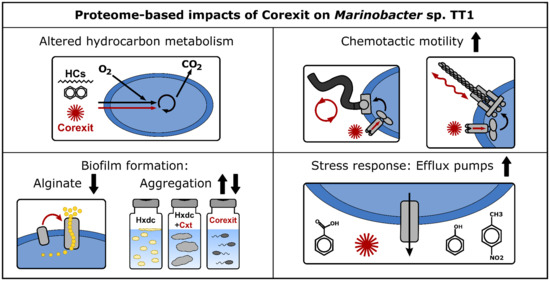
Comparative Proteomics of Marinobacter sp. TT1 Reveals Corexit Impacts on Hydrocarbon Metabolism, Chemotactic Motility, and Biofilm Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Culture Conditions
2.2. Experimental Design
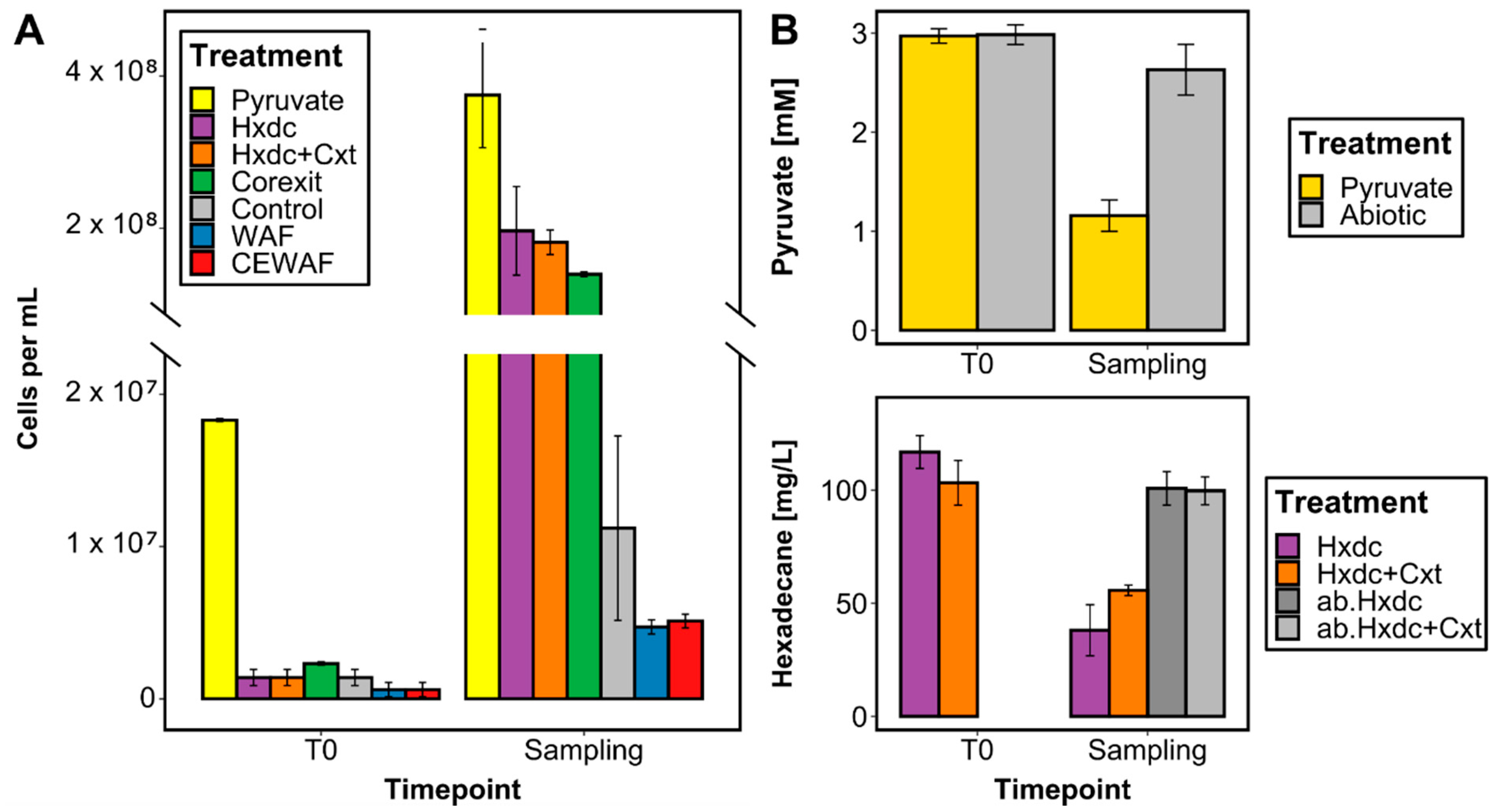
2.3. Cell Counts
2.4. Quantification of Pyruvate and Hydrocarbon Concentrations
2.5. Protein Extraction and Proteome Analysis
2.6. Data Analysis
3. Results
3.1. Growth and Substrate Utilization of Marinobacter sp. TT1
3.2. Overview of Proteomic Analysis
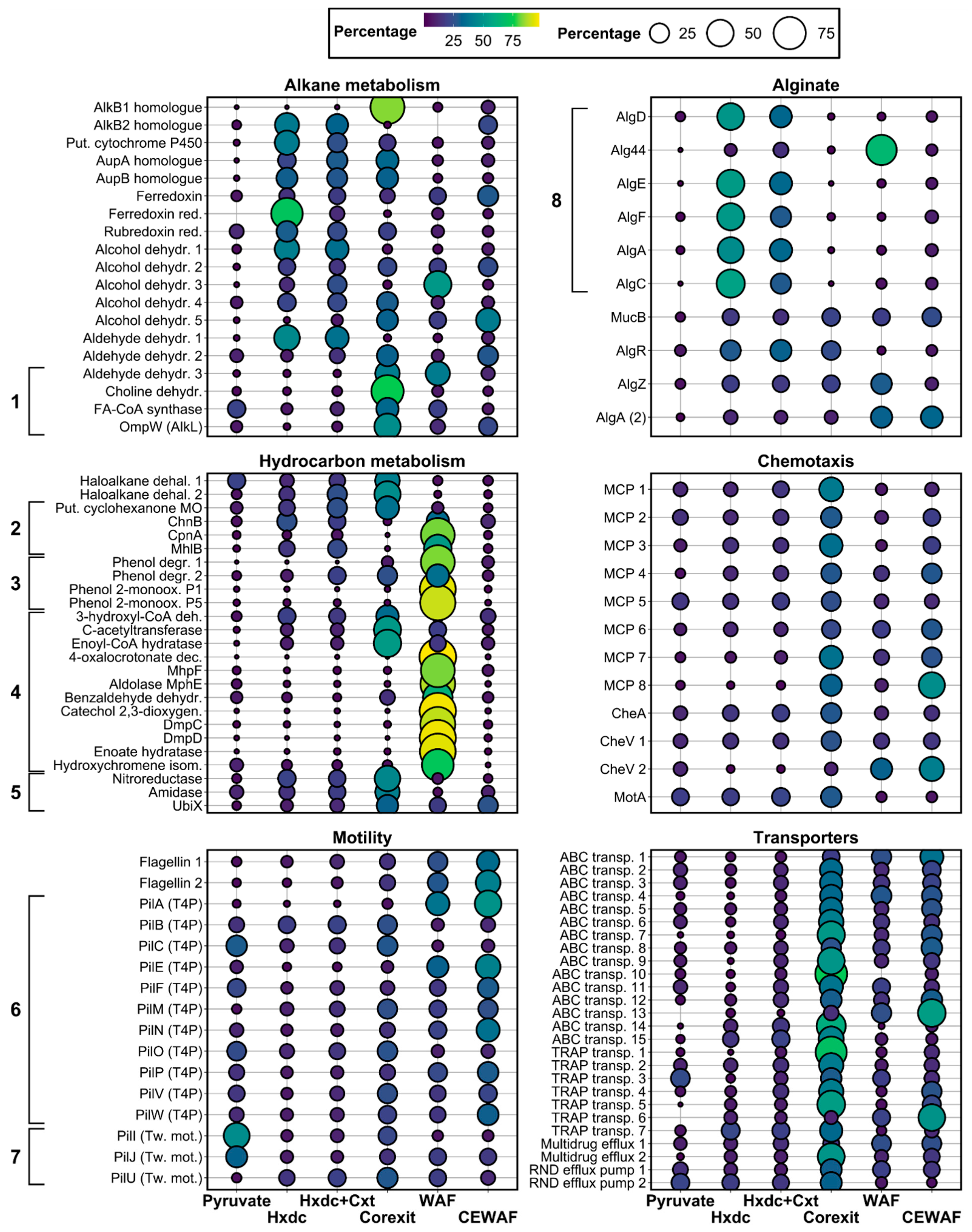
3.3. Proteome of Marinobacter sp. TT1 Grown on n-Hexadecane
3.4. Protein Profiles of Marinobacter sp. TT1 during Corexit Exposure
3.4.1. Corexit versus n-Hexadecane
3.4.2. n-Hexadecane/Corexit versus n-Hexadecane
3.4.3. CEWAF versus WAF
4. Discussion
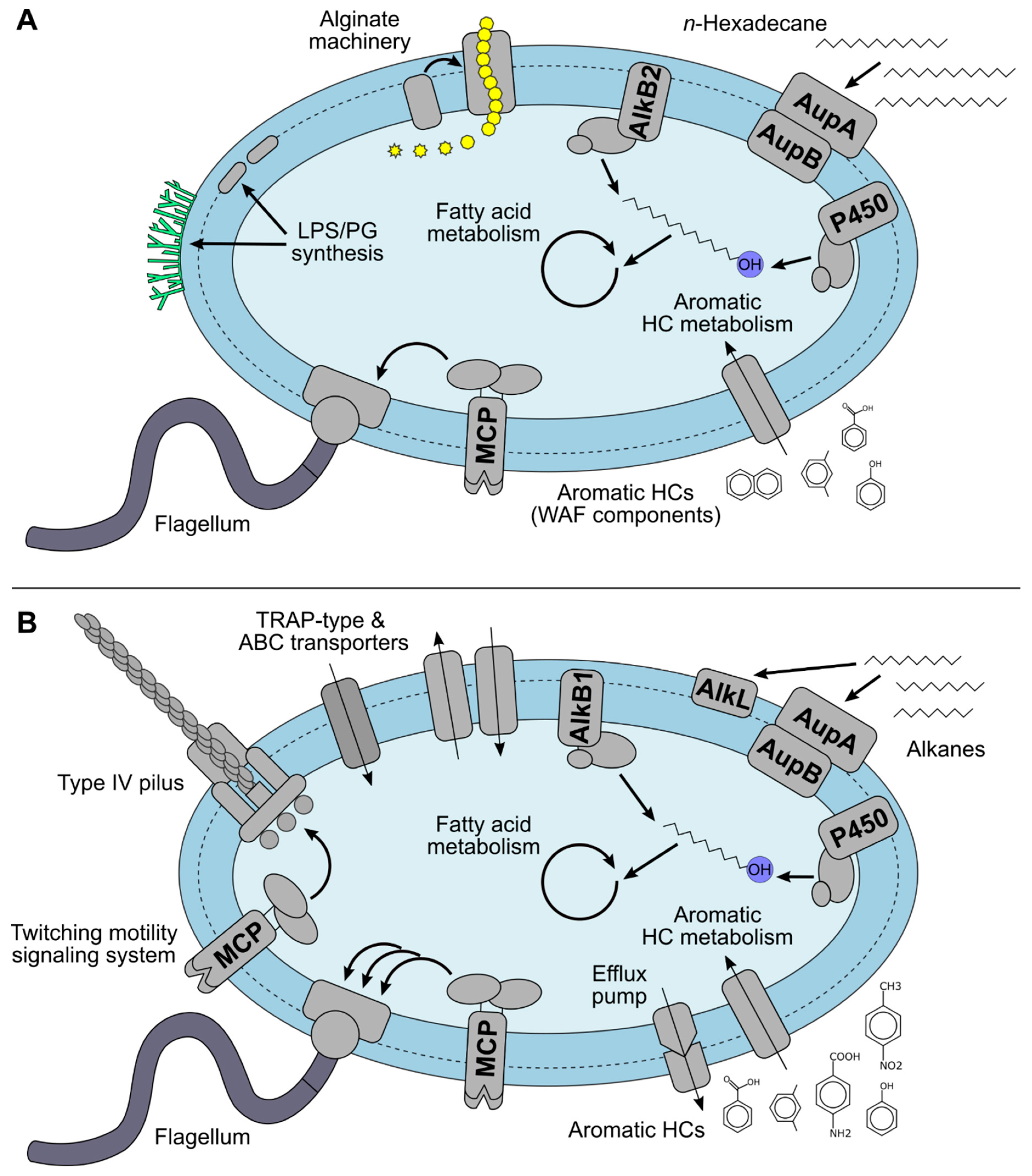
4.1. Hydrocarbon Metabolism of Marinobacter sp. TT1
4.2. Alginate Biosynthesis of Marinobacter sp. TT1
4.3. Biodegradation of Corexit Components by Marinobacter sp. TT1
4.3.1. Alkane Metabolism
4.3.2. Metabolism of Other Corexit Components
4.4. Additional Cellular Processes Affected by Corexit Exposure in Marinobacter sp. TT1
4.4.1. Transmembrane Transporter Systems
4.4.2. Chemotactic Motility and Biofilm Formation
4.5. Environmental Implications of Proteomic Findings
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McNutt, M.; Camilli, R.; Crone, T.J.; Guthrie, G.D.; Hsieh, P.A.; Ryerson, T.B.; Savas, O.; Shaffer, F. Review of flow rate estimates of the Deepwater Horizon oil spill. Proc. Natl. Acad. Sci. USA 2011, 109, 20260–20267. [Google Scholar] [CrossRef]
- US Nature Communications. Deepwater Horizon (DWH). The Use of Surface and Subsea Dispersants during the BP Deepwater Horizon Oil Spill; US Nature Communications: Washington, DC, USA, 2011. [Google Scholar]
- Doyle, S.M.; Whitaker, E.A.; De Pascuale, V.; Wade, T.L.; Knap, A.H.; Santschi, P.H.; Quigg, A.; Sylvan, J.B. Rapid formation of microbe-oil aggregates and changes in community composition in coastal surface water following exposure to oil and the dispersant Corexit. Front. Microbiol. 2018, 9, 689. [Google Scholar] [CrossRef]
- Kleindienst, S.; Seidel, M.; Ziervogel, K.; Grim, S.L.; Loftis, K.M.; Harrison, S.; Malkin, S.Y.; Perkins, M.J.; Field, J.A.; Sogin, M.L.; et al. Chemical dispersants can suppress the activity of natural oil-degrading microorganisms. Proc. Natl. Acad. Sci. USA 2015, 112, 14900–14905. [Google Scholar] [CrossRef]
- Suja, L.D.; Summers, S.; Gutierrez, T. Role of EPS, dispersant and nutrients on the microbial response and MOS formation in the subarctic northeast Atlantic. Front. Microbiol. 2017, 8, 676. [Google Scholar] [CrossRef]
- Sun, X.; Kostka, J.E. Hydrocarbon-degrading microbial communities are site specific, and their activity is limited by synergies in temperature and nutrient availability in surface ocean waters. Appl. Environ. Microbiol. 2019, 85, e00443-19. [Google Scholar] [CrossRef] [PubMed]
- Techtmann, S.M.; Zhuang, M.; Campo, P.; Holder, E.; Elk, M.; Hazen, T.C.; Conmy, R.; Domingo, J.S. Corexit 9500 enhances oil biodegradation and changes active bacterial community structure of oil-enriched microcosms. Appl. Environ. Microbiol. 2017, 83, e03462-16. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.; Fortin, N.; Elias, M.; Wasserscheid, J.; King, T.L.; Lee, K.; Greer, C.W. Metagenomic and metatranscriptomic responses of natural oil degrading bacteria in the presence of dispersants. Environ. Microbiol. 2019, 21, 2307–2319. [Google Scholar] [CrossRef] [PubMed]
- Baelum, J.; Borglin, S.; Chakraborty, R.; Fortney, J.L.; Lamendella, R.; Mason, O.U.; Auer, M.; Zemla, M.; Bill, M.; Conrad, M.E.; et al. Deep-sea bacteria enriched by oil and dispersant from the Deepwater Horizon spill. Environ. Microbiol. 2012, 14, 2405–2416. [Google Scholar] [CrossRef]
- Prince, R.C.; Kelley, B.A.; Butler, J.D. Three Widely-Available Dispersants Substantially Increase the Biodegradation of Otherwise Undispersed Oil. J. Mar. Sci. Res. Dev. 2015, 6, 183. [Google Scholar] [CrossRef]
- McFarlin, K.M.; Prince, R.C.; Perkins, R.; Leigh, M.B. Biodegradation of dispersed oil in Arctic seawater at −1 °C. PLoS ONE 2014, 9, e84297. [Google Scholar] [CrossRef]
- Hackbusch, S.; Noirungsee, N.; Viamonte, J.; Sun, X.; Bubenheim, P.; Kostka, J.E.; Müller, R.; Liese, A. Influence of pressure and dispersant on oil biodegradation by a newly isolated Rhodococcus strain from deep-sea sediments of the Gulf of Mexico. Mar. Pollut. Bull. 2020, 150, 110683. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, L.J.; Fulmer, P. Effects of COREXIT® EC9500A on bacteria from a beach oiled by the Deepwater Horizon spill. Aquat. Microb. Ecol. 2011, 63, 101–109. [Google Scholar] [CrossRef]
- Overholt, W.A.; Marks, K.P.; Romero, I.C.; Hollander, D.J.; Snell, T.W.; Kostka, J.E. Hydrocarbon-degrading bacteria exhibit a species-specific response to dispersed oil while moderating ecotoxicity. Appl. Environ. Microbiol. 2016, 82, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.; Yergeau, É.; Fortin, N.; Cobanli, S.; Elias, M.; King, T.L.; Lee, K.; Greer, C.W. Chemical dispersants enhance the activity of oil- and gas condensate-degrading marine bacteria. ISME J. 2017, 11, 2793–2808. [Google Scholar] [CrossRef] [PubMed]
- Duran, R. Marinobacter. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Heidelberg/Berlin, Germany, 2010; pp. 1725–1735. [Google Scholar]
- Rughöft, S.; Vogel, A.L.; Joye, S.B.; Gutierrez, T.; Kleindienst, S. Starvation-dependent inhibition of the hydrocarbon degrader Marinobacter sp. TT1 by a chemical dispersant. J. Mar. Sci. Eng. 2020, 8, 925. [Google Scholar] [CrossRef]
- Chakraborty, R.; Borglin, S.; Dubinsky, E.A.; Andersen, G.L.; Hazen, T.C. Microbial response to the MC-252 oil and Corexit 9500 in the Gulf of Mexico. Front. Microbiol. 2012, 3, 357. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shao, Z. Enzymes and genes involved in aerobic alkane degradation. Front. Microbiol. 2013, 4, 116. [Google Scholar] [CrossRef] [PubMed]
- Abbasian, F.; Lockington, R.; Megharaj, M.; Naidu, R. A review on the genetics of aliphatic and aromatic hydrocarbon degradation. Appl. Biochem. Biotechnol. 2015, 178, 224–250. [Google Scholar] [CrossRef]
- Choyke, S.; Ferguson, P.L. Molecular characterization of nonionic surfactant components of the Corexit® 9500 oil spill dispersant by high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 2019, 33, 1683–1694. [Google Scholar] [CrossRef]
- Place, B.J.; Perkins, M.J.; Sinclair, E.; Barsamian, A.L.; Blakemore, P.R.; Field, J. Trace analysis of surfactants in Corexit oil dispersant formulations and seawater. Deep. Sea Res. Part II Top. Studies Oceanogr. 2016, 129, 273–281. [Google Scholar] [CrossRef]
- Inácio, Â.S.; Domingues, N.S.; Nunes, A.; Martins, P.T.; Moreno, M.J.; Estronca, L.M.; Fernandes, R.; Moreno, A.J.M.; Borrego, M.J.; Gomes, J.P.; et al. Quaternary ammonium surfactant structure determines selective toxicity towards bacteria: Mechanisms of action and clinical implications in antibacterial prophylaxis. J. Antimicrob. Chemother. 2015, 71, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Partearroyo, M.A.; Ostolaza, H.; Goñi, F.M.; Barberá-Guillem, E. Surfactant-induced cell toxicity and cell lysis. A study using B16 melanoma cells. Biochem. Pharmacol. 1990, 40, 1323–1328. [Google Scholar] [CrossRef]
- Van der Werf, M.J.; Hartmans, S.; Van den Tweel, W.J.J. Permeabilization and lysis of Pseudomonas pseudoalcaligenes cells by Triton X-100 for efficient production of D-malate. Appl. Microbiol. Biotechnol. 1995, 43, 590–594. [Google Scholar] [CrossRef]
- Krell, T.; Lacal, J.; Guazzaroni, M.E.; Busch, A.; Silva-Jiménez, H.; Fillet, S.; Reyes-Darías, J.A.; Muñoz-Martínez, F.; Rico-Jiménez, M.; García-Fontana, C.; et al. Responses of Pseudomonas putida to toxic aromatic carbon sources. J. Biotechnol. 2012, 160, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Shitashiro, M.; Tanaka, H.; Hong, C.S.; Kuroda, A.; Takiguchi, N.; Ohtake, H.; Kato, J. Identification of chemosensory proteins for trichloroethylene in Pseudomonas aeruginosa. J. Biosci. Bioeng. 2005, 99, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Young, L.Y.; Mitchell, R. Negative chemotaxis of marine bacteria to toxic chemicals. Appl. Microbiol. 1973, 25, 972–975. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.L.; Duque, E.; Gallegos, M.-T.; Godoy, P.; Ramos-González, M.I.; Rojas, A.; Terán, W.; Segura, A. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 2002, 56, 743–768. [Google Scholar] [CrossRef]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [CrossRef]
- Radniecki, T.S.; Schneider, M.C.; Semprini, L. The influence of Corexit 9500A and weathering on Alaska North Slope crude oil toxicity to the ammonia oxidizing bacterium, Nitrosomonas europaea. Mar. Pollut. Bull. 2013, 68, 64–70. [Google Scholar] [CrossRef]
- Rico-Martinez, R.; Snell, T.W.; Shearer, T.L. Synergistic toxicity of Macondo crude oil and dispersant Corexit 9500A® to the Brachionus plicatilis species complex (Rotifera). Environ. Pollut. 2013, 173, 5–10. [Google Scholar] [CrossRef]
- Ozhan, K.; Parsons, M.L.; Bargu, S. How were phytoplankton affected by the Deepwater Horizon oil spill? Bioscience 2014, 64, 829–836. [Google Scholar] [CrossRef]
- Gutierrez, T.; Singleton, D.R.; E Berry, D.; Yang, T.; Aitken, M.D.; Teske, A. Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J. 2013, 7, 2091–2104. [Google Scholar] [CrossRef] [PubMed]
- Dyksterhouse, S.E.; Gray, J.P.; Herwig, R.P.; Lara, J.C.; Staley, J.T. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 1995, 45, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Haange, S.-B.; Jehmlich, N.; Hoffmann, M.; Weber, K.; Lehmann, J.; Von Bergen, M.; Slanina, U. Disease development is accompanied by changes in bacterial protein abundance and functions in a refined model of dextran sulfate sodium (dss)-induced colitis. J. Proteome Res. 2019, 18, 1774–1786. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- The UniProt Consortium UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- A Chen, I.-M.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R.; et al. IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019, 47, D666–D677. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Mounier, J.; Hakil, F.; Branchu, P.; Naïtali, M.; Goulas, P.; Sivadon, P.; Grimaud, R. AupA and AupB Are Outer and Inner Membrane Proteins Involved in Alkane Uptake in Marinobacter hydrocarbonoclasticus SP17. mBio 2018, 9, e00520-18. [Google Scholar] [CrossRef] [PubMed]
- Gacesa, P. Bacterial alginate biosynthesis—Recent progress and future prospects. Microbiology 1998, 144, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Schneiker, S.; Santos, V.A.P.M.D.; Bartels, D.; Bekel, T.; Brecht, M.; Buhrmester, J.; Chernikova, T.N.; Denaro, R.; Ferrer, M.; Gertler, C.; et al. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat. Biotechnol. 2006, 24, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Mounier, J.; Camus, A.; Mitteau, I.; Vaysse, P.-J.; Goulas, P.; Grimaud, R.; Sivadon, P. The marine bacterium Marinobacter hydrocarbonoclasticus SP17 degrades a wide range of lipids and hydrocarbons through the formation of oleolytic biofilms with distinct gene expression profiles. FEMS Microbiol. Ecol. 2014, 90, 816–831. [Google Scholar] [CrossRef] [PubMed]
- Gregson, B.H.; Metodieva, G.; Metodiev, M.V.; Golyshin, P.N.; McKew, B.A. Protein expression in the obligate hydrocarbon-degrading psychrophile Oleispira antarctica RB-8 during alkane degradation and cold tolerance. Environ. Microbiol. 2020, 22, 1870–1883. [Google Scholar] [CrossRef] [PubMed]
- Gregson, B.H.; Metodieva, G.; Metodiev, M.V.; McKew, B.A. Differential protein expression during growth on linear versus branched alkanes in the obligate marine hydrocarbon-degrading bacterium Alcanivorax borkumensis SK2 T. Environ. Microbiol. 2019, 21, 2347–2359. [Google Scholar] [CrossRef] [PubMed]
- Van Beilen, J.B.; Marin, M.M.; Smits, T.H.M.; Röthlisberger, M.; Franchini, A.G.; Witholt, B.; Rojo, F. Characterization of two alkane hydroxylase genes from the marine hydrocarbonoclastic bacterium Alcanivorax borkumensis. Environ. Microbiol. 2004, 6, 264–273. [Google Scholar] [CrossRef]
- Li, Y.P.; Pan, J.C.; Ma, Y. Elucidation of multiple alkane hydroxylase systems in biodegradation of crude oil n-alkane pollution by Pseudomonas aeruginosa DN1. J. Appl. Microbiol. 2019, 128, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, W.; Wu, Y.; Zhou, Z.; Lai, Q.; Shao, Z. Multiple alkane hydroxylase systems in a marine alkane degrader, Alcanivorax dieselolei B-5. Environ. Microbiol. 2011, 13, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Shin, B.; Jung, J.; Lee, Y.; Park, W. Metabolic and stress responses of Acinetobacter oleivorans DR1 during long-chain alkane degradation. Microb. Biotechnol. 2017, 10, 1809–1823. [Google Scholar] [CrossRef]
- Barbato, M.; Scoma, A.; Mapelli, F.; De Smet, R.; Banat, I.M.; Daffonchio, D.; Boon, N.; Borin, S. Hydrocarbonoclastic Alcanivorax Isolates Exhibit Different Physiological and Expression Responses to n-dodecane. Front. Microbiol. 2016, 7, 2056. [Google Scholar] [CrossRef]
- Ennouri, H.; d’Abzac, P.; Hakil, F.; Branchu, P.; Naïtali, M.; Lomenech, A.M.; Oueslati, R.; Desbrières, J.; Sivadon, P.; Grimaud, R. The extracellular matrix of the oleolytic biofilms of Marinobacter hydrocarbonoclasticus comprises cytoplasmic proteins and T2SS effectors that promote growth on hydrocarbons and lipids. Environ. Microbiol. 2017, 19, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Noh, J.; Park, W. Physiological and metabolic responses for hexadecane degradation in Acinetobacter oleivorans DR1. J. Microbiol. 2011, 49, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Attur, M.G.; Kanaya, S.; Morikawa, M. Alkane inducible proteins in Geobacillus thermoleovorans B23. BMC Microbiol. 2009, 9, 60. [Google Scholar] [CrossRef]
- Vaysse, P.J.; Sivadon, P.; Goulas, P.; Grimaud, R. Cells dispersed from Marinobacter hydrocarbonoclasticus sp17 biofilm exhibit a specific protein profile associated with a higher ability to reinitiate biofilm development at the hexadecane–water interface. Environ. Microbiol. 2011, 13, 737–746. [Google Scholar] [CrossRef]
- Bera, G.; Doyle, S.; Passow, U.; Kamalanathan, M.; Wade, T.L.; Sylvan, J.B.; Sericano, J.L.; Gold, G.; Quigg, A.; Knap, A.H. Biological response to dissolved versus dispersed oil. Mar. Pollut. Bull. 2020, 150, 110713. [Google Scholar] [CrossRef]
- Faksness, L.-G.; Brandvik, P.J.; Sydnes, L.K. Composition of the water accommodated fractions as a function of exposure times and temperatures. Mar. Pollut. Bull. 2008, 56, 1746–1754. [Google Scholar] [CrossRef]
- Bonin, P.; Vieira, C.; Grimaud, R.; Militon, C.; Cuny, P.; Lima, O.; Guasco, S.; Brussaard, C.P.; Michotey, V. Substrates specialization in lipid compounds and hydrocarbons of Marinobacter genus. Environ. Sci. Pollut. Res. 2015, 22, 15347–15359. [Google Scholar] [CrossRef]
- Dombrowski, N.; Donaho, J.A.; Gutierrez, T.; Seitz, K.W.; Teske, A.P.; Baker, B.J. Reconstructing metabolic pathways of hydrocarbon-degrading bacteria from the Deepwater Horizon oil spill. Nat. Microbiol. 2016, 1, 16057. [Google Scholar] [CrossRef] [PubMed]
- Urtuvia, V.; Maturana, N.; Acevedo, F.; Peña, C.; Díaz-Barrera, A. Bacterial alginate production: An overview of its biosynthesis and potential industrial production. World J. Microbiol. Biotechnol. 2017, 33, 198. [Google Scholar] [CrossRef] [PubMed]
- Hay, I.D.; Rehman, Z.U.; Moradali, M.F.; Wang, Y.; Rehm, B.H.A. Microbial alginate production, modification and its applications. Microb. Biotechnol. 2013, 6, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Gunasekera, T.S.; Striebich, R.C.; Mueller, S.S.; Strobel, E.M.; Ruiz, O.N. Transcriptional profiling suggests that multiple metabolic adaptations are required for effective proliferation of Pseudomonas aeruginosa in jet fuel. Environ. Sci. Technol. 2013, 47, 13449–13458. [Google Scholar] [CrossRef] [PubMed]
- Sabirova, J.S.; Becker, A.; Lünsdorf, H.; Nicaud, J.-M.; Timmis, K.N.; Golyshin, P.N. Transcriptional profiling of the marine oil-degrading bacterium Alcanivorax borkumensis during growth on n-alkanes. FEMS Microbiol. Lett. 2011, 319, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Ghafoor, A.; Hay, I.D.; Rehm, B.H.A. Role of Exopolysaccharides in Pseudomonas aeruginosa Biofilm Formation and Architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246. [Google Scholar] [CrossRef]
- Moradali, M.F.; Rehm, B.H.A. The role of alginate in bacterial biofilm formation. In Biologically-Inspired Systems; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2019; pp. 517–537. [Google Scholar]
- Manilla-Pérez, E.; Reers, C.; Baumgart, M.; Hetzler, S.; Reichelt, R.; Malkus, U.; Kalscheuer, R.; Wältermann, M.; Steinbüchel, A. Analysis of lipid export in hydrocarbonoclastic bacteria of the genus Alcanivorax: Identification of lipid export-negative mutants of Alcanivorax borkumensis SK2 and Alcanivorax jadensis T9. J. Bacteriol. 2009, 192, 643–656. [Google Scholar] [CrossRef]
- McFarlin, K.M.; Perkins, M.J.; Field, J.A.; Leigh, M.B. Biodegradation of crude oil and Corexit 9500 in Arctic seawater. Front. Microbiol. 2018, 9, 1788. [Google Scholar] [CrossRef]
- Word, J.Q.; Clark, J.R.; Word, L.S. Comparison of the acute toxicity of Corexit 9500 and household cleaning products. Hum. Ecol. Risk Assess. Int. J. 2014, 21, 707–725. [Google Scholar] [CrossRef]
- Grant, C.; Deszcz, D.; Wei, Y.-C.; Martínez-Torres, R.J.; Morris, P.; Folliard, T.; Sreenivasan, R.; Ward, J.M.; Dalby, P.A.; Woodley, J.M.; et al. Identification and use of an alkane transporter plug-in for applications in biocatalysis and whole-cell biosensing of alkanes. Sci. Rep. 2014, 4, srep05844. [Google Scholar] [CrossRef]
- Julsing, M.K.; Schrewe, M.; Cornelissen, S.; Hermann, I.; Schmid, A.; Bühler, B. Outer membrane protein alkL boosts biocatalytic oxyfunctionalization of hydrophobic substrates in Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 5724–5733. [Google Scholar] [CrossRef] [PubMed]
- Green, D.H.; Bowman, J.P.; Smith, E.A.; Gutierrez, T.; Bolch, C.J.S. Marinobacter algicola sp. nov., isolated from laboratory cultures of paralytic shellfish toxin-producing dinoflagellates. Int. J. Syst. Evol. Microbiol. 2006, 56, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.J.; Lopez-Perez, M.; Webb, H.K.; Gomez, D.; Sawabe, T.; Ryan, J.; Vyssotski, M.; Bizet, C.; Malherbe, F.; Mikhailov, V.V.; et al. Marinobacter Salarius sp. nov. and Marinobacter Similis sp. nov., Isolated from sea water. PLoS ONE 2014, 9, e106514. [Google Scholar] [CrossRef] [PubMed]
- Seidel, M.; Kleindienst, S.; Dittmar, T.; Joye, S.B.; Medeiros, P.M. Biodegradation of crude oil and dispersants in deep seawater from the Gulf of Mexico: Insights from ultra-high resolution mass spectrometry. Deep. Sea Res. Part II Top. Stud. Oceanogr. 2016, 129, 108–118. [Google Scholar] [CrossRef]
- García, M.T.; Campos, E.; Marsal, A.; Ribosa, I. Biodegradability and toxicity of sulphonate-based surfactants in aerobic and anaerobic aquatic environments. Water Res. 2009, 43, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Hales, S.G. Biodegradation of the Anionic Surfactant Dialkyl Sulphosuccinate. Environ. Toxicol. Chem. Int. J. 1993, 12, 1821–1828. [Google Scholar] [CrossRef]
- Kover, S.C.; Rosario-Ortiz, F.L.; Linden, K. Photochemical fate of solvent constituents of Corexit oil dispersants. Water Res. 2014, 52, 101–111. [Google Scholar] [CrossRef]
- Montenegro, T.D.; Kleindienst, S.; Allen, A.E.; Eren, A.M.; McCrow, J.; Calderon, J.D.; Arnold, J.; Joye, S.B. Colwellia and Marinobacter metapangenomes reveal species-specific responses to oil and dispersant exposure in deepsea microbial communities. bioRxiv 2020. [Google Scholar] [CrossRef]
- Rivers, A.R.; Sharma, S.; Tringe, S.G.; Martin, J.; Joye, S.B.; Moran, M.A. Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 2013, 7, 2315–2329. [Google Scholar] [CrossRef]
- Mulligan, C.; Fischer, M.; Thomas, G.H. Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol. Rev. 2011, 35, 68–86. [Google Scholar] [CrossRef]
- Rosa, L.T.; Bianconi, M.E.; Thomas, G.H.; Kelly, D.J. Tripartite ATP-Independent Periplasmic (TRAP) Transporters and Tripartite Tricarboxylate Transporters (TTT): From uptake to pathogenicity. Front. Cell. Infect. Microbiol. 2018, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Chae, J.-C.; Zylstra, G. 4-Chlorobenzoate uptake in Comamonas sp. Strain DJ-12 is mediated by a tripartite ATP-independent periplasmic transporter. J. Bacteriol. 2006, 188, 8407–8412. [Google Scholar] [CrossRef] [PubMed]
- Salmon, R.C.; Cliff, M.J.; Rafferty, J.B.; Kelly, D.J. The CouPSTU and TarPQM Transporters in Rhodopseudomonas palustris: Redundant, promiscuous uptake systems for lignin-derived aromatic substrates. PLoS ONE 2013, 8, e59844. [Google Scholar] [CrossRef] [PubMed]
- Aubé, J.; Senin, P.; Bonin, P.; Pringault, O.; Jeziorski, C.; Bouchez, O.; Klopp, C.; Guyoneaud, R.; Goñi-Urriza, M. Meta-omics provides insights into the impact of hydrocarbon contamination on microbial mat functioning. Microb. Ecol. 2020, 80, 286–295. [Google Scholar] [CrossRef]
- Xu, W.; You, Y.; Wang, Z.; Chen, W.; Zeng, J.; Zhao, X.; Su, Y. Dibutyl phthalate alters the metabolic pathways of microbes in black soils. Sci. Rep. 2018, 8, 2605. [Google Scholar] [CrossRef]
- Liu, S.; Guo, C.; Lin, W.; Wu, F.; Lu, G.; Lu, J.; Dang, Z. Comparative transcriptomic evidence for Tween80-enhanced biodegradation of phenanthrene by Sphingomonas sp. GY2B. Sci. Total. Environ. 2017, 609, 1161–1171. [Google Scholar] [CrossRef]
- Jones, P.M.; George, A.M. Mechanism of the ABC transporter ATPase domains: Catalytic models and the biochemical and biophysical record. Crit. Rev. Biochem. Mol. Biol. 2012, 48, 39–50. [Google Scholar] [CrossRef]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, Function, and Evolution of Bacterial ATP-Binding Cassette Systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef]
- Murínová, S.; Dercová, K. Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int. J. Microbiol. 2014, 2014, 1–16. [Google Scholar] [CrossRef]
- Nowak, A.; Mrozik, A. Facilitation of co-metabolic transformation and degradation of monochlorophenols by Pseudomonas sp. CF600 and changes in its fatty acid composition. Water Air Soil Pollut. 2016, 227, 83. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, Y.; Chen, X.; Diab, A.A.; Ruan, L.; He, J.; Wang, H.; He, Y. The multiple DSF-family QS signals are synthesized from carbohydrate and branched-chain amino acids via the FAS elongation cycle. Sci. Rep. 2015, 5, srep13294. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, L.-H.; Cámara, M.; He, Y. The DSF family of quorum sensing signals: Diversity, biosynthesis, and turnover. Trends Microbiol. 2017, 25, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Hosie, A.H.F.; Allaway, D.; Jones, M.A.; Walshaw, D.L.; Johnston, A.W.B.; Poole, P.S. Solute-binding protein-dependent ABC transporters are responsible for solute efflux in addition to solute uptake. Mol. Microbiol. 2001, 40, 1449–1459. [Google Scholar] [CrossRef] [PubMed]
- E Giuliani, S.; Frank, A.M.; Corgliano, D.M.; Seifert, C.; Hauser, L.; Collart, F.R. Environment sensing and response mediated by ABC transporters. BMC Genom. 2011, 12, S8. [Google Scholar] [CrossRef] [PubMed]
- Michalska, K.; Chang, C.; Mack, J.C.; Zerbs, S.; Joachimiak, A.; Collart, F.R. Characterization of transport proteins for aromatic compounds derived from lignin: Benzoate derivative binding proteins. J. Mol. Biol. 2012, 423, 555–575. [Google Scholar] [CrossRef] [PubMed]
- Noda, K.-I.; Watanabe, K.; Maruhashi, K. Isolation of the Pseudomonas aeruginosa gene affecting uptake of dibenzothiophene in n-tetradecane. J. Biosci. Bioeng. 2003, 95, 504–511. [Google Scholar] [CrossRef]
- Handbook of Hydrocarbon and Lipid Microbiology. In Handbook of Hydrocarbon and Lipid Microbiology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2010.
- Conrad, J.C.; Gibiansky, M.L.; Jin, F.; Gordon, V.D.; Motto, D.A.; Mathewson, M.A.; Stopka, W.G.; Zelasko, D.C.; Shrout, J.D.; Wong, G.C.L. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophys. J. 2011, 100, 1608–1616. [Google Scholar] [CrossRef]
- Wang, S.; Parsek, M.R.; Wozniak, D.J.; Ma, L.Z. A Spider web strategy of type IV pili-mediated migration to build a fibre-like psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2013, 15, 2238–2253. [Google Scholar] [CrossRef]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa Displays Multiple Phenotypes during Development as a Biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Wong, G.C.L. Sensational biofilms: Surface sensing in bacteria. Curr. Opin. Microbiol. 2016, 30, 139–146. [Google Scholar] [CrossRef]
- Passow, U. Formation of rapidly-sinking, oil-associated marine snow. Deep. Sea Res. Part II Top. Stud. Oceanogr. 2016, 129, 232–240. [Google Scholar] [CrossRef]
- Passow, U.; Ziervogel, K. Marine Snow Sedimented Oil Released During the Deepwater Horizon Spill. Oceanography 2016, 29, 118–125. [Google Scholar] [CrossRef]
- Joye, S.; Bracco, A.; Özgökmen, T.M.; Chanton, J.P.; Grosell, M.; Macdonald, I.R.; Cordes, E.E.; Montoya, J.P.; Passow, U. The Gulf of Mexico ecosystem, six years after the Macondo oil well blowout. Deep. Sea Res. Part II Top. Studies Oceanogr. 2016, 129, 4–19. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rughöft, S.; Jehmlich, N.; Gutierrez, T.; Kleindienst, S. Comparative Proteomics of Marinobacter sp. TT1 Reveals Corexit Impacts on Hydrocarbon Metabolism, Chemotactic Motility, and Biofilm Formation. Microorganisms 2021, 9, 3. https://doi.org/10.3390/microorganisms9010003
Rughöft S, Jehmlich N, Gutierrez T, Kleindienst S. Comparative Proteomics of Marinobacter sp. TT1 Reveals Corexit Impacts on Hydrocarbon Metabolism, Chemotactic Motility, and Biofilm Formation. Microorganisms. 2021; 9(1):3. https://doi.org/10.3390/microorganisms9010003
Chicago/Turabian StyleRughöft, Saskia, Nico Jehmlich, Tony Gutierrez, and Sara Kleindienst. 2021. "Comparative Proteomics of Marinobacter sp. TT1 Reveals Corexit Impacts on Hydrocarbon Metabolism, Chemotactic Motility, and Biofilm Formation" Microorganisms 9, no. 1: 3. https://doi.org/10.3390/microorganisms9010003
APA StyleRughöft, S., Jehmlich, N., Gutierrez, T., & Kleindienst, S. (2021). Comparative Proteomics of Marinobacter sp. TT1 Reveals Corexit Impacts on Hydrocarbon Metabolism, Chemotactic Motility, and Biofilm Formation. Microorganisms, 9(1), 3. https://doi.org/10.3390/microorganisms9010003