Dandelion (Taraxacum mongolicum Hand.-Mazz.) Supplementation-Enhanced Rumen Fermentation through the Interaction between Ruminal Microbiome and Metabolome
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Diet, and Experimental Design
2.2. Samples Collection and Measurement
2.3. Bioactive Compounds Certification of Dandelion
2.4. DNA Extraction and Sequencing
2.5. Metabolomics Analysis
2.6. Correlation and Statistical Analysis
3. Results
3.1. Chemical Composition of Dandelion
3.2. Rumen Fermentation Characteristics
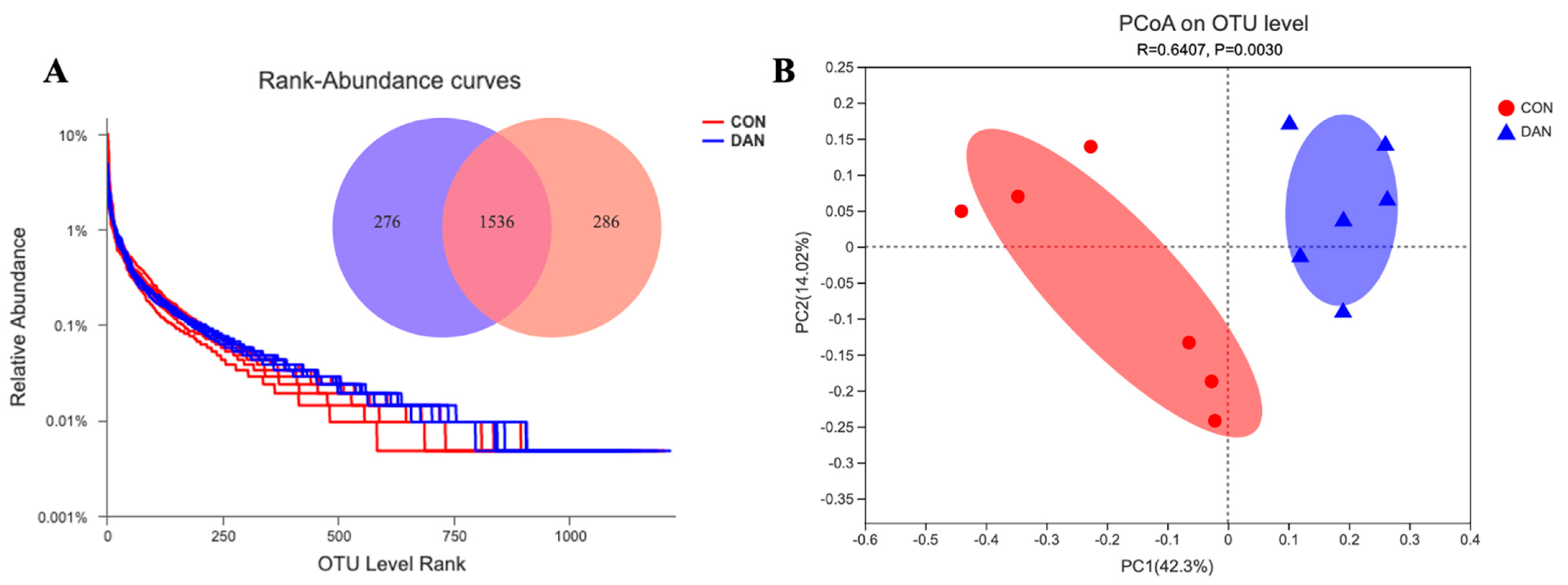
3.3. Changes in Ruminal Bacterial Communities
3.4. Correlation Analysis between the Ruminal Microbiome and Fermentation Parameters
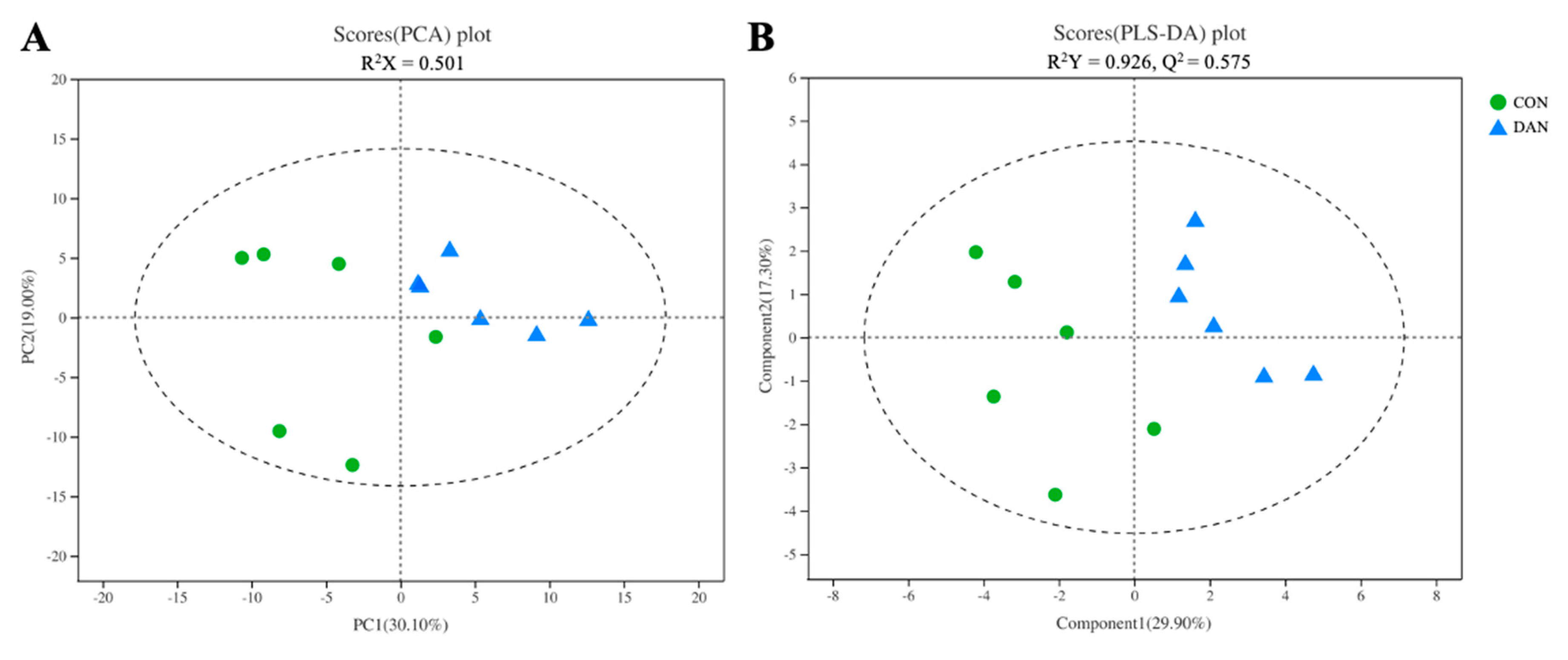
3.5. GC/MS Analysis of the Ruminal Fluid
3.6. Different Metabolites between CON and DAN Groups in Rumen Fluid
3.7. Characterization of Metabolic Pathways
3.8. Correlation Analysis between the Ruminal Metabolome and Microbiome
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- González-Castejón, M.; Visioli, F.; Rodriguez-Casado, A. Diverse biological activities of dandelion. Nutr. Rev. 2012, 70, 534–547. [Google Scholar] [CrossRef] [PubMed]
- Schütz, K.; Carle, R.; Schieber, A. Taraxacum—A review on its phytochemical and pharmacological profile. J. Ethnopharmacol. 2006, 107, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Kamra, D.N.; Patra, A.K.; Chatterjee, P.N.; Kumar, R.; Agarwal, N.; Chaudhary, L.C. Effect of plant extracts on methanogenesis and microbial profile of the rumen of buffalo: A brief overview. Aust. J. Exp. Agric. 2008, 48, 175–178. [Google Scholar] [CrossRef]
- Hristov, A.N.; Oh, J.; Firkins, J.L.; Dijkstra, J.; Kebreab, E.; Waghorn, G.; Makkar, H.P.S.; Adesogan, A.T.; Yang, W.; Lee, C.; et al. SPECIAL TOPICS—Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. J. Anim. Sci. 2013, 91, 5045–5069. [Google Scholar] [CrossRef] [PubMed]
- Mertenat, D.; Cero, M.D.; Vogl, C.R.; Ivemeyer, S.; Meier, B.; Maeschli, A.; Hamburger, M.; Walkenhorst, M. Ethnoveterinary knowledge of farmers in bilingual regions of Switzerland—Is there potential to extend veterinary options to reduce antimicrobial use? J. Ethnopharmacol. 2019, 246, 112184. [Google Scholar] [CrossRef] [PubMed]
- Huws, S.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing Global Ruminant Agricultural Challenges through Understanding the Rumen Microbiome: Past, Present, and Future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef]
- Sun, Z.; Yu, Z.; Wang, B. Perilla frutescens Leaf Alters the Rumen Microbial Community of Lactating Dairy Cows. Microorganisms 2019, 7, 562. [Google Scholar] [CrossRef]
- Wang, B.; Ma, M.P.; Diao, Q.Y.; Tu, Y. Saponin-Induced Shifts in the Rumen Microbiome and Metabolome of Young Cattle. Front. Microbiol. 2019, 10, 356. [Google Scholar] [CrossRef]
- Xue, M.-Y.; Sun, H.-Z.; Wu, X.-H.; Liu, J.; Guan, L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 1–19. [Google Scholar] [CrossRef]
- Mackie, R.I.; White, B.A. Recent advances in rumen microbial ecology and metabolism: Potential impact on nutrient output. J. Dairy Sci. 1990, 73, 2971–2995. [Google Scholar] [CrossRef]
- Shen, J.; Chai, Z.; Song, L.; Liu, J.; Wu, Y. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J. Dairy Sci. 2012, 95, 5978–5984. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.L.; Liu, J.X.; Ye, J.A.; Wu, Y.M.; Guo, Y.Q. Effect of tea saponin on rumen fermentation in vitro. Anim. Feed Sci. Technol. 2005, 120, 333–339. [Google Scholar] [CrossRef]
- Broderick, G.; Kang, J. Automated Simultaneous Determination of Ammonia and Total Amino Acids in Ruminal Fluid and In Vitro Media. J. Dairy Sci. 1980, 63, 64–75. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, Z.F.; Park, J.C.; Kim, I.H. Evaluation of Houttuynia cordata and Taraxacum officinale on Growth Performance, Nutrient Digestibility, Blood Characteristics, and Fecal Microbial Shedding in Diet for Weaning Pigs. Asian Australas. J. Anim. Sci. 2012, 25, 1439–1444. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Sun, Z.; Zhou, C.; Huang, Z.; Tan, L.; Xun, P.; Huang, Q.; Lin, H.; Ye, C.; Wang, A. Effects of dietary dandelion extract on intestinal morphology, antioxidant status, immune function and physical barrier function of juvenile golden pompano Trachinotus ovatus. Fish Shellfish Immunol. 2018, 73, 197–206. [Google Scholar] [CrossRef]
- Chabrol, E. L’action choléretique des Compositées. CR Soc. Biol. 1931, 108, 1100–1102. [Google Scholar]
- Blumenhal, M.; Busse, N.R.; Golddberg, A. The Complete Commission E. Monographs Therapeutic Guide to Herbal Medicine; Integrative Medicines Communications: Boston, MA, USA, 1998; pp. 80–81. [Google Scholar]
- Faber, K. The dandelion Taraxacum officinale Weber. Die Pharm. 1958, 13, 423. [Google Scholar]
- Chakŭrski, I.; Matev, M.; Koĭchev, A.; Angelova, I.; Stefanov, G. Treatment of chronic colitis with an herbal combination of Taraxacum officinale, Hipericum perforatum, Melissa officinaliss, Calendula officinalis and Foeniculum vulgare. Vutreshni Boles. 1981, 20, 51–54. [Google Scholar]
- Xue, M.; Sun, H.; Wu, X.; Guan, L.; Liu, J. Assessment of rumen bacteria in dairy cows with varied milk protein yield. J. Dairy Sci. 2019, 102, 5031–5041. [Google Scholar] [CrossRef]
- Zened, A.; Combes, S.; Cauquil, L.; Mariette, J.; Klopp, C.; Bouchez, O.; Troegeler-Meynadier, A.; Enjalbert, F. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol. Ecol. 2013, 83, 504–514. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; White, B.A.; Mizrahi, I. Potential Role of the Bovine Rumen Microbiome in Modulating Milk Composition and Feed Efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Saito, T.; Ohshima, T.; Nakagawa, Y.; Arita, T.; Yashima, A.; Makino, T.; Konnai, R.; Gomi, K.; Arai, T.; et al. Terminal RFLP analysis to determine the oral microbiota with hyposalivation. Arch. Microbiol. 2014, 196, 489–496. [Google Scholar] [CrossRef]
- Ahmad, A.A.; Yang, C.; Zhang, J.; Kalwar, Q.; Liang, Z.; Li, C.; Du, M.; Yan, P.; Long, R.; Han, J.; et al. Effects of Dietary Energy Levels on Rumen Fermentation, Microbial Diversity, and Feed Efficiency of Yaks (Bos grunniens). Front. Microbiol. 2020, 11, 625. [Google Scholar] [CrossRef]
- Krause, D.O.; Denman, S.E.; Mackie, R.I.; Morrison, M.; Rae, A.L.; Attwood, G.T.; McSweeney, C.S. Opportunities to improve fiber degradation in the rumen: Microbiology, ecology, and genomics. FEMS Microbiol. Rev. 2003, 27, 663–693. [Google Scholar] [CrossRef]
- Christopherson, M.R.; Dawson, J.A.; Stevenson, D.; Cunningham, A.C.; Bramhacharya, S.; Weimer, P.J.; Kendziorski, C.; Suen, G. Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis. BMC Genom. 2014, 15, 1066. [Google Scholar] [CrossRef]
- Kim, M.; Kim, J.; Kuehn, L.A.; Bono, J.L.; Berry, E.D.; Kalchayanand, N.; Freetly, H.C.; Benson, A.K.; Wells, J. Investigation of bacterial diversity in the feces of cattle fed different diets. J. Anim. Sci. 2014, 92, 683–694. [Google Scholar] [CrossRef]
- Chen, J.; Huang, C.; Wang, J.; Zhou, H.; Lu, Y.; Lou, L.; Zheng, J.; Tian, L.; Wang, X.; Cao, Z.; et al. Dysbiosis of intestinal microbiota and decrease in paneth cell antimicrobial peptide level during acute necrotizing pancreatitis in rats. PLoS ONE 2017, 12, e0176583. [Google Scholar] [CrossRef]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; Teh, K.H.; Lambie, S.C.; Cookson, A.L.; A Eloe-Fadrosh, E.; A Pavlopoulos, G.; Hadjithomas, M.; Varghese, N.J.; et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat. Biotechnol. 2018, 36, 359–367. [Google Scholar] [CrossRef]
- He, B.; Nohara, K.; Ajami, N.J.; Michalek, R.D.; Tian, X.; Wong, M.; Losee-Olson, S.H.; Petrosino, J.F.; Yoo, S.-H.; Shimomura, K.; et al. Transmissible microbial and metabolomic remodeling by soluble dietary fiber improves metabolic homeostasis. Sci. Rep. 2015, 5, 10604. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Huang, S.; Liu, T.; Geng, A. Bacterial xylose isomerases from the mammal gut Bacteroidetes cluster function in Saccharomyces cerevisiae for effective xylose fermentation. Microb. Cell Fact. 2015, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Grassian, A.R.; Parker, S.J.; Davidson, S.M.; Divakaruni, A.S.; Green, C.R.; Zhang, X.; Slocum, K.L.; Pu, M.; Lin, F.; Vickers, C.; et al. IDH1 Mutations Alter Citric Acid Cycle Metabolism and Increase Dependence on Oxidative Mitochondrial Metabolism. Cancer Res. 2014, 74, 3317–3331. [Google Scholar] [CrossRef] [PubMed]
- Antunes-Fernandes, E.; Van Gastelen, S.; Dijkstra, J.; Hettinga, K.A.; Vervoort, J. Milk metabolome relates enteric methane emission to milk synthesis and energy metabolism pathways. J. Dairy Sci. 2016, 99, 6251–6262. [Google Scholar] [CrossRef]
- Patra, K.C.; Hay, N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef]
- Stull, M.A.; Pai, V.; Vomachka, A.J.; Marshall, A.M.; Jacob, G.A.; Horseman, N.D. Mammary gland homeostasis employs serotonergic regulation of epithelial tight junctions. Proc. Natl. Acad. Sci. USA 2007, 104, 16708–16713. [Google Scholar] [CrossRef]
- Plaza, M.; Arruebo, M.P.; Murillo, M.D. 5-Hydroxytryptamine induces forestomach hypomotility in sheep through 5-HT4 receptors. Exp. Physiol. 1996, 81, 781–790. [Google Scholar] [CrossRef]
- Mao, S.; Huo, W.-J.; Zhu, W. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 2016, 18, 525–541. [Google Scholar] [CrossRef]
- Soest, P.J.V. Nutritional Ecology of the Ruminant; Cornell University Press: Ithaca, NY, USA, 1994; Volume 44, pp. 2552–2561. [Google Scholar]
- Solden, L.M.; Naas, A.E.; Roux, S.; Daly, R.A.; Collins, W.B.; Nicora, C.D.; Purvine, S.O.; Hoyt, D.W.; Schückel, J.; Jørgensen, B.; et al. Interspecies cross-feeding orchestrates carbon degradation in the rumen ecosystem. Nat. Microbiol. 2018, 3, 1274–1284. [Google Scholar] [CrossRef]





| Ingredient | % of DM 1 | Chemical Composition | % of DM |
|---|---|---|---|
| Corn silage | 30.36 | CP | 16.68 |
| Alfalfa hay | 13.68 | NDF | 37.89 |
| Corn grain | 12.70 | ADF | 19.81 |
| Orts hay | 3.26 | EE | 2.92 |
| Cotton seed | 3.26 | Ash | 7.22 |
| Beet pulp | 5.54 | NEL, Mcal/kg of DM | 1.68 |
| Soybean meal | 9.12 | ||
| DDGS 2 | 4.33 | ||
| Flaked corn | 9.77 | ||
| Rapeseed meal | 2.93 | ||
| Extruded soybean | 1.63 | ||
| Concentrate 3 | 3.42 |
| Bioactive Compounds | Proportion, % | Bioactive Compounds | Proportion, % |
|---|---|---|---|
| β-d-Glucopyranoside | 12.32 | Myristic acid | 0.42 |
| Caffeic acid | 9.57 | Tartaric acid | 0.38 |
| 7,8-Dihydroxyflavone | 8.61 | Methylsuccinic acid | 0.38 |
| Luteolin-4’-O-glucoside | 7.96 | Methyl palmitoleate | 0.27 |
| Rutin | 7.62 | Heptadecanoic acid | 0.16 |
| 9Z,11E-Linoleic acid | 5.89 | L-Valine | 0.11 |
| Choline | 5.60 | 2-Hexenal | 0.09 |
| Proline | 5.36 | Palmitic acid | 0.08 |
| Trigonelline HCl | 4.11 | Piperidine | 0.08 |
| Malic acid | 3.88 | l-Phenylalanine | 0.07 |
| Valine | 3.78 | Sucrose | 0.05 |
| Trans-Vaccenic acid | 3.32 | l-Glutamic Acid | 0.04 |
| Phenylalanine | 2.94 | Chlorogenic acid | 0.04 |
| Methyl vanillate | 2.65 | Aspartate | 0.04 |
| Quercetin | 2.58 | Nicotinamide | 0.04 |
| Stachydrine | 1.75 | Dehydrocostus lactone | 0.04 |
| l-Isoleucine | 1.63 | Phthalic anhydride | 0.04 |
| 9-Trans-Palmitelaidic acid | 1.54 | l-Carnitine | 0.04 |
| d-Gluconic acid | 1.53 | Isoimperatorin | 0.03 |
| Citrate | 1.28 | Tyrosine | 0.03 |
| Methyl Heptadecanoic acid | 0.98 | Formononetine | 0.03 |
| Mannitol | 0.86 | l-Theanine | 0.02 |
| Adenosine | 0.64 | Sarracenin | 0.02 |
| Nicotinic acid | 0.55 | p-Coumaraldehyde | 0.01 |
| l-Tryptophan | 0.53 | Decanoic acid | 0.01 |
| Items | Treatments | SEM | p-Value | |
|---|---|---|---|---|
| CON | DAN | |||
| pH | 6.85 | 6.73 | 0.010 | 0.92 |
| Total VFA (mM) | 93.78 | 99.65 | 1.982 | 0.09 |
| Acetate (mM) | 60.08 | 68.23 | 1.294 | 0.04 |
| Propionate (mM) | 19.10 | 18.93 | 0.812 | 0.85 |
| Butyrate (mM) | 7.16 | 9.24 | 0.237 | 0.05 |
| Valerate (mM) | 1.79 | 1.85 | 0.051 | 0.39 |
| Isobutyrate (mM) | 0.62 | 0.69 | 0.042 | 0.09 |
| Isovalerate (mM) | 1.59 | 1.66 | 0.026 | 0.08 |
| NH3-N (mg/dL) | 10.74 | 14.30 | 0.841 | 0.03 |
| AA/PA 1 | 3.14 | 3.60 | 0.103 | 0.07 |
| Items | Treatments | SEM | p-Value | |
|---|---|---|---|---|
| CON | DAN | |||
| Sobs | 1039 | 1156 | 31.1 | 0.06 |
| Ace | 1313 | 1424 | 31.4 | 0.07 |
| Chao | 1321 | 1454 | 30.6 | 0.02 |
| Shannon | 5.40 | 5.66 | 0.089 | 0.16 |
| Simpson | 0.01 | 0.01 | 0.001 | 0.15 |
| Coverage | 0.99 | 0.98 | 0.001 | 0.25 |
| Metabolites | Similarity | VIP 1 | FC 2 | p-Value |
|---|---|---|---|---|
| Ribulose-5-phosphate | 99.40 | 1.66 | 1.96 | 0.02 |
| Glycerate | 99.00 | 1.30 | 1.74 | 0.05 |
| d-glucose | 99.90 | 1.30 | 1.72 | 0.02 |
| 3-(Methylthio)-propylamine | 72.60 | 2.54 | 1.11 | <0.01 |
| Trans caftaric acid | 62.10 | 2.26 | 1.09 | 0.01 |
| Psicose | 91.50 | 2.24 | 1.08 | 0.01 |
| β-ionone | 78.90 | 1.91 | 1.07 | 0.01 |
| Acetanilide | 73.90 | 1.76 | 1.06 | <0.01 |
| Galactosamine | 98.90 | 1.65 | 1.05 | 0.01 |
| Benzoylformic acid | 80.40 | 1.40 | 1.05 | 0.04 |
| Threonic acid | 98.60 | 1.45 | 1.04 | 0.02 |
| Xylose | 88.80 | 1.35 | 1.04 | 0.04 |
| Phenanthrene | 61.10 | 1.26 | 1.04 | 0.02 |
| Arabinose | 99.90 | 1.40 | 1.03 | 0.04 |
| l-threonine | 98.10 | 1.48 | 1.03 | <0.01 |
| Methyl-β-d-galactopyranoside | 95.20 | 1.39 | 1.03 | 0.04 |
| Fructose-1,6-diphosphate | 77.90 | 1.11 | 1.03 | 0.04 |
| Hydrocinnamic acid | 99.30 | 1.13 | 1.02 | 0.01 |
| 2,3-Dihydroxybiphenyl | 96.10 | 1.30 | 1.02 | 0.03 |
| 2-Aminoethanethiol | 68.20 | 1.04 | 1.02 | <0.01 |
| Palmitic acid | 98.80 | 1.00 | 0.99 | 0.03 |
| Antiarol | 74.50 | 1.22 | 0.98 | 0.01 |
| l-(+) lactic acid | 98.00 | 1.44 | 0.97 | <0.01 |
| 1,3-Propanediol | 95.90 | 1.67 | 0.97 | <0.01 |
| 2-Methoxyxanthone | 64.50 | 1.31 | 0.97 | <0.01 |
| 3-Hydroxyflavone | 82.20 | 1.77 | 0.96 | <0.01 |
| Vanillin | 70.50 | 1.34 | 0.96 | <0.01 |
| l-mimosine | 69.50 | 1.47 | 0.96 | 0.01 |
| N-methylalanine | 88.70 | 1.82 | 0.95 | <0.01 |
| 5,6-Dihydro-5-methyluracil | 79.90 | 1.67 | 0.95 | 0.03 |
| Acetohydroxamic acid | 95.60 | 1.94 | 0.94 | 0.01 |
| Quinic acid | 68.00 | 1.77 | 0.94 | 0.01 |
| N-omega-acetylhistamine | 68.50 | 1.92 | 0.93 | <0.01 |
| Phosphoric acid | 69.20 | 2.14 | 0.92 | 0.01 |
| Serotonin | 98.40 | 1.40 | 0.67 | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Lv, M.; Wang, J.; Tian, Z.; Yu, B.; Wang, B.; Liu, J.; Liu, H. Dandelion (Taraxacum mongolicum Hand.-Mazz.) Supplementation-Enhanced Rumen Fermentation through the Interaction between Ruminal Microbiome and Metabolome. Microorganisms 2021, 9, 83. https://doi.org/10.3390/microorganisms9010083
Li Y, Lv M, Wang J, Tian Z, Yu B, Wang B, Liu J, Liu H. Dandelion (Taraxacum mongolicum Hand.-Mazz.) Supplementation-Enhanced Rumen Fermentation through the Interaction between Ruminal Microbiome and Metabolome. Microorganisms. 2021; 9(1):83. https://doi.org/10.3390/microorganisms9010083
Chicago/Turabian StyleLi, Yan, Mei Lv, Jiaqi Wang, Zhonghong Tian, Bo Yu, Bing Wang, Jianxin Liu, and Hongyun Liu. 2021. "Dandelion (Taraxacum mongolicum Hand.-Mazz.) Supplementation-Enhanced Rumen Fermentation through the Interaction between Ruminal Microbiome and Metabolome" Microorganisms 9, no. 1: 83. https://doi.org/10.3390/microorganisms9010083
APA StyleLi, Y., Lv, M., Wang, J., Tian, Z., Yu, B., Wang, B., Liu, J., & Liu, H. (2021). Dandelion (Taraxacum mongolicum Hand.-Mazz.) Supplementation-Enhanced Rumen Fermentation through the Interaction between Ruminal Microbiome and Metabolome. Microorganisms, 9(1), 83. https://doi.org/10.3390/microorganisms9010083







