Abstract
Antimicrobial resistance (AMR) in Helicobacter pylori is increasing and can result in treatment failure and inappropriate antibiotic usage. This study used whole genome sequencing (WGS) to comprehensively analyze the H. pylori resistome and phylogeny in order to characterize Israeli H. pylori. Israeli H. pylori isolates (n = 48) underwent antimicrobial susceptibility testing (AST) against five antimicrobials and WGS analysis. Literature review identified 111 mutations reported to correlate with phenotypic resistance to these antimicrobials. Analysis was conducted via our in-house bioinformatics pipeline targeting point mutations in the relevant genes (pbp1A, 23S rRNA, gyrA, rdxA, frxA, and rpoB) in order to assess genotype-to-phenotype correlation. Resistance rates of study isolates were as follows: clarithromycin 54%, metronidazole 31%, amoxicillin 10%, rifampicin 4%, and levofloxacin 2%. Genotype-to-phenotype correlation was inconsistent; for every analyzed gene at least one phenotypically susceptible isolate was found to have a mutation previously associated with resistance. This was also observed regarding mutations commonly used in commercial kits to diagnose AMR in H. pylori cases. Furthermore, 11 novel point mutations associated with a resistant phenotype were detected. Analysis of a unique set of H. pylori isolates demonstrates that inferring resistance phenotypes from WGS in H. pylori remains challenging and should be optimized further.
1. Introduction
Antimicrobial resistance (AMR) is increasingly being reported among clinical Helicobacter pylori isolates worldwide [1,2]. In various parts of the world, disparate H. pylori antimicrobial resistance rates have been observed, suggesting geographical variation [3,4]. In Israel alone, six papers dealing with the prevalence of antimicrobial resistance among H. pylori isolates from the last decade have observed differing resistance rates [2,5,6,7,8,9], suggesting that within-country rates may vary among ethnic and social populations. Given these findings as well as the overall dearth of H. pylori surveillance, geographic characterization of antibiotic resistance is extremely important in order to adjust treatment appropriately [10].
However, antimicrobial surveillance data for H. pylori are limited as the use of antimicrobial susceptibility testing (AST) is frequently only implemented after initial treatment failure [10]. To address these challenges of variable resistance rates, as well as other logistical challenges involved in AST of H. pylori isolates, culture-independent techniques have been implemented. Several PCR-based commercial kits are commonly used to identify genotypic resistance from clinical samples [11], usually by targeting three to seven point mutations in the 23S rRNA (A2142C, A2142G, and A2143G) and gyrA (N87K, D91G, D91N, D91Y) genes that are generally considered to correlate with resistance [11,12]. However, these tests are limited due to imperfect correlations between these mutations and resistance [13,14,15,16], the numerous other mutations associated with resistance [17], and the possibility of co-infection with other organisms like Epstein-Barr Virus [18].
With the advent of next-generation sequencing (NGS) technology, utilizing genomics to investigate AMR in bacteria of public health importance is a growing area of research and combining whole genome sequencing (WGS) with traditional phenotypic resistance data can produce powerful results. Given the dearth of H. pylori AMR surveillance overall, the challenges involved in traditional H. pylori AST, as well as the potential for WGS to provide needed data, more research is needed in this regard. In order to address this need, we used WGS in order to conduct a comprehensive genotypic-phenotypic comparison among a set of Israeli H. pylori isolates.
2. Materials and Methods
2.1. H. pylori Culture and Phenotypic Antimicrobial Susceptibility Testing (AST)
Forty-eight unique patient H. pylori isolates were recovered from routine, clinical samples in the Baruch Padeh Medical Center Poriya in Northern Israel between 2015–2019. All biopsy specimens were placed in sterile Eppendorf tubes containing 1 mL of sterile physiological solution (0.9% NaCl) and were sent to the Clinical Microbiology Laboratory in Poriya Medical Center. Biopsies were minced manually with a sterile scalpel and seeded on Modified BD Helicobacter Agar (BD Diagnostics, Sparks, MD, USA) and blood agar (Hy-lab, Rehovot, Israel). The plates were incubated at 37 °C in a micro-aerophilic atmosphere produced by a gas generating system adapted for Campylobacter (Campy-Gen™, Oxoid, Basingstoke, UK) and inspected for growth at 7 and 10 days. H. pylori was identified based on Gram-staining followed by oxidase, catalase, and urease tests. Final identification was performed using MALDI-TOF MS (Bruker Daltonics, Bremen, Germany). For AST and DNA extraction one colony of the bacterial isolate was picked and re-grown on a selective agar under the conditions mentioned above. H. pylori suspensions were adjusted to a McFarland standard of 3.0 and subject to AST as part of a routine, clinical diagnostics procedure for five antimicrobials (amoxicillin, clarithromycin, levofloxacin, metronidazole, and rifampicin) using the gradient diffusion E-test method (bioMerieux, Mercy l’Etoile, France) and minimum inhibitory concentration (MIC) Test Strip (MTS™) (Liofilchem, Roseto degli Abruzzi, Italy). Testing was performed according to manufacturers’ instructions using Mueller-Hinton agar plates with 5% sheep’s [19,20]. Following 72h of incubation, minimum inhibitory concentration (MIC) values were determined and interpreted according to British Society for Antimicrobial Chemotherapy (BSAC) breakpoints [21]. The clinical laboratory is ISO-accredited and maintains a fully operational quality assurance and quality control scheme. Borderline MIC values were repeated and confirmed. H. pylori ATCC 43,504 was used for quality control. This study was approved by the Ethics Committee of the Baruch Padeh Medical Center Poriya (#23-13-POR).
2.2. DNA Extraction, Library Preparation, and WGS of H. pylori Isolates
DNA was extracted from isolates via a physical lysis process utilizing beating with acid-washed glass beads (Precellys Evolution, Bertin Instruments, Montigny-le-Bretonneux, France). High-quality DNA was subject to library preparation using the Nextera Flex kit (Illumina, San Diego, CA, USA) according to the manufacturer’s recommendations, followed by paired-end sequencing on Illumina iSeq 100 or Miseq platforms. Raw WGS data (FASTQ files) underwent quality control (QC), filtering, trimming, and de novo assembly using shovill (spades, v3.12; using the parameters ‘--trim’ and ‘--opts “--sc”’) (https://github.com/tseemann/shovill). Assemblies were annotated using the tool prokka (v 1.14.0; using the H. pylori 26,695 reference strain proteins in the parameter ‘--proteins GCF_000008525.1_ASM852v1_protein.faa’). Further details are available in Table S1. Sequence data have been deposited to BioProject PRJEB37854.
2.3. Antimicrobial Resistance-Associated Mutations Analysis
A literature review was conducted to devise a list of mutations at the genes pbp1A (amoxicillin-Resistance (R)), 23S rRNA (clarithromycin-R), gyrA (levofloxacin-R), rdxA and frxA (metronidazole-R), and rpoB (rifampicin-R) (Table S2). These genes were extracted and aligned with the tool MAFFT (v7.397) [22]. The 23S rRNA genes were identified using rnammer (v1.2) since rnammer provides more complete 23S rRNA sequences. Only perfectly-aligned gene sequences were analyzed for the presence of the abovementioned mutations (number of analysed alignments for each gene is shown in Tables S3–S8). The isolates then underwent further analysis to compare genotypic and phenotypic resistance data and describe genotype-to-phenotype correlations. Phenotypically-resistant isolates were analyzed for any novel point mutations. Subsequently, the remaining susceptible isolates were interrogated for these novel mutations. For the MIC plots and mutations clustering analysis, R version 3.4.2 [23] was used with the following packages: tidyr_0.8.3 [24], data.table_1.12.0 [25], cowplot_0.9.4 [26], ggplot2_3.1.0 [27], pheatmap_1.0.12 [28], RColorBrewer_1.1-2 [29], gplots_3.0.1 [30] and ggtree_1.15.6 [31,32]. The tools csvtk (https://github.com/shenwei356/csvtk) and seqkit (https://github.com/shenwei356/seqkit) were also used [33]. Clustering analysis was performed on the genes in which point mutations were identified, in order to identify correlation between observed mutations and MIC values.
3. Results
3.1. Phenotypic Antimicrobial Susceptibility Testing (AST) of Study Isolates
AST was performed on all 48 isolates to assess susceptibility to the five antibiotics most commonly used for H. pylori treatment. The distribution of MIC values for tested agents is shown in Figure S1. Full susceptibility to all tested antimicrobials was observed in 10 isolates (20.8%). Resistance was observed with the following rates: amoxicillin 10.4%, clarithromycin 54.2%, levofloxacin 2.0%, metronidazole 31.1% and rifampicin 4.2%. Only clarithromycin has an intermediately-resistant category (0.25–0.5 mg/L), and eight isolates (16.7%) were observed to be intermediate. Resistance to more than one antimicrobial drug was observed in nine isolates, 18.8% (clarithromycin-metronidazole n = 3; clarithromycin-amoxicillin n = 2; clarithromycin-rifampicin n = 1; clarithromycin-metronidazole-levofloxacin n = 1; clarithromycin-metronidazole-rifampicin n = 1).
3.2. Resistome Analysis and Genotype-to-Phenotype Correlation of Resistance
Mutations previously associated with phenotypic resistance were compared to AST results, and 51 such mutations were detected as shown in Figure 1. Of these, 25 mutations were found among both phenotypically-susceptible and -resistant isolates, while 24 mutations were found exclusively among phenotypically-susceptible isolates. The mutations A2142G at the 23s rRNA gene, mutations R16H, R16C, and the variant R90S in the rdxA gene were found exclusively in resistant isolates. The new variant T593S at the pbp1A gene was detected in both susceptible and resistant isolates. Seventeen isolates had the A2143G mutation at the 23S rRNA gene, of which seven were resistant, four were intermediately-resistant and six were susceptible. The gyrA mutations N87K, and D91N were found among 4.2% of isolates, in both resistant and susceptible isolates. No clear correlation was observed between resistant phenotypes and co-existing mutations after clustering analysis (Figure 2).
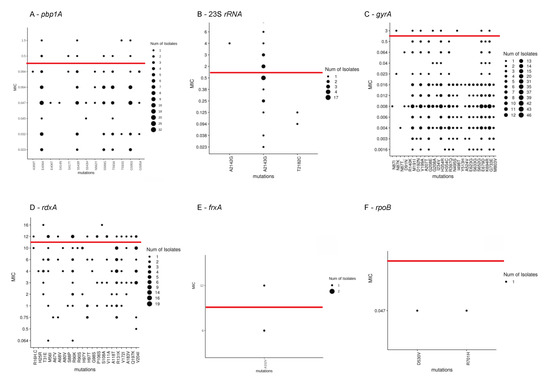
Figure 1.
Concordance between minimum inhibitory concentration (MIC) values and genotypic mutations in (A) pbp1A, (B) 23S rRNA, (C) gyrA, (D) rdxA, (E) frxA, and (F) rpoB detected in Israeli isolates (n = 48). Red lines reflect the resistance breakpoint per British Society for Anti-Microbial Chemotherapy (BSAC) guidelines.
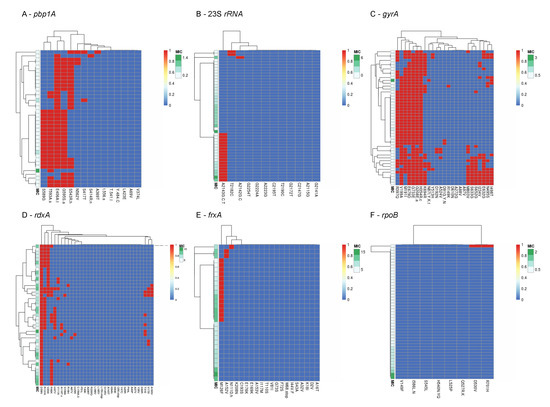
Figure 2.
Visualization of clustering of mutations in (A) pbp1A, (B) 23S rRNA, (C) gyrA, (D) rdxA, (E) frxA, and (F) rpoB likely associated with phenotypic resistance among Israeli isolates, compared to the phenotype (MIC values). The clustering of mutations (top of each panel) reflects their co-existence in single genomes. Presence of mutations is denoted in red and absence in denoted in blue. MIC values are shown on a green scale.
3.3. Novel Point Mutations
Novel point mutations discovered among phenotypically-resistant isolates are summarized in Tables S2–S9. The mutations G94E at the pbp1A gene, C2173T and G2212A at the 23S rRNA gene, T239M at the gyrA gene, G122R at the rdxA gene, A70T and A138V at the frxA gene were found among resistant isolates only.
4. Discussion
Previous resistome analysis of H. pylori via WGS has highlighted the role of point mutations as a mechanism of antibiotic resistance [34]. Furthermore, preliminary phenotypic-genotypic analysis utilizing WGS has strengthened the knowledge base of the association between 23S rRNA, gyrA, and rpoB mutations and resistance to macrolides, fluoroquinolones, and rifampicin but the practical implications deserve further study [34,35,36]. For example, Lauener et al. [35] noted a strong correlation between certain mutations in the 23 rRNA gene (A2146C, A2146G, and A2147G) and resistance, however these mutations were not observed in our study. This further supports the need for future research on various mutations and their phenotypic correlation. Our results add to these findings by demonstrating a complicated relationship between genotype and phenotype where mutations previously associated with resistance were found among phenotypically-susceptible isolates. For example, we observed discrepancies among mutations frequently associated with resistance including the A2143G mutation [12]; 10 out of 17 (59%) isolates harboring that mutation were susceptible (n = 6) or intermediately-resistant (n = 4). While this mutation is highly utilized in PCR-based kits, the discrepancy observed in this study has been observed, at differing rates, in previous PCR-based studies [13,14,15,16,37,38]. Similarly, further inconsistency was observed among 48 other mutations. These findings suggest that further research is needed to accurately assess phenotype-genotype correlation in this microorganism. It also bears noting that recent research has found that H. pylori can sometimes transform from the expected spiral form to a coccoid form, which could result in an inaccurate AST if the bacteria are in a viable-but-nonculturable state. This transformation could result in treatment failure without the presence of ARGs, further complicating the picture of AMR in H. pylori [39]. While WGS may be useful in overcoming the limitations of studies by allowing analysis of the full resistome, the relative contribution of each mutation to the MIC as well as the effect of combinations of mutations and shape transformation remains to be elucidated.
Regarding phenotypic antimicrobial resistance, we observed that resistance rates are higher for clarithromycin (54.2%) and metronidazole (31.1%) than amoxicillin (10.4%), rifampicin (4.2%) and levofloxacin (2%). When comparing the results of this study to previous studies of resistance rates in H. pylori in Israel, findings varied based on antibiotic as well as ethnic groups. Amoxicillin resistance rates were consistent with the other studies [2,8,9]. Clarithromycin resistance rates were similar to the rate observed among an Arab-Israeli population in Yeganeh et al. [8] article and higher compared to the Jewish-Israeli population in the same study as well as other studies [2,5,7,8,9]. Levofloxacin resistance rates in our study were lower than those previously observed by Peretz et al. [6] but similar to those of Pastukh et al. [2]. Observed metronidazole resistance rates were similar to those of Zevit et al. [5] and are within the range of rates observed among all studies [2,7,8,9]. For rifampicin, resistance rates are closer to those of the Arab-Israeli patient population than the Jewish-Israeli patient population in the Yeganeh et al. [8] article. It is not surprising that several of our findings are similar to previous Israeli findings, as some of the previous studies were carried out in the same hospital which contributed isolates to the current study, and therefore are likely from a similar patient pool experiencing similar exposures and antibiotic pressures. The instances where our rates differ are not substantial, and this variation may be due to factors including but not limited to sample size, differential antibiotic exposure among different ethnic and social groups, as well as acquisition of different H. pylori strains during international travel. A large proportion of Israeli citizens and residents are immigrants from other countries, including Eastern Europe, and research has noted that prevalence of H. pylori is high in immigrants from high-prevalence countries [40]. Patient ethnicity was not collected for this study, however the patient pool of the hospital contains both Jewish- and Arab-Israelis.
When comparing the resistance rates of the studied Israeli isolates to other regions of the WHO, similar resistance rates for amoxicillin were observed in American regions and equivalent resistance rates for metronidazole were observed in European countries. However, the Israeli clarithromycin rates are higher than those of all regions and levofloxacin rates are lower than those of all regions [4].
The main limitation of this study is the relatively small sample size and possible limited generalizability both in the Middle East and globally, as isolates originate from a single hospital in Israel. Furthermore, the isolates were de-identified, and as such we were unable to characterize isolates on demographic factors like age, sex, ethnicity, or immigration status. Moreover, all isolates were recovered as part of routine diagnosis, likely from patients who failed several treatment lines, and also were not tested by reference dilution methods. No information about first line treatment antimicrobial resistance rates was collected, since primary diagnosis was made with non-invasive tests (e.g., urea breath test (UBT) or stool antigen test). Additionally, as H. pylori is a fastidious bacterium, it is possible that the cultured Israeli isolates are somehow different from those which failed to be cultured in a clinical laboratory setting. Thus, further studies should corroborate our findings.
5. Conclusions
In conclusion, in light of the growing interest in using WGS for routine microbiological diagnosis [41], and the future possibility of harnessing metagenomic NGS for analyzing infections caused by fastidious bacteria such as H. pylori, the challenges in inferring phenotypic AST from genomic analysis shown in our resistome analysis should be addressed in order to underpin the future integration of NGS in the study of this microorganism in the clinical setting.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/1/2/s1, Figure S1: Minimum inhibitory concentration (MIC) value distributions for the 48 Israeli isolates for amoxicillin, clarithromycin, levofloxacin, metronidazole, and rifampicin. Blue bars indicate MIC values which render the isolate susceptible to the particular agent, whereas red bars indicate resistant isolates per BSAC breakpoints. Table S1: Metadata of isolate whole genome sequencing. Table S2: List of previously described mutations associated with phenotypic resistance in H. pylori and mutations observed for the first time in this study. Table S3: Genotype to phenotype concordance of Israeli isolates (n = 48) with respect to published mutations known to confer resistance and novel point mutations. Table S4: Mutations observed in the pbp1A gene (previously described and novel mutations). After imperfect alignments were omitted, 37 of the total 48 sequences remained for analysis. Table S5: Mutations observed in the 23S rRNA gene (previously described in the literature and novel mutations). After imperfect alignments were omitted, 45 of the total 48 sequences remained for analysis. Table S6: Mutations observed in the gyrA gene (previously described and novel mutations). After imperfect alignments were omitted, 47 of the total 48 sequences remained for analysis. Table S7: Mutations observed in the rdxA gene (previously described and novel mutations). Number of alignments remaining after imperfect alignments were removed is represented in the “N” column. Table S8: Mutations observed in the frxA gene (previously described and novel mutations). After imperfect alignments were omitted, 30 of the total 48 sequences remained for analysis. Table S9: Mutations observed in the rpoB gene (previously described and novel mutations). After imperfect alignments were omitted, 46 of the total 48 sequences remained for analysis.
Author Contributions
Conceptualization, J.M.-G., A.P.; methodology, J.M.-G., A.P., Y.M.; formal analysis, T.D.-A., Y.M.; investigation, T.D.-A., B.K., Y.M., H.A.C.; data curation, Y.M.; writing—original draft preparation, T.D.-A., H.A.C.; writing—review and editing, T.D.-A., Y.M., B.K., A.P., H.A.C., J.M.-G.; visualization, T.D.-A., Y.M.; supervision, J.M.-G., H.A.C., Y.M.; funding acquisition, J.M.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ghotaslou, R.; Leylabadlo, H.E.; Asl, Y.M. Prevalence of antibiotic resistance in Helicobacter pylori: A recent literature review. World J. Methodol. 2015, 5, 164–174. [Google Scholar] [CrossRef]
- Pastukh, N.; Peretz, A.; Brodsky, D.; Isakovich, N.; Azrad, M.; On, A. Antimicrobial susceptibility of Helicobacter pylori strains isolated from children in Israel. J. Glob. Antimicrob. Resist. 2018, 12, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Thung, I.; Aramin, H.; Vavinskaya, V.; Gupta, S.; Park, J.Y.; Crowe, S.E.; Valasek, M.A. Review article: The global emergence of Helicobacter pylori antibiotic resistance. Aliment. Pharmacol. Ther. 2016, 43, 514–533. [Google Scholar] [CrossRef] [PubMed]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382.e17. [Google Scholar] [CrossRef] [PubMed]
- Zevit, N.; Levy, I.; Shmuely, H.; Samra, Z.; Yahav, J. Antibiotic resistance of Helicobacter pylori in Israeli children. Scand. J. Gastroenterol. 2010, 45, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Peretz, A.; Paritsky, M.; Dinisman-Zavulunov, E.; Pastukh, N.; Glyatman, T.; On, A. Susceptibility of Helicobacter pylori to Levofloxacin and Rifampicin in Israel. Microb. Drug Resist. 2015, 21, 448–451. [Google Scholar] [CrossRef]
- Kori, M.; Yahav, J.; Berdinstein, R.; Shmuely, H. Primary and Secondary Antibiotic Resistance of Helicobacter pylori in Israeli Children and Adolescents. IMAJ 2017, 19, 747–750. [Google Scholar] [PubMed]
- Yeganeh, M.; Paritsky, M.; On, A.; Azrad, M.; Roshrosh, H.; Moalem, R.; Peretz, A. Characteristics of Antibiotic Resistance of Helicobacter pylori Among Adult Arab and Jewish Populations in Northern Israel. Microb. Drug Resist. 2018, 25, 103–107. [Google Scholar] [CrossRef]
- Peretz, A.; Paritsky, M.; Nasser, O.; Brodsky, D.; Glyatman, T.; Segal, S.; On, A. Resistance of Helicobacter pylori to tetracycline, amoxicillin, clarithromycin and metronidazole in Israeli children and adults. J. Antibiot. 2014, 67, 555–557. [Google Scholar] [CrossRef]
- Chey, W.; Leontiadis, G.; Howden, C.; Moss, S. ACG Clinical Guideline: Treatment of Helicobacter pylori Infection. Am. J. Gastroenterol. 2017, 112, 212–239. [Google Scholar] [CrossRef]
- Pohl, D.; Keller, P.M.; Bordier, V.; Wagner, K. Review of current diagnostic methods and advances in Helicobacter pylori diagnostics in the era of next generation sequencing. World J. Gastroenterol. 2019, 25, 4629–4660. [Google Scholar] [CrossRef]
- Redondo, J.J.; Keller, P.M.; Zbinden, B.; Wagner, K. A novel RT-PCR for the detection of Helicobacter pylori and identification of clarithromycin resistance mediated by mutations in the 23S rRNA gene. ScienceDirect 2018, 90, 1–6. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, V.; Zullo, A.; Ierardi, E.; Giorgio, F.; Perna, F.; Hassan, C.; Morini, S.; Panella, C.; Vaira, D. Phenotypic and genotypic Helicobacter pylori clarithromycin resistance and therapeutic outcome: Benefits and limits. J. Antimicrob. Chemother. 2010, 65, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Liou, J.-M.; Chang, C.-Y.; Sheng, W.-H.; Wang, Y.-C.; Chen, M.-J.; Lee, Y.-C.; Hung, H.-W.; Chian, H.; Chang, S.-C.; Wu, M.-S.; et al. Genotypic Resistance in Helicobacter pylori Strains Correlates with Susceptibility Test and Treatment Outcomes after Levofloxacin-And Clarithromycin-Based Therapies. Antimicrob. Agents Chemother. 2011, 55, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Wang, L.; Zhu-Ge, L.; Zhao, R.; Wu, S.; Wang, Y.; An, Y.; Xie, Y. A systematic review and meta-analysis of genotypic methods for detecting antibiotic resistance in Helicobacter pylori. Helicobacter 2018, 23, e12467. [Google Scholar] [CrossRef] [PubMed]
- Binyamin, D.; Pastukh, N.; On, A.; Paritsky, M.; Peretz, A. Phenotypic and genotypic correlation as expressed in Helicobacter pylori resistance to clarithromycin and fluoroquinolones. Gut Pathog. 2017, 9, 48. [Google Scholar] [CrossRef]
- Jenks, P.J.; Ferrero, R.L.; Labigne, A. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J. Antimicrob. Chemother. 1999, 43, 753–758. [Google Scholar] [CrossRef][Green Version]
- Fasciana, T.; Capra, G.; Cala, C.; Zambuto, S.; Mascarella, C.; Colomba, C.; Di Carlo, P.; Giammanco, A. Helicobacter Pylori and Epstein–Barr Co-Infection in Gastric Disease. Phamacol. Online 2017, 1, 73–82. [Google Scholar]
- BioMérieux. E-Test Application Guide. Available online: https://www.biomerieux-usa.com/sites/subsidiary_us/files/supplementary_inserts_-_16273_-_b_-_en_-_eag_-_etest_application_guide-3.pdf (accessed on 16 December 2020).
- Liofilchem. MTS Application Guide. Available online: http://www.liofilchem.net/login.area.mic/technical_sheets/MTS21.pdf (accessed on 16 December 2020).
- British Society for Antimicrobial Chemotheraphy. Antimicrobial Susceptibility Testing Standing Committee Guidance; Susceptibility Testing of Helicobacter Pylori; British Society for Antimicrobial Chemotheraphy: Birmingham, UK, 2014. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- R Development Core Team. A Language and Environment for Statistical Computing. Place of Publication: R Foundation for Statistical Computing; R Development Core Team: Vienna, Austria, 2017; Available online: https://www.R-project.org/ (accessed on 20 December 2020).
- Wickham, H.; Henry, L. Tidyr: Easily Tidy Data with “Spread()” and “Gather()” Functions. R Package Version 0.8.3. 2019. Available online: https://cran.r-project.org/web/packages/tidyr/index.html (accessed on 20 December 2020).
- Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Stetsenko, P.; Short, T.; Lianoglou, S.; Antonyan, E.; Bonsch, M.; Parsonage, H.; et al. Data.Table: Extension of “Data. Frame”. 2020. Available online: https://cran.r-project.org/web/packages/data.table/index.html (accessed on 20 December 2020).
- Wilke, C.O. Cowplot: Streamlined Plot Theme and Plot Annotations for “Ggplot2”. R Package Version 0.9.4. 2019. Available online: https://cran.r-project.org/web/packages/cowplot/index.html (accessed on 20 December 2020).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://cran.r-project.org/web/packages/cowplot/index.html (accessed on 20 December 2020).
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. 2019. Available online: https://rdrr.io/cran/pheatmap/ (accessed on 20 December 2020).
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes. R Package Version 1.1–2. 2014. Available online: https://cran.r-project.org/web/packages/RColorBrewer/index.html (accessed on 20 December 2020).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data. R Package Version 3.0.1. 2016. Available online: https://rdrr.io/cran/gplots/ (accessed on 20 December 2020).
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T. Ggtree: An R Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Yu, G.; Lam, T.T.; Zhu, H.; Guan, Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 3041–30433. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-Platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Tuan, V.P.; Narith, D.; Tshibangu-Kabamba, E.; Dung, H.D.Q.; Viet, P.T.; Sokomoth, S.; Binh, T.T.; Sokhem, S.; Tri, T.D.; Ngov, S.; et al. A Next-Generation Sequencing-Based Approach to Identify Genetic Determinants of Antibiotic Resistance in Cambodian Helicobacter pylori Clinical Isolates. JCM 2019, 8, 858. [Google Scholar] [CrossRef] [PubMed]
- Lauener, F.N.; Imkamp, F.; Lehours, P.; Buissonnière, A.; Benejat, L.; Zbinden, R.; Keller, P.M.; Wagner, K. Genetic Determinants and Prediction of Antibiotic Resistance Phenotypes in Helicobacter pylori. J. Clin. Med. 2019, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Tshibangu-Kabamba, E.; Ngoma-Kisoko, P.D.J.; Tuan, V.P.; Matsumoto, T.; Akada, J.; Kido, Y.; Tshimpi-Wola, A.; Tshiamala-Kashala, P.; Ahuka-Mundeke, S.; Mumba Ngoy, D.; et al. Next-Generation Sequencing of the Whole Bacterial Genome for Tracking Molecular Insight into the Broad-Spectrum Antimicrobial Resistance of Helicobacter pylori Clinical Isolates from the Democratic Republic of Congo. Microorganisms 2020, 8, 887. [Google Scholar] [CrossRef]
- De Francesco, V.; Zullo, A.; Giorgio, F.; Saracino, I.; Zaccaro, C.; Hassan, C.; Ierardi, E.; Di Leo, A.; Fiorini, G.; Castelli, V.; et al. Change of point mutations in Helicobacter pylori rRNA associated with clarithromycin resistance in Italy. J. Med. Microbiol. 2014, 63, 453–457. [Google Scholar] [CrossRef]
- Trespalacios, A.A.; Otero, W.; Caminos, J.E.; Mercado, M.M.; Avila, J.; Rosero, L.E.; Arévalo, A.; Poutou-Piñales, R.A.; Graham, D.Y. Phenotypic and genotypic analysis of clarithromycin-resistant Helicobacter pylori from Bogotá, D.C., Colombia. J. Microbiol. 2013, 51, 448–452. [Google Scholar] [CrossRef]
- Krzyżek, P.; Grande, R. Transformation of Helicobacter pylori into Coccoid Forms as a Challenge for Research Determining Activity of Antimicrobial Substances. Pathogens 2020, 9, 184. [Google Scholar] [CrossRef]
- Eusebi, L.H.; Zagari, R.M.; Bazzoli, F. Epidemiology of Helicobacter pylori infection. Helicobacter 2014, 19, 1–5. [Google Scholar] [CrossRef]
- Besser, J.; Carleton, H.A.; Gerner-Smidt, P.; Lindsey, R.L.; Trees, E. Next-Generation Sequencing Technologies and their Application to the Study and Control of Bacterial Infections. Clin. Microbiol. Infect. 2018, 24, 335–341. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).