Regulation of Serum Exosomal MicroRNAs in Mice Infected with Orientia tsutsugamushi
Abstract
1. Introduction
2. Materials and Methods
2.1. Mouse Inoculation and Sample Collection
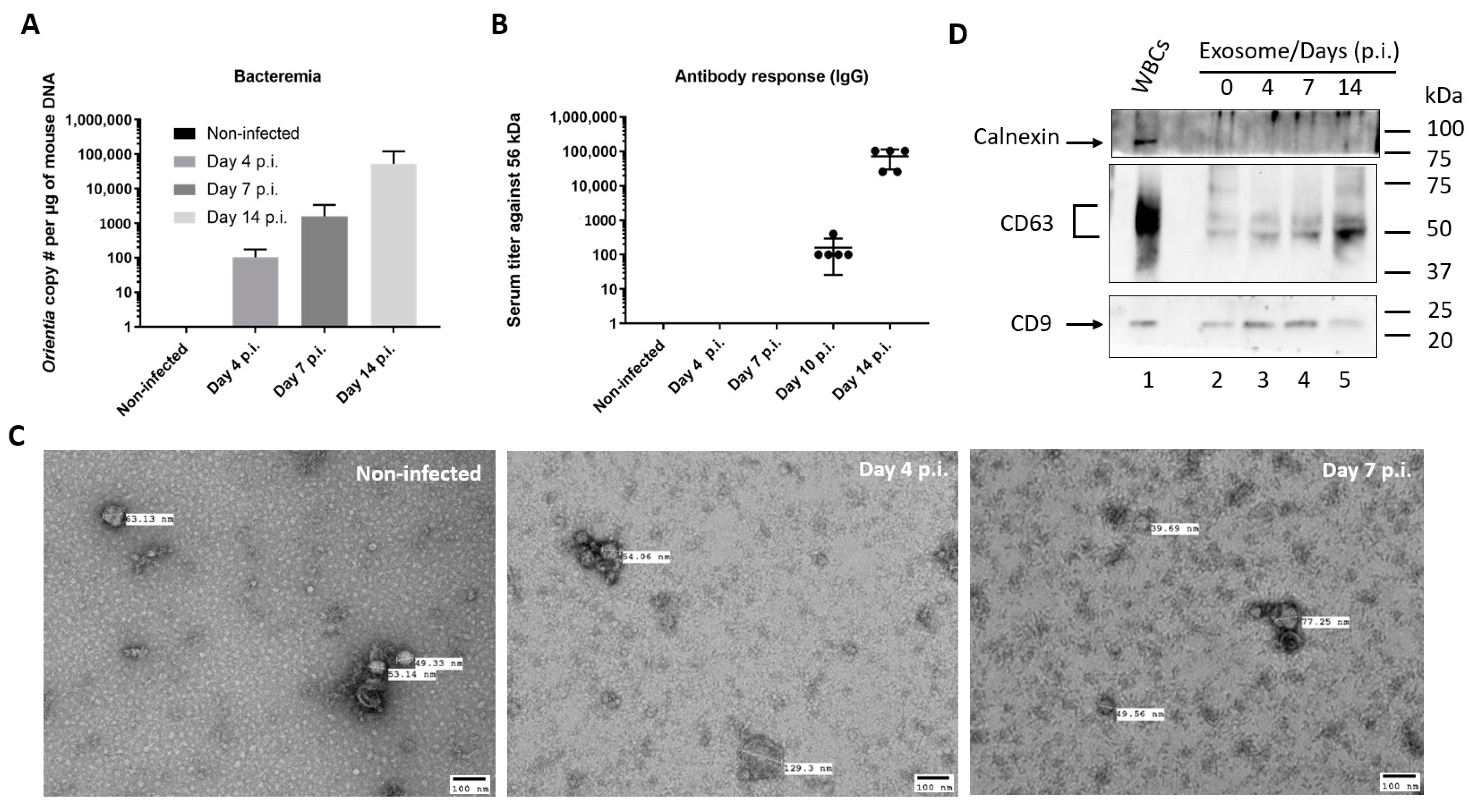
2.2. Determination of Bacteremia and Antibody Titer
2.3. Isolation of Exosomes
2.4. Characterization of Exosomes by Electron Microscopy and Western Blot
2.5. Nanoparticle Tracking Analysis
2.6. MicroRNA Profiling
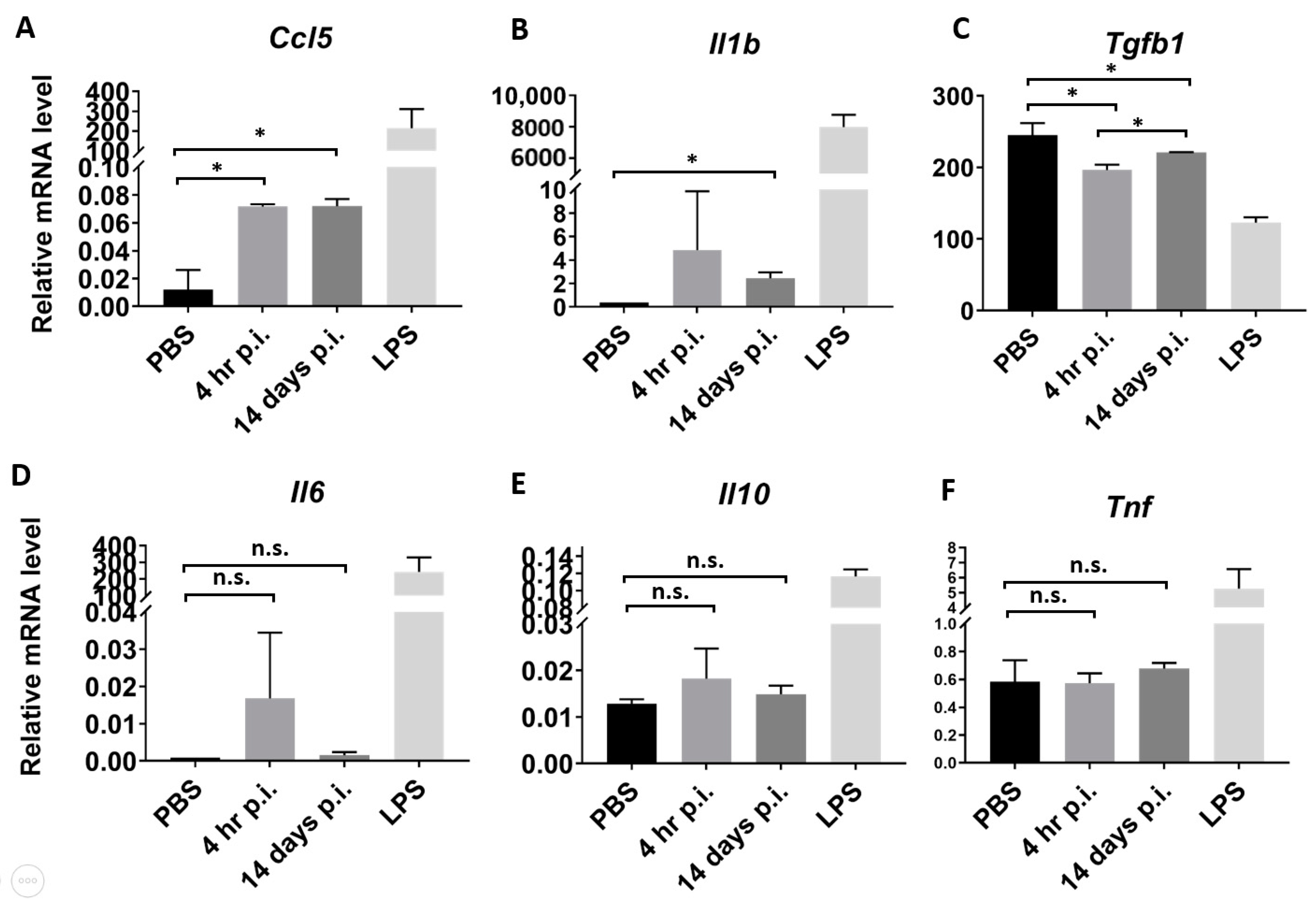
2.7. Treatment of Bone-Marrow Derived Macrophages and mRNA Quantification
2.8. Statistical Analysis
3. Results
3.1. Characterization of Serum Exosomes Isolated from Orientia Tsutsugamushi-Infected Mice
3.2. Profile of Serum Exosomal microRNAs
3.3. Serum Exosomal microRNAs Are Regulated during Orientia Tsutsugamushi Infection
3.4. Members of Let-7 Family of microRNAs Are Dynamically Regulated during Orientia Tsutsugamushi Infection
3.5. Serum Exosomes from Orientia Tsutsugamushi-Infected Mice Are Proinflammatory
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Watt, G.; Parola, P. Scrub typhus and tropical rickettsioses. Curr. Opin. Infect. Dis. 2003, 16, 429–436. [Google Scholar] [CrossRef]
- Jiang, J.; Richards, A.L. Scrub Typhus: No Longer Restricted to the Tsutsugamushi Triangle. Trop. Med. Infect. Dis. 2018, 3, 11. [Google Scholar] [CrossRef]
- Paris, D.H.; Shelite, T.R.; Day, N.P.; Walker, D.H. Unresolved problems related to scrub typhus: A seriously neglected life-threatening disease. Am. J. Trop. Med. Hyg. 2013, 89, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Paris, D.H.; Phetsouvanh, R.; Tanganuchitcharnchai, A.; Jones, M.; Jenjaroen, K.; Vongsouvath, M.; Ferguson, D.P.; Blacksell, S.D.; Newton, P.N.; Day, N.P.; et al. Orientia tsutsugamushi in human scrub typhus eschars shows tropism for dendritic cells and monocytes rather than endothelium. PLoS Negl. Trop. Dis. 2012, 6, e1466. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, M.; Fukazawa, M.; Tamura, A.; Nakamura, T.; Urakami, H. Survival of two Orientia tsutsugamushi bacterial strains that infect mouse macrophages with varying degrees of virulence. Microb. Pathog. 2005, 39, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Cheong, T.C.; Ha, N.Y.; Ko, Y.; Cho, C.H.; Jeon, J.H.; So, I.; Kim, I.K.; Choi, M.S.; Kim, I.S.; et al. Orientia tsutsugamushi subverts dendritic cell functions by escaping from autophagy and impairing their migration. PLoS Negl. Trop. Dis. 2013, 7, e1981. [Google Scholar] [CrossRef] [PubMed]
- Hauptmann, M.; Kolbaum, J.; Lilla, S.; Wozniak, D.; Gharaibeh, M.; Fleischer, B.; Keller, C.A. Protective and Pathogenic Roles of CD8+ T Lymphocytes in Murine Orientia tsutsugamushi Infection. PLoS Negl. Trop. Dis. 2016, 10, e0004991. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Okoye, I.S.; Coomes, S.M.; Pelly, V.S.; Czieso, S.; Papayannopoulos, V.; Tolmachova, T.; Seabra, M.C.; Wilson, M.S. MicroRNA-Containing T-Regulatory-Cell-Derived Exosomes Suppress Pathogenic T Helper 1 Cells. Immunity 2014, 41, 503. [Google Scholar] [CrossRef]
- Wahlund, C.J.E.; Gucluler, G.; Hiltbrunner, S.; Veerman, R.E.; Naslund, T.I.; Gabrielsson, S. Exosomes from antigen-pulsed dendritic cells induce stronger antigen-specific immune responses than microvesicles in vivo. Sci. Rep. 2017, 7, 17095. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, X.; Bao, J.; Wang, Y.; Liu, H.; Tang, L. Exosomes in Pathogen Infections: A Bridge to Deliver Molecules and Link Functions. Front. Immunol. 2018, 9, 90. [Google Scholar] [CrossRef]
- Ni, Y.S.; Chan, T.C.; Chao, C.C.; Richards, A.L.; Dasch, G.A.; Ching, W.M. Protection against scrub typhus by a plasmid vaccine encoding the 56-KD outer membrane protein antigen gene. Am. J. Trop. Med. Hyg. 2005, 73, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.C.; Jiang, J.; Temenak, J.J.; Richards, A.L. Development of a rapid method for determining the infectious dose (ID)50 of Orientia tsutsugamushi in a scrub typhus mouse model for the evaluation of vaccine candidates. Vaccine 2003, 21, 4550–4554. [Google Scholar] [CrossRef]
- Jiang, J.; Chan, T.C.; Temenak, J.J.; Dasch, G.A.; Ching, W.M.; Richards, A.L. Development of a quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi. Am. J. Trop. Med. Hyg. 2004, 70, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.C.; Belinskaya, T.; Zhang, Z.; Jiang, L.; Ching, W.M. Assessment of a Sensitive qPCR Assay Targeting a Multiple-Copy Gene to Detect Orientia tsutsugamushi DNA. Trop. Med. Infect. Dis. 2019, 4, 113. [Google Scholar] [CrossRef] [PubMed]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef]
- Zhou, X.; Jiao, Z.; Ji, J.; Li, S.; Huang, X.; Lu, X.; Zhao, H.; Peng, J.; Chen, X.; Ji, Q.; et al. Characterization of mouse serum exosomal small RNA content: The origins and their roles in modulating inflammatory response. Oncotarget 2017, 8, 42712–42727. [Google Scholar] [CrossRef]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Zhang, X.; Goncalves, R.; Mosser, D.M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 2008. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.W.; Zhang, Z.; Huber, E.; Mutumanje, E.; Chao, C.C.; Ching, W.M. Kinetics and magnitude of antibody responses against the conserved 47-kilodalton antigen and the variable 56-kilodalton antigen in scrub typhus patients. Clin. Vaccine Immunol. 2011, 18, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.; Hu, R.; Runtsch, M.C.; Kagele, D.A.; Mosbruger, T.L.; Tolmachova, T.; Seabra, M.C.; Round, J.L.; Ward, D.M.; O’Connell, R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015, 6, 7321. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.; Sampey, G.; Chung, M.C.; Bailey, C.; van Hoek, M.L.; Kashanchi, F.; Hakami, R.M. The carrying pigeons of the cell: Exosomes and their role in infectious diseases caused by human pathogens. Pathog. Dis. 2014, 71, 109–120. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Teng, G.G.; Wang, W.H.; Dai, Y.; Wang, S.J.; Chu, Y.X.; Li, J. Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting Toll-like receptor 4. PLoS ONE 2013, 8, e56709. [Google Scholar] [CrossRef]
- Schulte, L.N.; Eulalio, A.; Mollenkopf, H.J.; Reinhardt, R.; Vogel, J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011, 30, 1977–1989. [Google Scholar] [CrossRef]
- Kumar, M.; Sahu, S.K.; Kumar, R.; Subuddhi, A.; Maji, R.K.; Jana, K.; Gupta, P.; Raffetseder, J.; Lerm, M.; Ghosh, Z.; et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappaB pathway. Cell Host Microbe 2015, 17, 345–356. [Google Scholar] [CrossRef]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef]
- Jiang, S.; Yan, W.; Wang, S.E.; Baltimore, D. Dual mechanisms of posttranscriptional regulation of Tet2 by Let-7 microRNA in macrophages. Proc. Natl. Acad. Sci. USA 2019, 116, 12416–12421. [Google Scholar] [CrossRef]
- Arango Duque, G.; Descoteaux, A. Macrophage cytokines: Involvement in immunity and infectious diseases. Front. Immunol. 2014, 5, 491. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Shinagawa, K.; Castellino, F.J.; Schorey, J.S. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood 2007, 110, 3234–3244. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Kwon, H.Y.; Im, J.H.; Baek, J.H.; Kang, J.S.; Lee, J.S. Identification of Outer Membrane Vesicles Derived from Orientia tsutsugamushi. J. Korean Med. Sci. 2015, 30, 866–870. [Google Scholar] [CrossRef] [PubMed]
- Nik Mohamed Kamal, N.; Shahidan, W.N.S. Non-Exosomal and Exosomal Circulatory MicroRNAs: Which Are More Valid as Biomarkers? Front. Pharmacol. 2019, 10, 1500. [Google Scholar] [CrossRef] [PubMed]
- Palviainen, M.; Saraswat, M.; Varga, Z.; Kitka, D.; Neuvonen, M.; Puhka, M.; Joenvaara, S.; Renkonen, R.; Nieuwland, R.; Takatalo, M.; et al. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo-Implications for biomarker discovery. PLoS ONE 2020, 15, e0236439. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.J.; Gray, W.D.; Hayek, S.S.; Ko, Y.A.; Thomas, S.; Rooney, K.; Awad, M.; Roback, J.D.; Quyyumi, A.; Searles, C.D. Platelets confound the measurement of extracellular miRNA in archived plasma. Sci. Rep. 2016, 6, 32651. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Jiang, S. Recent findings regarding let-7 in immunity. Cancer Lett. 2018, 434, 130–131. [Google Scholar] [CrossRef]
- Swaminathan, S.; Suzuki, K.; Seddiki, N.; Kaplan, W.; Cowley, M.J.; Hood, C.L.; Clancy, J.L.; Murray, D.D.; Mendez, C.; Gelgor, L.; et al. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J. Immunol. 2012, 188, 6238–6246. [Google Scholar] [CrossRef]
- Chao, C.C.; Yang, R.; Zhang, Z.; Belinskaya, T.; Chan, C.T.; Miller, S.A.; Hammamieh, R.; Jett, M.; Ching, W.M. Temporal analysis of mRNA expression profiles in Orientia infected C3HeB/FeJ mouse. BMC Microbiol. 2020, 20, 3. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Belinskaya, T.; Zhang, Z.; Chan, T.-C.; Ching, W.-M.; Chao, C.-C. Regulation of Serum Exosomal MicroRNAs in Mice Infected with Orientia tsutsugamushi. Microorganisms 2021, 9, 80. https://doi.org/10.3390/microorganisms9010080
Jiang L, Belinskaya T, Zhang Z, Chan T-C, Ching W-M, Chao C-C. Regulation of Serum Exosomal MicroRNAs in Mice Infected with Orientia tsutsugamushi. Microorganisms. 2021; 9(1):80. https://doi.org/10.3390/microorganisms9010080
Chicago/Turabian StyleJiang, Le, Tatyana Belinskaya, Zhiwen Zhang, Teik-Chye Chan, Wei-Mei Ching, and Chien-Chung Chao. 2021. "Regulation of Serum Exosomal MicroRNAs in Mice Infected with Orientia tsutsugamushi" Microorganisms 9, no. 1: 80. https://doi.org/10.3390/microorganisms9010080
APA StyleJiang, L., Belinskaya, T., Zhang, Z., Chan, T.-C., Ching, W.-M., & Chao, C.-C. (2021). Regulation of Serum Exosomal MicroRNAs in Mice Infected with Orientia tsutsugamushi. Microorganisms, 9(1), 80. https://doi.org/10.3390/microorganisms9010080



