer, R.L.; Gurung, M.; Vasquez-Perez, S.; Peremyslova, E.; Sosnovtsev, S.; Quezado, M.; Yao, M.; et al. CVID enteropathy is characterized by exceeding low mucosal IgA levels and interferon-driven inflammation possibly related to the presence of a pathobiont. Clin. Immunol. 2018, 197, 139–153. [Google Scholar] [CrossRef]
- Neu, J.; Walker, W.A. Necrotizing enterocolitis. N. Engl. J. Med. 2011, 364, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Gephart, S.M.; McGrath, J.M.; Effken, J.A.; Halpern, M.D. Necrotizing enterocolitis risk: State of the science. Adv. Neonatal. Care 2012, 12, 77–87. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef]
- Zhou, Y.; Shan, G.; Sodergren, E.; Weinstock, G.; Walker, W.A.; Gregory, K.E. Longitudinal analysis of the premature infant intestinal microbiome prior to necrotizing enterocolitis: A case-control study. PLoS ONE 2015, 10, e0118632. [Google Scholar] [CrossRef]
- Alexander, V.N.; Northrup, V.; Bizzarro, M.J. Antibiotic exposure in the newborn intensive care unit and the risk of necrotizing enterocolitis. J. Pediatr. 2011, 159, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.N.; Gong, H.; Good, M.; McElroy, S.J. Dithizone-induced Paneth cell disruption significantly decreases intestinal perfusion in the murine small intestine. J. Pediatr. Surg. 2019, 54, 2402–2407. [Google Scholar] [CrossRef]
- Lueschow, S.R.; Stumphy, J.; Gong, H.; Kern, S.L.; Elgin, T.G.; Underwood, M.A.; Kalanetra, K.M.; Mills, D.A.; Wong, M.H.; Meyerholz, D.K.; et al. Loss of murine Paneth cell function alters the immature intestinal microbiome and mimics changes seen in neonatal necrotizing enterocolitis. PLoS ONE 2018, 13, e0204967. [Google Scholar] [CrossRef]
- White, J.R.; Gong, H.; Pope, B.; Schlievert, P.; McElroy, S.J. Paneth-cell-disruption-induced necrotizing enterocolitis in mice requires live bacteria and occurs independently of TLR4 signaling. Dis. Model. Mech. 2017, 10, 727–736. [Google Scholar] [CrossRef]
- Musemeche, C.A.; Kosloske, A.M.; Bartow, S.A.; Umland, E.T. Comparative effects of ischemia, bacteria, and substrate on the pathogenesis of intestinal necrosis. J. Pediatr. Surg. 1986, 21, 536–538. [Google Scholar] [CrossRef]
- Itani, T.; Ayoub Moubareck, C.; Melki, I.; Rousseau, C.; Mangin, I.; Butel, M.J.; Karam-Sarkis, D. Preterm infants with necrotising enterocolitis demonstrate an unbalanced gut microbiota. Acta Paediatr. 2018, 107, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Paveglio, S.; Ledala, N.; Rezaul, K.; Lin, Q.; Zhou, Y.; Provatas, A.A.; Bennett, E.; Lindberg, T.; Caimano, M.; Matson, A.P. Cytotoxin-producing Klebsiella oxytoca in the preterm gut and its association with necrotizing enterocolitis. Emerg. Microbes Infect. 2020, 9, 1321–1329. [Google Scholar] [CrossRef]
- Olm, M.R.; Bhattacharya, N.; Crits-Christoph, A.; Firek, B.A.; Baker, R.; Song, Y.S.; Morowitz, M.J.; Banfield, J.F. Necrotizing enterocolitis is preceded by increased gut bacterial replication, Klebsiella, and fimbriae-encoding bacteria. Sci. Adv. 2019, 5, eaax5727. [Google Scholar] [CrossRef]
- Shaw, A.G.; Sim, K.; Rose, G.; Wooldridge, D.J.; Li, M.S.; Misra, R.V.; Gharbia, S.; Kroll, J.S. Premature neonatal gut microbial community patterns supporting an epithelial TLR-mediated pathway for necrotizing enterocolitis. BMC Microbiol. 2021, 21, 225. [Google Scholar] [CrossRef] [PubMed]
- McMurtry, V.E.; Gupta, R.W.; Tran, L.; Blanchard, E.E.T.; Penn, D.; Taylor, C.M.; Ferris, M.J. Bacterial diversity and Clostridia abundance decrease with increasing severity of necrotizing enterocolitis. Microbiome 2015, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, P.S.; Warner, B.B.; Zhou, Y.; Weinstock, G.M.; Sodergren, E.; Hall-Moore, C.M.; Stevens, H.J.; Bennett, W.E., Jr.; Shaikh, N.; Linneman, L.A.; et al. Patterned progression of bacterial populations in the premature infant gut. Proc. Natl. Acad. Sci. USA 2014, 111, 12522–12527. [Google Scholar] [CrossRef]
- Lueschow, S.R.; Kern, S.L.; Gong, H.; Grobe, J.L.; Segar, J.L.; Carlson, S.J.; McElroy, S.J. Feeding Formula Eliminates the Necessity of Bacterial Dysbiosis and Induces Inflammation and Injury in the Paneth Cell Disruption Murine NEC Model in an Osmolality-Dependent Manner. Nutrients 2020, 12, 900. [Google Scholar] [CrossRef]
- Cortez, J.; Makker, K.; Kraemer, D.F.; Neu, J.; Sharma, R.; Hudak, M.L. Maternal milk feedings reduce sepsis, necrotizing enterocolitis and improve outcomes of premature infants. J. Perinatol. 2018, 38, 71–74. [Google Scholar] [CrossRef]
- Parr, E.L.; Bozzola, J.J.; Parr, M.B. Purification and measurement of secretory IgA in mouse milk. J. Immunol. Methods 1995, 180, 147–157. [Google Scholar] [CrossRef]
- Gopalakrishna, K.P.; Macadangdang, B.R.; Rogers, M.B.; Tometich, J.T.; Firek, B.A.; Baker, R.; Ji, J.; Burr, A.H.P.; Ma, C.; Good, M.; et al. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat. Med. 2019, 25, 1110–1115. [Google Scholar] [CrossRef]
- Mirpuri, J.; Raetz, M.; Sturge, C.R.; Wilhelm, C.L.; Benson, A.; Savani, R.C.; Hooper, L.V.; Yarovinsky, F. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 2014, 5, 28–39. [Google Scholar] [CrossRef]
- Eibl, M.M.; Wolf, H.M.; Furnkranz, H.; Rosenkranz, A. Prevention of necrotizing enterocolitis in low-birth-weight infants by IgA-IgG feeding. N. Engl. J. Med. 1988, 319, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Dunsmore, G.; Koleva, P.; Elloumi, Y.; Wu, R.Y.; Sutton, R.T.; Ambrosio, L.; Hotte, N.; Nguyen, V.; Madsen, K.L.; et al. The Profile of Human Milk Metabolome, Cytokines, and Antibodies in Inflammatory Bowel Diseases Versus Healthy Mothers, and Potential Impact on the Newborn. J. Crohns Colitis 2019, 13, 431–441. [Google Scholar] [CrossRef]
- Brawner, K.M.; Yeramilli, V.A.; Kennedy, B.A.; Patel, R.K.; Martin, C.A. Prenatal stress increases IgA coating of offspring microbiota and exacerbates necrotizing enterocolitis-like injury in a sex-dependent manner. Brain Behav. Immun. 2020, 89, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, X.; Lv, A.; Fan, S.; Zhang, J. Saccharomyces boulardii modulates necrotizing enterocolitis in neonatal mice by regulating the sirtuin 1/NFkappaB pathway and the intestinal microbiota. Mol. Med. Rep. 2020, 22, 671–680. [Google Scholar] [CrossRef]
- Morgan, R.L.; Preidis, G.A.; Kashyap, P.C.; Weizman, A.V.; Sadeghirad, B.; McMaster Probiotic, P.; Synbiotic Work, G. Probiotics Reduce Mortality and Morbidity in Preterm, Low-Birth-Weight Infants: A Systematic Review and Network Meta-analysis of Randomized Trials. Gastroenterology 2020, 159, 467–480. [Google Scholar] [CrossRef]
- Isani, M.; Bell, B.A.; Delaplain, P.T.; Bowling, J.D.; Golden, J.M.; Elizee, M.; Illingworth, L.; Wang, J.; Gayer, C.P.; Grishin, A.V.; et al. Lactobacillus murinus HF12 colonizes neonatal gut and protects rats from necrotizing enterocolitis. PLoS ONE 2018, 13, e0196710. [Google Scholar] [CrossRef]
- Colombel, J.F.; Mahadevan, U. Inflammatory Bowel Disease 2017: Innovations and Changing Paradigms. Gastroenterology 2017, 152, 309–312. [Google Scholar] [CrossRef]
- Alatab, S.; Sepanlou, S.G.; Ikuta, K.; Vahedi, H.; Bisignano, C.; Safiri, S.; Sadeghi, A.; Nixon, M.R.; Abdoli, A.; Abolhassani, H.; et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar]
- Hernandez-Chirlaque, C.; Aranda, C.J.; Ocon, B.; Capitan-Canadas, F.; Ortega-Gonzalez, M.; Carrero, J.J.; Suarez, M.D.; Zarzuelo, A.; Sanchez de Medina, F.; Martinez-Augustin, O. Germ-free and Antibiotic-treated Mice are Highly Susceptible to Epithelial Injury in DSS Colitis. J. Crohns Colitis 2016, 10, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Jangid, A.; Fukuda, S.; Seki, M.; Horiuchi, T.; Suzuki, Y.; Taylor, T.D.; Ohno, H.; Prakash, T. Association of colitis with gut-microbiota dysbiosis in clathrin adapter AP-1B knockout mice. PLoS ONE 2020, 15, e0228358. [Google Scholar] [CrossRef] [PubMed]
- Selvanantham, T.; Lin, Q.; Guo, C.X.; Surendra, A.; Fieve, S.; Escalante, N.K.; Guttman, D.S.; Streutker, C.J.; Robertson, S.J.; Philpott, D.J.; et al. NKT Cell-Deficient Mice Harbor an Altered Microbiota That Fuels Intestinal Inflammation during Chemically Induced Colitis. J. Immunol. 2016, 197, 4464–4472. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, Y.; Wang, J.; Wu, G.; Long, W.; Xue, Z.; Wang, L.; Zhang, X.; Pang, X.; Zhao, Y.; et al. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci. Rep. 2016, 6, 27572. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S.; Gallini, C.A.; Yatsunenko, T.; Michaud, M.; DuBois, A.; Delaney, M.L.; Punit, S.; Karlsson, M.; Bry, L.; Glickman, J.N.; et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 2010, 8, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 119–129. [Google Scholar] [CrossRef]
- Imhann, F.; Vich Vila, A.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; van Dullemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018, 67, 108–119. [Google Scholar] [CrossRef]
- Reikvam, D.H.; Derrien, M.; Islam, R.; Erofeev, A.; Grcic, V.; Sandvik, A.; Gaustad, P.; Meza-Zepeda, L.A.; Jahnsen, F.L.; Smidt, H.; et al. Epithelial-microbial crosstalk in polymeric Ig receptor deficient mice. Eur. J. Immunol. 2012, 42, 2959–2970. [Google Scholar] [CrossRef]
- Landuyt, A.E.; Klocke, B.J.; Duck, L.W.; Kemp, K.M.; Muir, R.Q.; Jennings, M.S.; Blum, S.I.; Tse, H.M.; Lee, G.; Morrow, C.D.; et al. ICOS ligand and IL-10 synergize to promote host-microbiota mutualism. Proc. Natl. Acad. Sci. USA 2021, 118, e2018278118. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, X.; Yang, Q.; Deng, H.; Liu, Y.; Zhou, P.; Xu, H.; Chen, D.; Feng, D.; Zhang, H.; et al. Critical Role of Intestinal Microbiota in ATF3-Mediated Gut Immune Homeostasis. J. Immunol. 2020, 205, 842–852. [Google Scholar] [CrossRef]
- Cao, Y.; Yang, Q.; Deng, H.; Tang, J.; Hu, J.; Liu, H.; Zhi, M.; Ye, L.; Zou, B.; Liu, Y.; et al. Transcriptional factor ATF3 protects against colitis by regulating follicular helper T cells in Peyer’s patches. Proc. Natl. Acad. Sci. USA 2019, 116, 6286–6291. [Google Scholar] [CrossRef] [PubMed]
- Kubinak, J.L.; Petersen, C.; Stephens, W.Z.; Soto, R.; Bake, E.; O’Connell, R.M.; Round, J.L. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 2015, 17, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Charbonnier, L.M.; Noval Rivas, M.; Georgiev, P.; Li, N.; Gerber, G.; Bry, L.; Chatila, T.A. MyD88 Adaptor-Dependent Microbial Sensing by Regulatory T Cells Promotes Mucosal Tolerance and Enforces Commensalism. Immunity 2015, 43, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Itoiz, M.A.; Pena-Cearra, A.; Martin-Ruiz, I.; Lavin, J.L.; Simo, C.; Rodriguez, H.; Atondo, E.; Flores, J.M.; Carreras-Gonzalez, A.; Tomas-Cortazar, J.; et al. The mitochondrial negative regulator MCJ modulates the interplay between microbiota and the host during ulcerative colitis. Sci. Rep. 2020, 10, 572. [Google Scholar] [CrossRef] [PubMed]
- Harrington, L.; Srikanth, C.V.; Antony, R.; Rhee, S.J.; Mellor, A.L.; Shi, H.N.; Cherayil, B.J. Deficiency of indoleamine 2,3-dioxygenase enhances commensal-induced antibody responses and protects against Citrobacter rodentium-induced colitis. Infect. Immun. 2008, 76, 3045–3053. [Google Scholar] [CrossRef]
- Van der Waaij, L.A.; Kroese, F.G.; Visser, A.; Nelis, G.F.; Westerveld, B.D.; Jansen, P.L.; Hunter, J.O. Immunoglobulin coating of faecal bacteria in inflammatory bowel disease. Eur. J. Gastroenterol. Hepatol. 2004, 16, 669–674. [Google Scholar] [CrossRef]
- Palm, N.W.; de Zoete, M.R.; Cullen, T.W.; Barry, N.A.; Stefanowski, J.; Hao, L.; Degnan, P.H.; Hu, J.; Peter, I.; Zhang, W.; et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014, 158, 1000–1010. [Google Scholar] [CrossRef]
- Jackson, M.A.; Pearson, C.; Ilott, N.E.; Huus, K.E.; Hegazy, A.N.; Webber, J.; Finlay, B.B.; Macpherson, A.J.; Powrie, F.; Lam, L.H. Accurate identification and quantification of commensal microbiota bound by host immunoglobulins. Microbiome 2021, 9, 33. [Google Scholar] [CrossRef]
- Shapiro, J.M.; de Zoete, M.R.; Palm, N.W.; Laenen, Y.; Bright, R.; Mallette, M.; Bu, K.; Bielecka, A.A.; Xu, F.; Hurtado-Lorenzo, A.; et al. Immunoglobulin A Targets a Unique Subset of the Microbiota in Inflammatory Bowel Disease. Cell Host Microbe 2021, 29, 83–93.e83. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Chen, H.; Shu, W.; Sun, M.; Fang, L.; Shi, Y.; Pang, Z.; Wu, W.; Liu, Z. Clinical significance of soluble immunoglobulins A and G and their coated bacteria in feces of patients with inflammatory bowel disease. J. Transl. Med. 2018, 16, 359. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Basu, S.; Bal, V.; Rath, S.; George, A. Gut IgA abundance in adult life is a major determinant of resistance to dextran sodium sulfate-colitis and can compensate for the effects of inadequate maternal IgA received by neonates. Immunology 2019, 158, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Rochereau, N.; Roblin, X.; Michaud, E.; Gayet, R.; Chanut, B.; Jospin, F.; Corthesy, B.; Paul, S. NOD2 deficiency increases retrograde transport of secretory IgA complexes in Crohn’s disease. Nat. Commun. 2021, 12, 261. [Google Scholar] [CrossRef]
- Zhang, T.; Ding, C.; Zhao, M.; Dai, X.; Yang, J.; Li, Y.; Gu, L.; Wei, Y.; Gong, J.; Zhu, W.; et al. Sodium Butyrate Reduces Colitogenic Immunoglobulin A-Coated Bacteria and Modifies the Composition of Microbiota in IL-10 Deficient Mice. Nutrients 2016, 8, 728. [Google Scholar] [CrossRef]
- Okai, S.; Usui, F.; Ohta, M.; Mori, H.; Kurokawa, K.; Matsumoto, S.; Kato, T.; Miyauchi, E.; Ohno, H.; Shinkura, R. Intestinal IgA as a modulator of the gut microbiota. Gut Microbes 2017, 8, 486–492. [Google Scholar] [CrossRef]
- Rogier, E.W.; Frantz, A.L.; Bruno, M.E.; Wedlund, L.; Cohen, D.A.; Stromberg, A.J.; Kaetzel, C.S. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc. Natl. Acad. Sci. USA 2014, 111, 3074–3079. [Google Scholar] [CrossRef]
- Ramanan, D.; Sefik, E.; Galvan-Pena, S.; Wu, M.; Yang, L.; Yang, Z.; Kostic, A.; Golovkina, T.V.; Kasper, D.L.; Mathis, D.; et al. An Immunologic Mode of Multigenerational Transmission Governs a Gut Treg Setpoint. Cell 2020, 181, 1276–1290.e1213. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Chalkias, A.; Nikotian, G.; Koutsovasilis, A.; Bramis, J.; Manouras, A.; Mystrioti, D.; Katergiannakis, V. Patients with colorectal cancer are characterized by increased concentration of fecal hb-hp complex, myeloperoxidase, and secretory IgA. Am. J. Clin. Oncol. 2011, 34, 561–566. [Google Scholar] [CrossRef]
- Chen, M.; Lin, X.; Zhang, L.; Yu, L.; Wu, Q.; Zhang, S.; Xue, F.; Huang, Y. Development of a panel of serum IgG and IgA autoantibodies for early diagnosis of colon cancer. Int. J. Med. Sci 2020, 17, 2744–2750. [Google Scholar] [CrossRef]
- De Chiara, L.; Paez de la Cadena, M.; Rodriguez-Berrocal, J.; Alvarez-Pardinas, M.C.; Pardinas-Anon, M.C.; Varela-Calvino, R.; Cordero, O.J. CD26-Related Serum Biomarkers: sCD26 Protein, DPP4 Activity, and Anti-CD26 Isotype Levels in a Colorectal Cancer-Screening Context. Dis. Markers 2020, 2020, 4347936. [Google Scholar] [CrossRef]
- Butvilovskaya, V.I.; Popletaeva, S.B.; Chechetkin, V.R.; Zubtsova, Z.I.; Tsybulskaya, M.V.; Samokhina, L.O.; Vinnitskii, L.I.; Ragimov, A.A.; Pozharitskaya, E.I.; Grigor Eva, G.A.; et al. Multiplex determination of serological signatures in the sera of colorectal cancer patients using hydrogel biochips. Cancer Med. 2016, 5, 1361–1372. [Google Scholar] [CrossRef]
- Staff, C.; Magnusson, C.G.; Hojjat-Farsangi, M.; Mosolits, S.; Liljefors, M.; Frodin, J.E.; Wahren, B.; Mellstedt, H.; Ullenhag, G.J. Induction of IgM, IgA and IgE antibodies in colorectal cancer patients vaccinated with a recombinant CEA protein. J. Clin. Immunol. 2012, 32, 855–865. [Google Scholar] [CrossRef]
- Kurt, M.; Yumuk, Z. Diagnostic accuracy of Fusobacterium nucleatum IgA and IgG ELISA test in colorectal cancer. Sci. Rep. 2021, 11, 1608. [Google Scholar] [CrossRef] [PubMed]
- Magat, E.M.; Balanag, G.A.; CariNo, A.M.; Fellizar, A.; Ortin, T.S.; Guevarra, L., Jr.; Albano, P.M. Clostridioides difficile antibody response of colorectal cancer patients versus clinically healthy individuals. Biosci. Microbiota Food Health 2020, 39, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; Li, L.F.; Guo, S.H.; Zeng, Q.Y.; Ning, F.; Liu, W.L.; Zhang, G. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci. Rep. 2016, 6, 33440. [Google Scholar] [CrossRef] [PubMed]
- Mion, F.; Vetrano, S.; Tonon, S.; Valeri, V.; Piontini, A.; Burocchi, A.; Petti, L.; Frossi, B.; Gulino, A.; Tripodo, C.; et al. Reciprocal influence of B cells and tumor macro and microenvironments in the Apc(Min/+) model of colorectal cancer. Oncoimmunology 2017, 6, e1336593. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhong, Y.; Zhuang, Z.; Xie, J.; Lu, Y.; Huang, C.; Sun, Y.; Wu, L.; Yin, J.; Yu, H.; et al. Multiregion single-cell sequencing reveals the transcriptional landscape of the immune microenvironment of colorectal cancer. Clin. Transl. Med. 2021, 11, e253. [Google Scholar] [PubMed]
- Hale, L.P. Deficiency of activation-induced cytidine deaminase in a murine model of ulcerative colitis. PLoS ONE 2020, 15, e0239295. [Google Scholar] [CrossRef]
- Muthuswamy, R.V.; Sundstrom, P.; Borjesson, L.; Gustavsson, B.; Quiding-Jarbrink, M. Impaired migration of IgA-secreting cells to colon adenocarcinomas. Cancer Immunol. Immunother 2013, 62, 989–997. [Google Scholar] [CrossRef]
- Malik, A.; Sharma, D.; Zhu, Q.; Karki, R.; Guy, C.S.; Vogel, P.; Kanneganti, T.D. IL-33 regulates the IgA-microbiota axis to restrain IL-1alpha-dependent colitis and tumorigenesis. J. Clin. Investig. 2016, 126, 4469–4481. [Google Scholar] [CrossRef]
- Garrett, W.S. The gut microbiota and colon cancer. Science 2019, 364, 1133–1135. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Hinglais, N. [Intercapillary deposits of IgA-IgG]. J. Urol. Nephrol. 1968, 74, 694–695. [Google Scholar]
- Tomana, M.; Novak, J.; Julian, B.A.; Matousovic, K.; Konecny, K.; Mestecky, J. Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J. Clin. Investig. 1999, 104, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Yaneva, H.; Nabarra, B.; Barbanel, C. Recurrence of mesangial deposition of IgA after renal transplantation. Kidney Int 1975, 7, 232–241. [Google Scholar] [CrossRef]
- Berger, J. IgA glomerular deposits in renal disease. Transplant. Proc. 1969, 1, 939–944. [Google Scholar] [PubMed]
- Zhang, Y.M.; Zhang, H. Insights into the Role of Mucosal Immunity in IgA Nephropathy. Clin. J. Am. Soc. Nephrol 2018, 13, 1584–1586. [Google Scholar] [CrossRef]
- Rodrigues, J.C.; Haas, M.; Reich, H.N. IgA Nephropathy. Clin. J. Am. Soc. Nephrol. 2017, 12, 677–686. [Google Scholar] [CrossRef]
- Jarrick, S.; Lundberg, S.; Welander, A.; Carrero, J.J.; Hoijer, J.; Bottai, M.; Ludvigsson, J.F. Mortality in IgA Nephropathy: A Nationwide Population-Based Cohort Study. J. Am. Soc. Nephrol. 2019, 30, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Sallustio, F.; Curci, C.; Chaoul, N.; Fonto, G.; Lauriero, G.; Picerno, A.; Divella, C.; Di Leo, V.; De Angelis, M.; Ben Mkaddem, S.; et al. High levels of gut-homing immunoglobulin A+ B lymphocytes support the pathogenic role of intestinal mucosal hyperresponsiveness in immunoglobulin A nephropathy patients. Nephrol. Dial. Transplant. 2021, 36, 452–464. [Google Scholar] [CrossRef]
- Kamata, T.; Nogaki, F.; Fagarasan, S.; Sakiyama, T.; Kobayashi, I.; Miyawaki, S.; Ikuta, K.; Muso, E.; Yoshida, H.; Sasayama, S.; et al. Increased frequency of surface IgA-positive plasma cells in the intestinal lamina propria and decreased IgA excretion in hyper IgA (HIGA) mice, a murine model of IgA nephropathy with hyperserum IgA. J. Immunol. 2000, 165, 1387–1394. [Google Scholar] [CrossRef]
- Kiryluk, K.; Moldoveanu, Z.; Sanders, J.T.; Eison, T.M.; Suzuki, H.; Julian, B.A.; Novak, J.; Gharavi, A.G.; Wyatt, R.J. Aberrant glycosylation of IgA1 is inherited in both pediatric IgA nephropathy and Henoch-Schonlein purpura nephritis. Kidney Int. 2011, 80, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Gharavi, A.G.; Moldoveanu, Z.; Wyatt, R.J.; Barker, C.V.; Woodford, S.Y.; Lifton, R.P.; Mestecky, J.; Novak, J.; Julian, B.A. Aberrant IgA1 glycosylation is inherited in familial and sporadic IgA nephropathy. J. Am. Soc. Nephrol. 2008, 19, 1008–1014. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.L.; Zhu, L.; Shi, S.F.; Liu, L.J.; Lv, J.C.; Zhang, H. Increased APRIL Expression Induces IgA1 Aberrant Glycosylation in IgA Nephropathy. Medicine 2016, 95, e3099. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Q.; Li, M.; Zhang, H.; Low, H.Q.; Wei, X.; Wang, J.Q.; Sun, L.D.; Sim, K.S.; Li, Y.; Foo, J.N.; et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat. Genet. 2011, 44, 178–182. [Google Scholar] [CrossRef]
- McCarthy, D.D.; Kujawa, J.; Wilson, C.; Papandile, A.; Poreci, U.; Porfilio, E.A.; Ward, L.; Lawson, M.A.; Macpherson, A.J.; McCoy, K.D.; et al. Mice overexpressing BAFF develop a commensal flora-dependent, IgA-associated nephropathy. J. Clin. Investig. 2011, 121, 3991–4002. [Google Scholar] [CrossRef]
- McCarthy, D.D.; Chiu, S.; Gao, Y.; Summers-deLuca, L.E.; Gommerman, J.L. BAFF induces a hyper-IgA syndrome in the intestinal lamina propria concomitant with IgA deposition in the kidney independent of LIGHT. Cell Immunol. 2006, 241, 85–94. [Google Scholar] [CrossRef]
- Roos, A.; Rastaldi, M.P.; Calvaresi, N.; Oortwijn, B.D.; Schlagwein, N.; van Gijlswijk-Janssen, D.J.; Stahl, G.L.; Matsushita, M.; Fujita, T.; van Kooten, C.; et al. Glomerular activation of the lectin pathway of complement in IgA nephropathy is associated with more severe renal disease. J. Am. Soc. Nephrol. 2006, 17, 1724–1734. [Google Scholar] [CrossRef]
- Roos, A.; Bouwman, L.H.; van Gijlswijk-Janssen, D.J.; Faber-Krol, M.C.; Stahl, G.L.; Daha, M.R. Human IgA activates the complement system via the mannan-binding lectin pathway. J. Immunol. 2001, 167, 2861–2868. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.Q.; Raska, M.; Stewart, T.J.; Reily, C.; King, R.G.; Crossman, D.K.; Crowley, M.R.; Hargett, A.; Zhang, Z.; Suzuki, H.; et al. Somatic Mutations Modulate Autoantibodies against Galactose-Deficient IgA1 in IgA Nephropathy. J. Am. Soc. Nephrol. 2016, 27, 3278–3284. [Google Scholar] [CrossRef]
- Kiryluk, K.; Li, Y.; Scolari, F.; Sanna-Cherchi, S.; Choi, M.; Verbitsky, M.; Fasel, D.; Lata, S.; Prakash, S.; Shapiro, S.; et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat. Genet. 2014, 46, 1187–1196. [Google Scholar] [CrossRef]
- He, J.W.; Zhou, X.J.; Li, Y.F.; Wang, Y.N.; Liu, L.J.; Shi, S.F.; Xin, X.H.; Li, R.S.; Falchi, M.; Lv, J.C.; et al. Associations of Genetic Variants Contributing to Gut Microbiota Composition in Immunoglobin A Nephropathy. mSystems 2021, 6, e00819-20. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Bai, M.; Zhao, J.; Wang, D.; Ning, X.; Sun, S. A Comparative Study of the Gut Microbiota Associated With Immunoglobulin a Nephropathy and Membranous Nephropathy. Front. Cell Infect. Microbiol 2020, 10, 557368. [Google Scholar] [CrossRef] [PubMed]
- Chai, L.; Luo, Q.; Cai, K.; Wang, K.; Xu, B. Reduced fecal short-chain fatty acids levels and the relationship with gut microbiota in IgA nephropathy. BMC Nephrol. 2021, 22, 209. [Google Scholar] [CrossRef]
- Chemouny, J.M.; Gleeson, P.J.; Abbad, L.; Lauriero, G.; Boedec, E.; Le Roux, K.; Monot, C.; Bredel, M.; Bex-Coudrat, J.; Sannier, A.; et al. Modulation of the microbiota by oral antibiotics treats immunoglobulin A nephropathy in humanized mice. Nephrol. Dial. Transplant. 2019, 34, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Lamm, M.E.; Emancipator, S.N.; Robinson, J.K.; Yamashita, M.; Fujioka, H.; Qiu, J.; Plaut, A.G. Microbial IgA protease removes IgA immune complexes from mouse glomeruli in vivo: Potential therapy for IgA nephropathy. Am. J. Pathol 2008, 172, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Pillebout, E. IgA Vasculitis and IgA Nephropathy: Same Disease? J. Clin. Med. 2021, 10, 2310. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, L.; Wang, Y.; Liu, X.; Zhang, H.; Liu, Y.; Shen, N.; Yang, J.; Gai, Z. Gut microbiota dysbiosis is associated with Henoch-Schonlein Purpura in children. Int. Immunopharmacol. 2018, 58, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, G.; Nie, X.; Zeng, Y.; Chen, Y.; Qian, Y.; Chen, G.; Huang, J.; Wang, C.; Zhang, C.; et al. Differences in Manifestations and Gut Microbiota Composition Between Patients With Different Henoch-Schonlein Purpura Phenotypes. Front. Cell Infect. Microbiol. 2021, 11, 641997. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Li, X.A.; Law, B.; U, K.I.; Pan, B.Q.; Lei, C.; Hsiao, W.W. Correlation of gut microbial compositions to the development of Kawasaki disease vasculitis in children. Future Microbiol. 2020, 15, 591–600. [Google Scholar] [CrossRef]
- Ye, Z.; Zhang, N.; Wu, C.; Zhang, X.; Wang, Q.; Huang, X.; Du, L.; Cao, Q.; Tang, J.; Zhou, C.; et al. A metagenomic study of the gut microbiome in Behcet’s disease. Microbiome 2018, 6, 135. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, J.; Kubota, T.; Takada, E.; Takai, K.; Fujiwara, N.; Arimitsu, N.; Ueda, Y.; Wakisaka, S.; Suzuki, T.; Suzuki, N. Bifidobacteria Abundance-Featured Gut Microbiota Compositional Change in Patients with Behcet’s Disease. PLoS ONE 2016, 11, e0153746. [Google Scholar] [CrossRef]
- Consolandi, C.; Turroni, S.; Emmi, G.; Severgnini, M.; Fiori, J.; Peano, C.; Biagi, E.; Grassi, A.; Rampelli, S.; Silvestri, E.; et al. Behcet’s syndrome patients exhibit specific microbiome signature. Autoimmun Rev. 2015, 14, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Ajmera, A.; Shabbir, N. Salmonella; StatPearls: Treasure Island, FL, USA, 2021. [Google Scholar]
- Boore, A.L.; Hoekstra, R.M.; Iwamoto, M.; Fields, P.I.; Bishop, R.D.; Swerdlow, D.L. Salmonella enterica Infections in the United States and Assessment of Coefficients of Variation: A Novel Approach to Identify Epidemiologic Characteristics of Individual Serotypes, 1996–2011. PLoS ONE 2015, 10, e0145416. [Google Scholar] [CrossRef] [PubMed]
- Wotzka, S.Y.; Nguyen, B.D.; Hardt, W.D. Salmonella Typhimurium Diarrhea Reveals Basic Principles of Enteropathogen Infection and Disease-Promoted DNA Exchange. Cell Host Microbe 2017, 21, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.D.; Ghori, N.; Falkow, S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J. Exp. Med. 1994, 180, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Hashizume-Takizawa, T.; Kobayashi, R.; Tsuzukibashi, O.; Saito, M.; Kurita-Ochiai, T. CCR7-deficient mice exhibit a delayed antigen-specific mucosal IgA antibody response to an oral recombinant Salmonella strain. Pathog. Dis. 2019, 77, ftz024. [Google Scholar] [CrossRef]
- Martinoli, C.; Chiavelli, A.; Rescigno, M. Entry route of Salmonella typhimurium directs the type of induced immune response. Immunity 2007, 27, 975–984. [Google Scholar] [CrossRef]
- Vazquez-Torres, A.; Jones-Carson, J.; Baumler, A.J.; Falkow, S.; Valdivia, R.; Brown, W.; Le, M.; Berggren, R.; Parks, W.T.; Fang, F.C. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 1999, 401, 804–808. [Google Scholar] [CrossRef]
- Man, A.L.; Gicheva, N.; Regoli, M.; Rowley, G.; De Cunto, G.; Wellner, N.; Bassity, E.; Gulisano, M.; Bertelli, E.; Nicoletti, C. CX3CR1+ Cell-Mediated Salmonella Exclusion Protects the Intestinal Mucosa during the Initial Stage of Infection. J. Immunol. 2017, 198, 335–343. [Google Scholar] [CrossRef]
- Chami, B.; Yeung, A.; Buckland, M.; Liu, H.; Fong, G.M.; Tao, K.; Bao, S. CXCR3 plays a critical role for host protection against Salmonellosis. Sci. Rep. 2017, 7, 10181. [Google Scholar] [CrossRef]
- Hashizume-Takizawa, T.; Shibata, N.; Kurashima, Y.; Kiyono, H.; Kurita-Ochiai, T.; Fujihashi, K. Distinct roles for Peyer’s patch B cells for induction of antigen-specific IgA antibody responses in mice administered oral recombinant Salmonella. Int. Immunol. 2019, 31, 531–541. [Google Scholar] [CrossRef]
- Hashizume, T.; Togawa, A.; Nochi, T.; Igarashi, O.; Kweon, M.N.; Kiyono, H.; Yamamoto, M. Peyer’s patches are required for intestinal immunoglobulin A responses to Salmonella spp. Infect. Immun. 2008, 76, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Richards, A.F.; Doering, J.E.; Lozito, S.A.; Varrone, J.J.; Willsey, G.G.; Pauly, M.; Whaley, K.; Zeitlin, L.; Mantis, N.J. Inhibition of invasive salmonella by orally administered IgA and IgG monoclonal antibodies. PLoS Negl. Trop. Dis. 2020, 14, e0007803. [Google Scholar] [CrossRef] [PubMed]
- Richards, A.F.; Baranova, D.E.; Pizzuto, M.S.; Jaconi, S.; Willsey, G.G.; Torres-Velez, F.J.; Doering, J.E.; Benigni, F.; Corti, D.; Mantis, N.J. Recombinant Human Secretory IgA Induces Salmonella Typhimurium Agglutination and Limits Bacterial Invasion into Gut-Associated Lymphoid Tissues. ACS Infect. Dis. 2021, 7, 1221–1235. [Google Scholar] [CrossRef]
- Bioley, G.; Monnerat, J.; Lotscher, M.; Vonarburg, C.; Zuercher, A.; Corthesy, B. Plasma-Derived Polyreactive Secretory-Like IgA and IgM Opsonizing Salmonella enterica Typhimurium Reduces Invasion and Gut Tissue Inflammation through Agglutination. Front. Immunol. 2017, 8, 1043. [Google Scholar] [CrossRef] [PubMed]
- Corthesy, B.; Monnerat, J.; Lotscher, M.; Vonarburg, C.; Schaub, A.; Bioley, G. Oral Passive Immunization with Plasma-Derived Polyreactive Secretory-Like IgA/M Partially Protects Mice Against Experimental Salmonellosis. Front. Immunol. 2018, 9, 2970. [Google Scholar] [CrossRef] [PubMed]
- Betz, K.J.; Maier, E.A.; Amarachintha, S.; Wu, D.; Karmele, E.P.; Kinder, J.M.; Steinbrecher, K.A.; McNeal, M.M.; Luzader, D.H.; Hogan, S.P.; et al. Enhanced survival following oral and systemic Salmonella enterica serovar Typhimurium infection in polymeric immunoglobulin receptor knockout mice. PLoS ONE 2018, 13, e0198434. [Google Scholar] [CrossRef] [PubMed]
- Wijburg, O.L.; Uren, T.K.; Simpfendorfer, K.; Johansen, F.E.; Brandtzaeg, P.; Strugnell, R.A. Innate secretory antibodies protect against natural Salmonella typhimurium infection. J. Exp. Med. 2006, 203, 21–26. [Google Scholar] [CrossRef]
- Uren, T.K.; Wijburg, O.L.; Simmons, C.; Johansen, F.E.; Brandtzaeg, P.; Strugnell, R.A. Vaccine-induced protection against gastrointestinal bacterial infections in the absence of secretory antibodies. Eur. J. Immunol. 2005, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zeng, X.; Dai, Q.; Hou, Y.; Zhu, D.; Wang, M.; Jia, R.; Chen, S.; Liu, M.; Yang, Q.; et al. Immunogenicity and protection efficacy of a Salmonella enterica serovar Typhimurium fnr, arcA and fliC mutant. Vaccine 2021, 39, 588–595. [Google Scholar] [CrossRef]
- Endt, K.; Stecher, B.; Chaffron, S.; Slack, E.; Tchitchek, N.; Benecke, A.; Van Maele, L.; Sirard, J.C.; Mueller, A.J.; Heikenwalder, M.; et al. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 2010, 6, e1001097. [Google Scholar] [CrossRef]
- LaRusso, N.F. Proteins in bile: How they get there and what they do. Am. J. Physiol. 1984, 247, G199–G205. [Google Scholar] [CrossRef]
- Vuitton, D.A.; Seilles, E.; Claude, P.; Sava, P.; Delacroix, D.L. Gall bladder: The predominant source of bile IgA in man? Clin. Exp. Immunol. 1985, 62, 185–192. [Google Scholar] [PubMed]
- Wu, C.T.; Davis, P.A.; Luketic, V.A.; Gershwin, M.E. A review of the physiological and immunological functions of biliary epithelial cells: Targets for primary biliary cirrhosis, primary sclerosing cholangitis and drug-induced ductopenias. Clin. Dev. Immunol. 2004, 11, 205–213. [Google Scholar] [CrossRef]
- Brown, W.R.; Kloppel, T.M. The role of the liver in translocation of IgA into the gastrointestinal tract. Immunol. Investig. 1989, 18, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Chandy, K.G.; Hubscher, S.G.; Elias, E.; Berg, J.; Khan, M.; Burnett, D. Dual role of the liver in regulating circulating polymeric IgA in man: Studies on patients with liver disease. Clin. Exp. Immunol. 1983, 52, 207–218. [Google Scholar] [PubMed]
- Sakisaka, S.; Gondo, K.; Yoshitake, M.; Harada, M.; Sata, M.; Kobayashi, K.; Tanikawa, K. Functional differences between hepatocytes and biliary epithelial cells in handling polymeric immunoglobulin A2 in humans, rats, and guinea pigs. Hepatology 1996, 24, 398–406. [Google Scholar] [CrossRef]
- Russell, M.W.; Brown, T.A.; Claflin, J.L.; Schroer, K.; Mestecky, J. Immunoglobulin A-mediated hepatobiliary transport constitutes a natural pathway for disposing of bacterial antigens. Infect. Immun. 1983, 42, 1041–1048. [Google Scholar] [CrossRef]
- Russell, M.W.; Brown, T.A.; Mestecky, J. Role of serum IgA. Hepatobiliary transport of circulating antigen. J. Exp. Med. 1981, 153, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Counihan, N.A.; Anderson, D.A. Specific IgA Enhances the Transcytosis and Excretion of Hepatitis A Virus. Sci. Rep. 2016, 6, 21855. [Google Scholar] [CrossRef] [PubMed]
- Haswell-Elkins, M.R.; Sithithaworn, P.; Mairiang, E.; Elkins, D.B.; Wongratanacheewin, S.; Kaewkes, S.; Mairiang, P. Immune responsiveness and parasite-specific antibody levels in human hepatobiliary disease associated with Opisthorchis viverrini infection. Clin. Exp. Immunol. 1991, 84, 213–218. [Google Scholar] [CrossRef]
- Jacob, C.O.; Vaerman, J.P. Induction of rat secretory IgA antibodies against cholera toxin by a synthetic peptide. Immunology 1986, 59, 129–133. [Google Scholar] [PubMed]
- Sharma, A.W.; Mayrhofer, G. Biliary antibody response in rats infected with rodent Giardia duodenalis isolates. Parasite Immunol. 1988, 10, 181–191. [Google Scholar] [CrossRef]
- Verdon, R.; Polianski, J.; Grodet, A.; Garry, L.; Carbon, C. Cryptosporidium parvum biliary tract infection in adult immunocompetent and immunosuppressed mice. J. Med. Microbiol. 1998, 47, 71–77. [Google Scholar] [CrossRef]
- Wongratanacheewin, S.; Bunnag, D.; Vaeusorn, N.; Sirisinha, S. Characterization of humoral immune response in the serum and bile of patients with opisthorchiasis and its application in immunodiagnosis. Am. J. Trop Med. Hyg. 1988, 38, 356–362. [Google Scholar] [CrossRef]
- Aagaard, B.D.; Heyworth, M.F.; Oesterle, A.L.; Jones, A.L.; Way, L.W. Intestinal immunisation with Escherichia coli protects rats against Escherichia coli induced cholangitis. Gut 1996, 39, 136–140. [Google Scholar] [CrossRef][Green Version]
- Yio, X.Y.; Jin, B.W.; Yin, F.Z.; Li, X.J. Bile secretory immunoglobulin A in biliary infection and cholelithiasis. Gastroenterology 1992, 102, 1000–1008. [Google Scholar] [CrossRef]
- James, S.P.; Jones, E.A.; Schafer, D.F.; Hoofnagle, J.H.; Varma, R.R.; Strober, W. Selective immunoglobulin A deficiency associated with primary biliary cirrhosis in a family with liver disease. Gastroenterology 1986, 90, 283–288. [Google Scholar] [CrossRef]
- Danon, Y.L.; Dinari, G.; Garty, B.Z.; Horodniceanu, C.; Nitzan, M.; Grunebaum, M. Cholelithiasis in children with immunoglobulin A deficiency: A new gastroenterologic syndrome. J. Pediatr. Gastroenterol. Nutr. 1983, 2, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Iwata, K.; Komatsu, T.; Watanabe, M.; Nishiya, H.; Kunii, O. Significance of test for antibody-coated bacteria in biliary tract infection. Jpn. J. Exp. Med. 1983, 53, 59–63. [Google Scholar] [PubMed]
- Moro-Sibilot, L.; Blanc, P.; Taillardet, M.; Bardel, E.; Couillault, C.; Boschetti, G.; Traverse-Glehen, A.; Defrance, T.; Kaiserlian, D.; Dubois, B. Mouse and Human Liver Contain Immunoglobulin A-Secreting Cells Originating From Peyer’s Patches and Directed Against Intestinal Antigens. Gastroenterology 2016, 151, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhang, Z.; Liu, B.; Hou, D.; Liang, Y.; Zhang, J.; Shi, P. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics 2013, 14, 669. [Google Scholar] [CrossRef]
- Saltykova, I.V.; Petrov, V.A.; Logacheva, M.D.; Ivanova, P.G.; Merzlikin, N.V.; Sazonov, A.E.; Ogorodova, L.M.; Brindley, P.J. Biliary Microbiota, Gallstone Disease and Infection with Opisthorchis felineus. PLoS Negl. Trop. Dis. 2016, 10, e0004809. [Google Scholar] [CrossRef]
- Petrov, V.A.; Fernandez-Peralbo, M.A.; Derks, R.; Knyazeva, E.M.; Merzlikin, N.V.; Sazonov, A.E.; Mayboroda, O.A.; Saltykova, I.V. Biliary Microbiota and Bile Acid Composition in Cholelithiasis. Biomed. Res. Int. 2020, 2020, 1242364. [Google Scholar] [CrossRef]
- Plieskatt, J.L.; Deenonpoe, R.; Mulvenna, J.P.; Krause, L.; Sripa, B.; Bethony, J.M.; Brindley, P.J. Infection with the carcinogenic liver fluke Opisthorchis viverrini modifies intestinal and biliary microbiome. FASEB J. 2013, 27, 4572–4584. [Google Scholar] [CrossRef]
- Baumann, U.; Miescher, S.; Vonarburg, C. Immunoglobulin replacement therapy in antibody deficiency syndromes: Are we really doing enough? Clin. Exp. Immunol. 2014, 178 (Suppl. 1), 83–85. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.J.; Benani, D.J. A review of blinatumomab, a novel immunotherapy. J. Oncol. Pharm. Pract. 2016, 22, 639–645. [Google Scholar] [CrossRef]
- Romero, D. Amivantamab is effective in NSCLC harbouring EGFR exon 20 insertions. Nat. Rev. Clin. Oncol 2021, 18, 604. [Google Scholar] [CrossRef] [PubMed]
- Sedykh, S.E.; Prinz, V.V.; Buneva, V.N.; Nevinsky, G.A. Bispecific antibodies: Design, therapy, perspectives. Drug Des. Devel. Ther. 2018, 12, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Deyev, S.M.; Lebedenko, E.N. Modern Technologies for Creating Synthetic Antibodies for Clinical application. Acta Naturae 2009, 1, 32–50. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Pi, X.; Jiang, Y.; Ren, G.; Liu, Z.; Liu, H.; Wang, M.; Sun, W.; Li, S.; Gao, Z.; et al. An immuno-blocking agent targeting IL-1beta and IL-17A reduces the lesion of DSS-induced ulcerative colitis in mice. Inflammation 2021, 44, 1724–1736. [Google Scholar] [CrossRef] [PubMed]
- Peyrin-Biroulet, L.; Demarest, S.; Nirula, A. Bispecific antibodies: The next generation of targeted inflammatory bowel disease therapies. Autoimmun. Rev. 2019, 18, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Vandenbroucke, K.; de Haard, H.; Beirnaert, E.; Dreier, T.; Lauwereys, M.; Huyck, L.; Van Huysse, J.; Demetter, P.; Steidler, L.; Remaut, E.; et al. Orally administered L. lactis secreting an anti-TNF Nanobody demonstrate efficacy in chronic colitis. Mucosal. Immunol. 2010, 3, 49–56. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abokor, A.A.; McDaniel, G.H.; Golonka, R.M.; Campbell, C.; Brahmandam, S.; Yeoh, B.S.; Joe, B.; Vijay-Kumar, M.; Saha, P. Immunoglobulin A, an Active Liaison for Host-Microbiota Homeostasis. Microorganisms 2021, 9, 2117. https://doi.org/10.3390/microorganisms9102117
Abokor AA, McDaniel GH, Golonka RM, Campbell C, Brahmandam S, Yeoh BS, Joe B, Vijay-Kumar M, Saha P. Immunoglobulin A, an Active Liaison for Host-Microbiota Homeostasis. Microorganisms. 2021; 9(10):2117. https://doi.org/10.3390/microorganisms9102117
Chicago/Turabian StyleAbokor, Ahmed A., Grant H. McDaniel, Rachel M. Golonka, Connor Campbell, Sreya Brahmandam, Beng San Yeoh, Bina Joe, Matam Vijay-Kumar, and Piu Saha. 2021. "Immunoglobulin A, an Active Liaison for Host-Microbiota Homeostasis" Microorganisms 9, no. 10: 2117. https://doi.org/10.3390/microorganisms9102117
APA StyleAbokor, A. A., McDaniel, G. H., Golonka, R. M., Campbell, C., Brahmandam, S., Yeoh, B. S., Joe, B., Vijay-Kumar, M., & Saha, P. (2021). Immunoglobulin A, an Active Liaison for Host-Microbiota Homeostasis. Microorganisms, 9(10), 2117. https://doi.org/10.3390/microorganisms9102117






