Production Optimization, Structural Analysis, and Prebiotic- and Anti-Inflammatory Effects of Gluco-Oligosaccharides Produced by Leuconostoc lactis SBC001
Abstract
1. Introduction
2. Materials and Methods
2.1. Lactobacillus Strains and Growth Conditions
2.2. Optimization of Oligosaccharide Production from Leu. lactis SBC001 Using Experimental Design
2.3. Purification of Oligosaccharides
2.4. Determination of Oligosaccharide Structure
2.4.1. Thin-layer Chromatography
2.4.2. HPAEC-PAD Analysis
2.4.3. Size exclusion HPLC Analysis
2.4.4. Analysis of Monosaccharide Composition
2.4.5. Analysis of the Composition of Glycosidic Bonds by Enzymatic Analysis
2.4.6. Analysis of Linkage Ratio by 1H-NMR Spectroscopy
2.5. Prebiotic Effects of Purified Oligosaccharides
2.5.1. Bacterial and Yeast Strains and Culture Conditions
2.5.2. Media for Bacterial and Yeast Strains for the Determination of Prebiotic Effect
2.5.3. Determination of Viable Cell Number of Bacterial and Yeast Strains
2.6. Anti-inflammatory Activity of Oligosaccharides
2.6.1. Cell Culture
2.6.2. Cytotoxicity on RAW 264.7 Cells
2.6.3. Measurement of Nitric Oxide Production
2.6.4. cDNA Synthesis and Real-time PCR
2.6.5. Western Blotting
2.7. Statistical Analysis
3. Results and Discussion
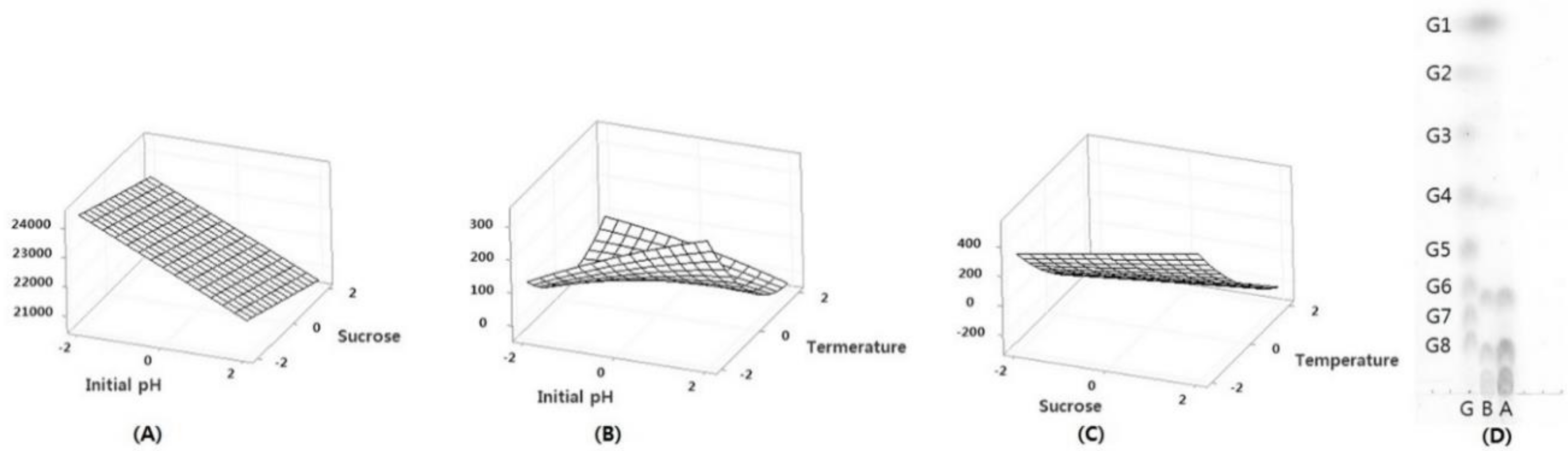
3.1. Optimization of Oligosaccharide Production
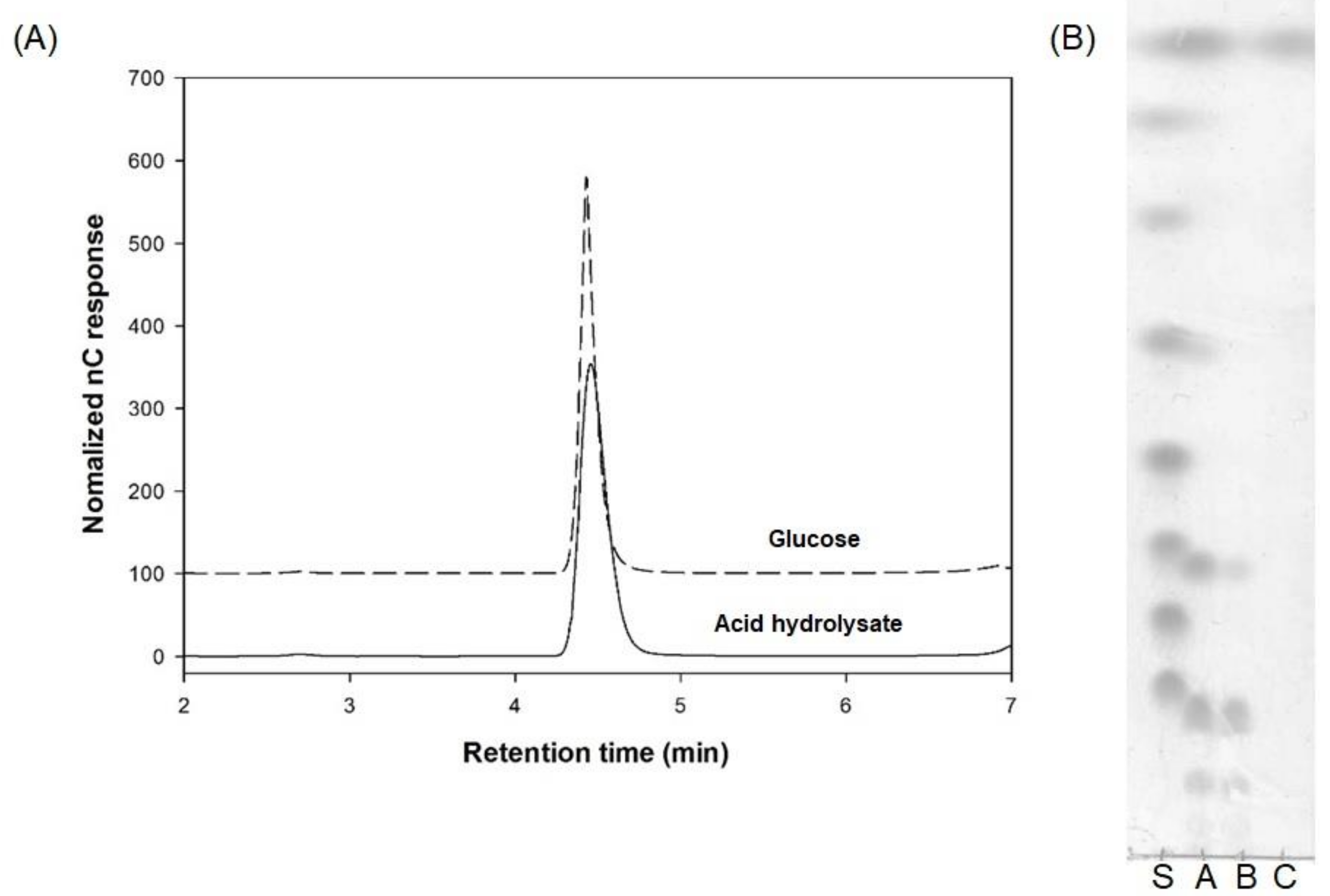
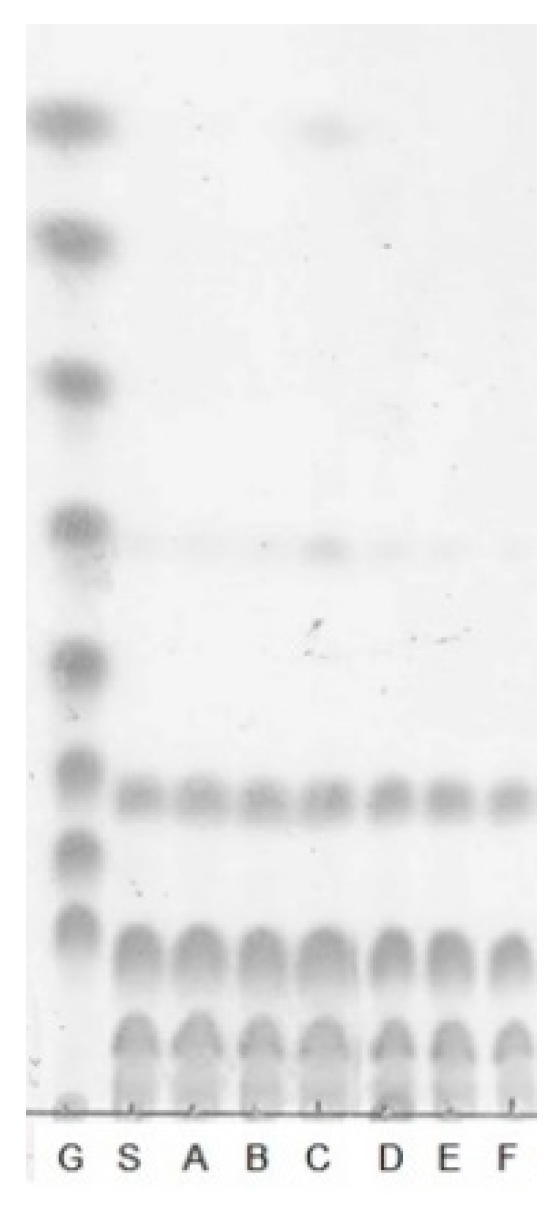
3.2. Structural Analysis of the Oligosaccharides Produced by Leu. lactis SBC001
3.2.1. Analysis of the Monosaccharide Composition of Oligosaccharides
3.2.2. Enzymatic Hydrolysis of SBC-Oligosaccharides
3.2.3. 1H-NMR Analysis
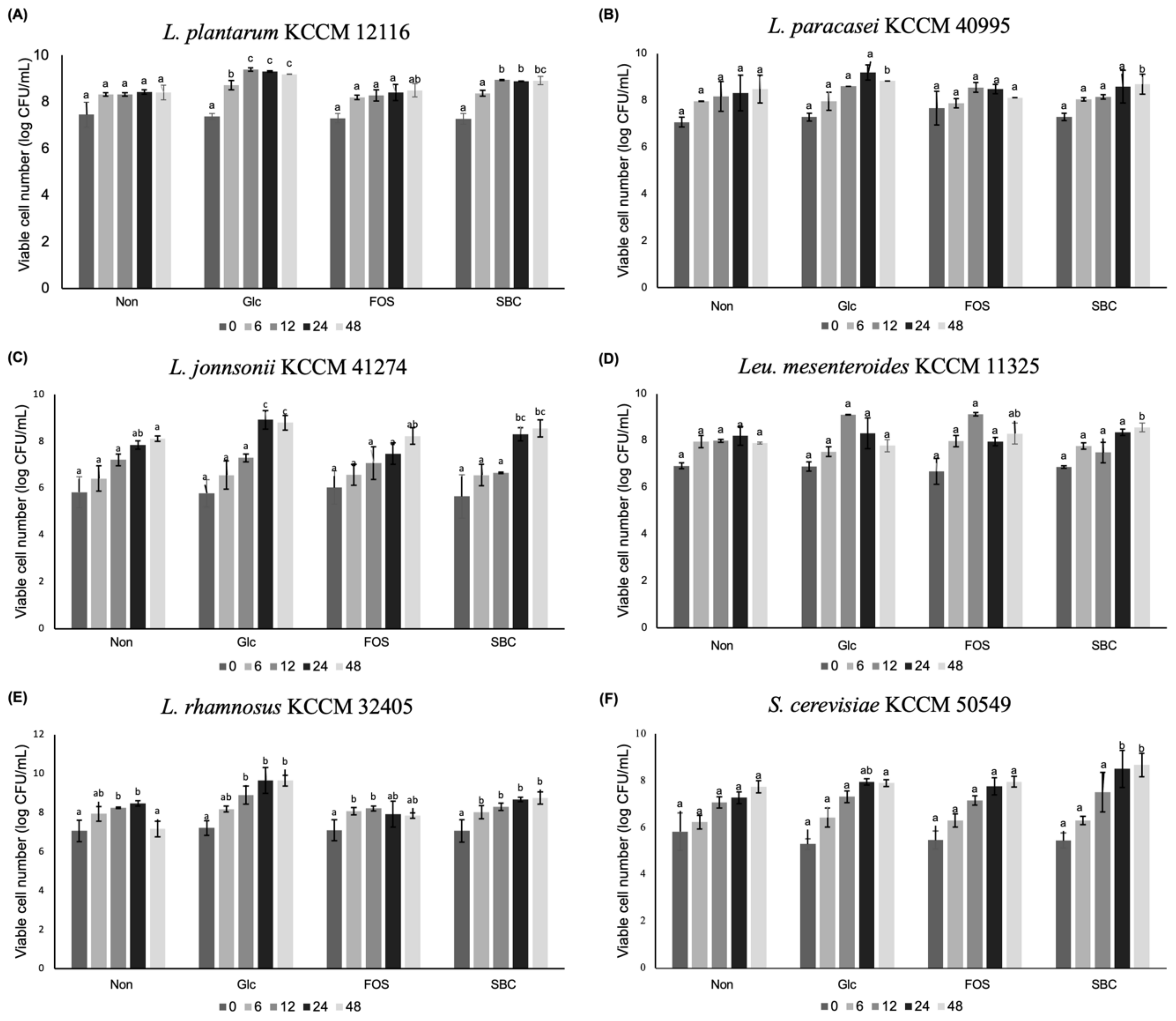
3.3. Prebiotic Effect of Oligosaccharides on the Growth of Bacterial and Yeast Strains
3.4. Anti-Inflammatory Effect of Oligosaccharides
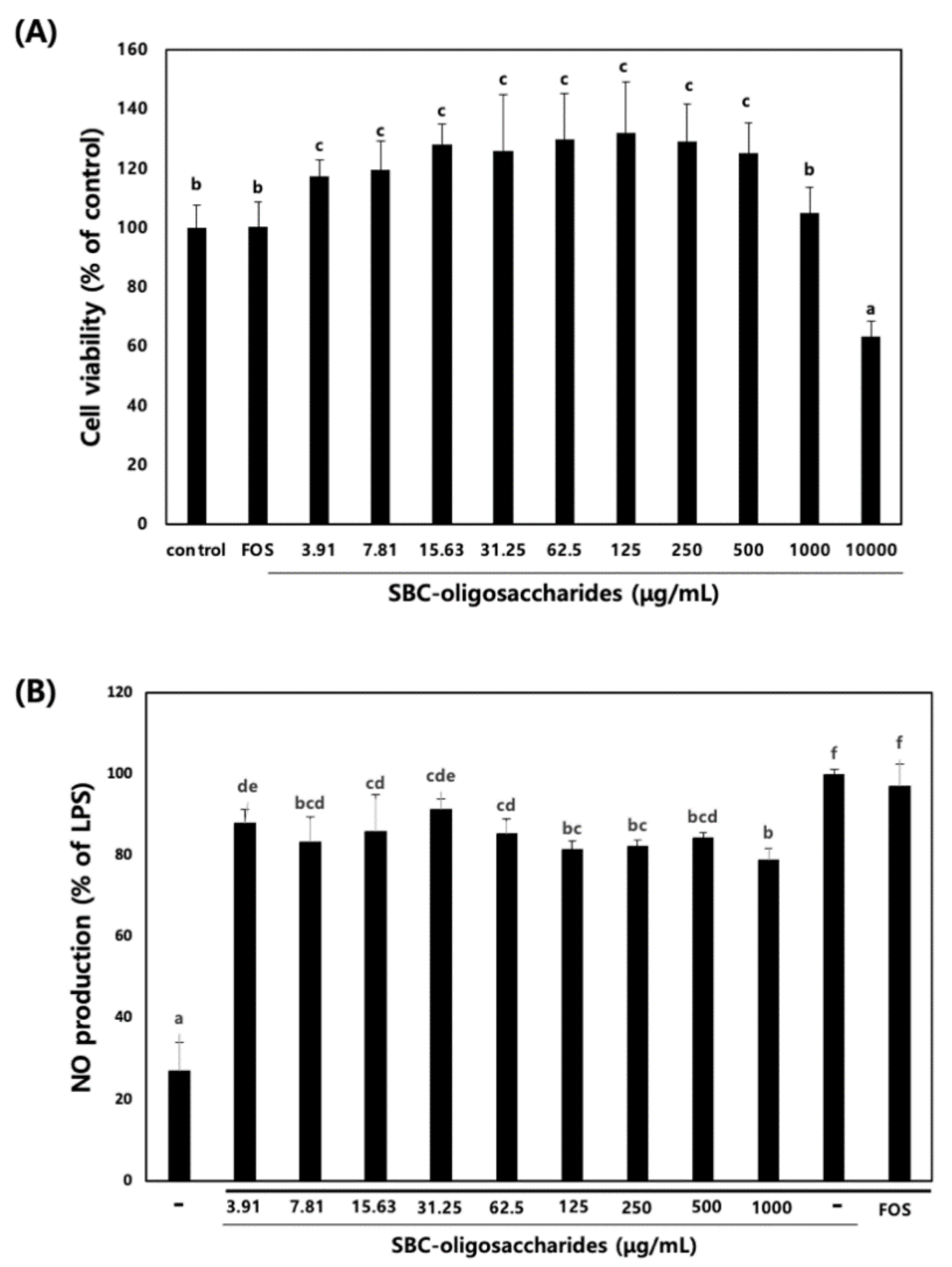
3.4.1. Effect of Oligosaccharides on RAW 264.7 Cell Viability and NO Production
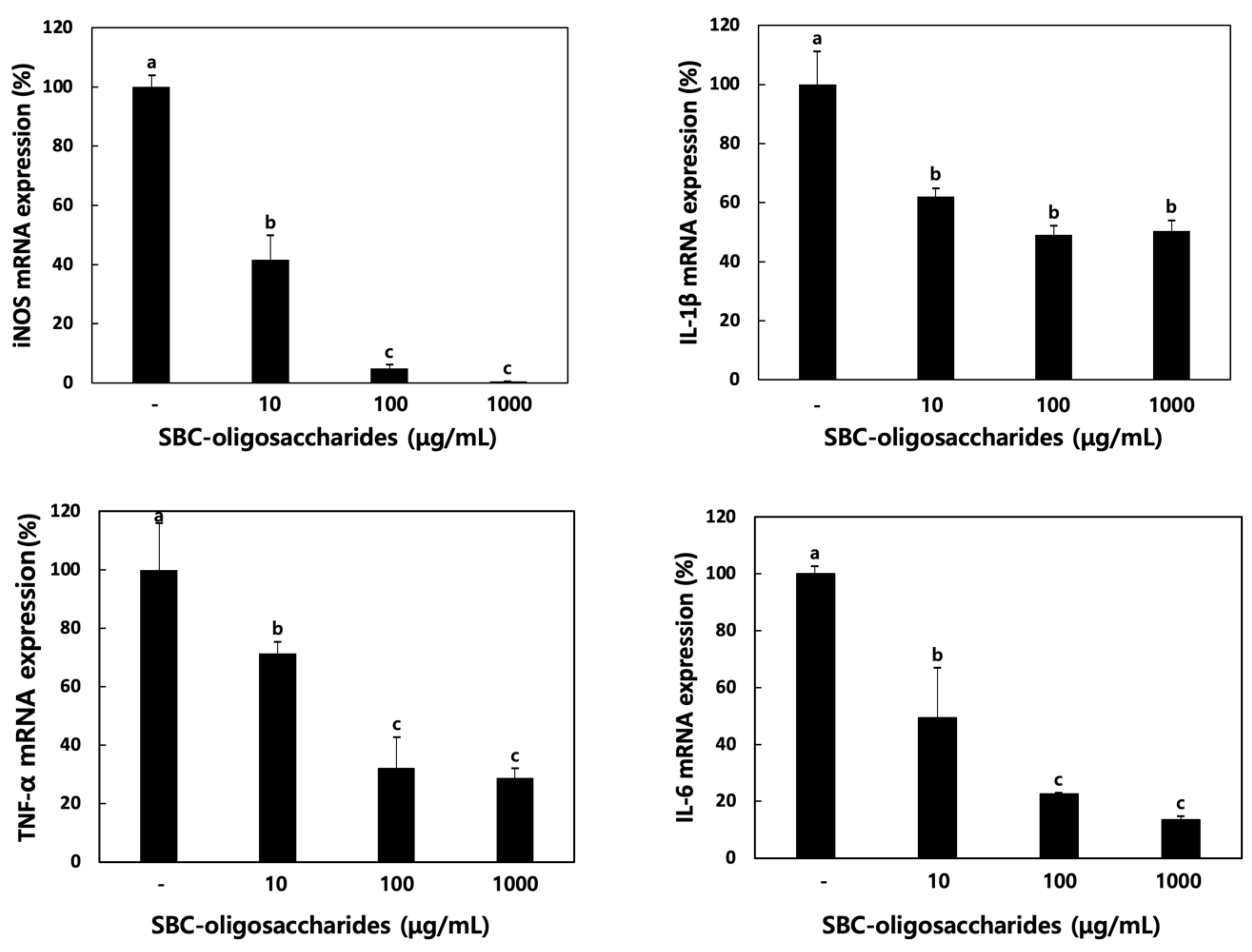
3.4.2. Effect of Oligosaccharides on Cytokine Production
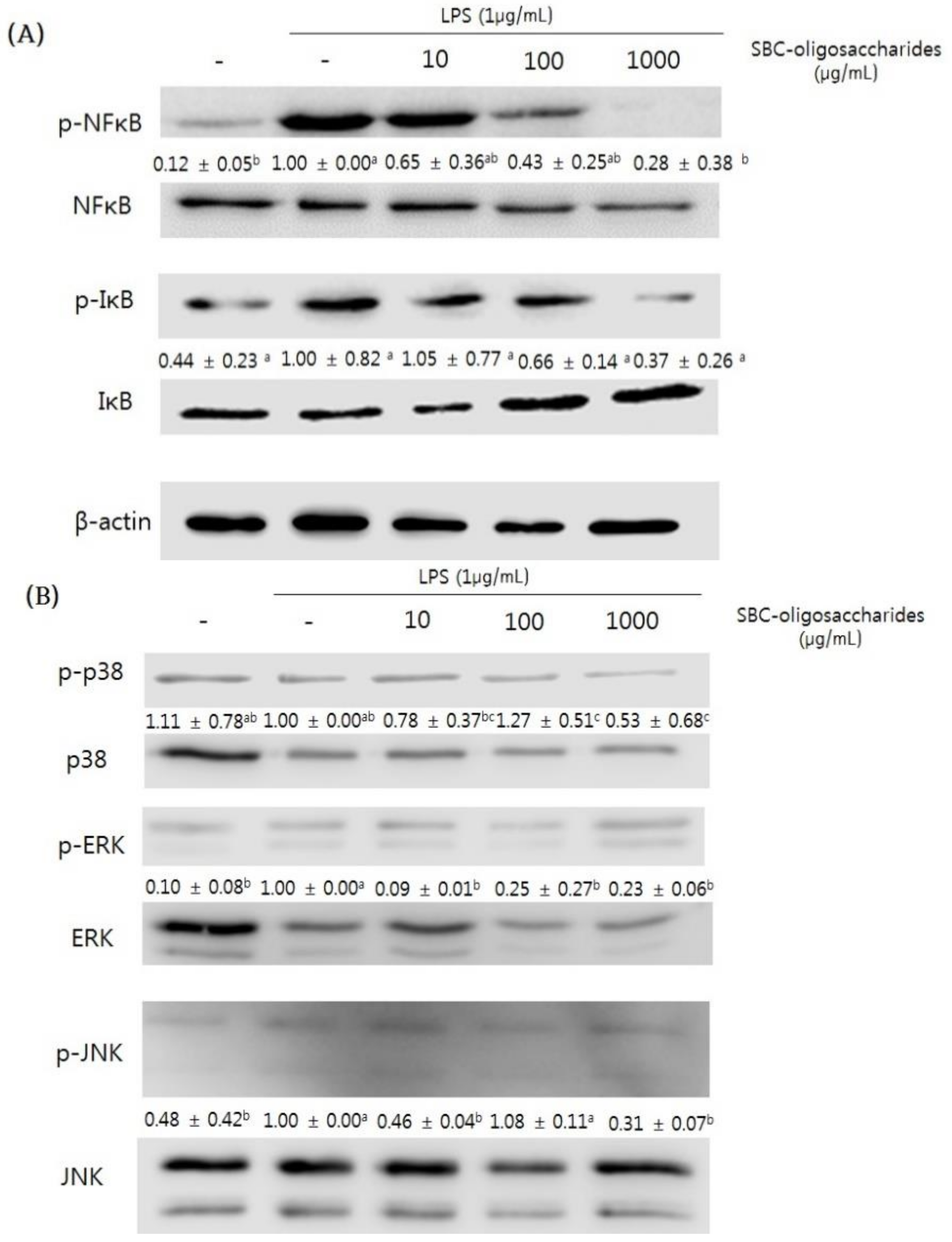
3.4.3. Effect of Oligosaccharides on the MAPK Signaling Pathway of RAW 264.7 Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, E.; Yang, H.; Zou, Y.; Wang, H.; Hu, T.; Li, Q.; Liao, S.J.L. In-vitro digestion by simulated gastrointestinal juices of Lactobacillus rhamnosus cultured with mulberry oligosaccharides and subsequent fermentation with human fecal inocula. LWT 2019, 101, 61–68. [Google Scholar] [CrossRef]
- Akhter, N.; Wu, B.; Memon, A.M.; Mohsin, M.J.F. Probiotics and prebiotics associated with aquaculture: A review. Fish Shellfish Immunol. 2015, 45, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Fuller, R. Probiotics in man and animals. J. Appl. Bacteriol. 1989, 66, 365–378. [Google Scholar] [PubMed]
- Balcázar, J.L.; de Blas, I.; Ruiz-Zarzuela, I.; Cunningham, D.; Vendrell, D.; Múzquiz, J.L. The role of probiotics in aquaculture. Vet. Microbiol. 2006, 114, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Vandenplas, Y.; Zakharova, I.; Dmitrieva, Y. Oligosaccharides in infant formula: More evidence to validate the role of prebiotics. Br. J. Nutr. 2015, 113, 1339–1344. [Google Scholar] [CrossRef]
- Kothari, D.; Patel, S.; Goyal, A. Therapeutic spectrum of nondigestible oligosaccharides: Overview of current state and prospect. J. Food Sci. 2014, 79, R1491–R1498. [Google Scholar] [CrossRef]
- Chapla, D.; Pandit, P.; Shah, A. Production of xylooligosaccharides from corncob xylan by fungal xylanase and their utilization by probiotics. Bioresour. Technol. 2012, 115, 215–221. [Google Scholar] [CrossRef]
- Ben, X.M.; Li, J.; Feng, Z.T.; Shi, S.Y.; Lu, Y.D.; Chen, R.; Zhou, X.Y. Low level of galacto-oligosaccharide in infant formula stimulates growth of intestinal Bifidobacteria and Lactobacilli. World J. Gastroenterol. 2008, 14, 6564–6568. [Google Scholar] [CrossRef]
- Chen, G.; Chen, X.; Yang, B.; Yu, Q.; Wei, X.; Ding, Y.; Kan, J. New insight into bamboo shoot (Chimonobambusa quadrangularis) polysaccharides: Impact of extraction processes on its prebiotic activity. Food Hydrocoll. 2019, 95, 367–377. [Google Scholar] [CrossRef]
- Nawaz, A.; Irshad, S.; Hoseinifar, S.H.; Xiong, H. The functionality of prebiotics as immunostimulant: Evidences from trials on terrestrial and aquatic animals. Fish Shellfish Immunol. 2018, 76, 272–278. [Google Scholar] [CrossRef]
- Costabile, A.; Fava, F.; Röytiö, H.; Forssten, S.D.; Olli, K.; Klievink, J.; Rowland, I.R.; Ouwehand, A.C.; Rastall, R.A.; Gibson, G.R.; et al. Impact of polydextrose on the faecal microbiota: A double–blind, crossover, placebo–controlled feeding study in healthy human subjects. Br. J. Nutr. 2012, 108, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Putaala, H.; Mäkivuokko, H.; Tiihonen, K.; Rautonen, N. Simulated colon fiber metabolome regulates genes involved in cell cycle, apoptosis, and energy metabolism in human colon cancer cells. Mol. Cell. Biochem. 2011, 357, 235–245. [Google Scholar] [CrossRef]
- Lee, S.; Park, G.G.; Jang, J.K.; Park, Y.S. Optimization of oligosaccharide production from Leuconostoc lactis using a response surface methodology and the immunostimulating effects of these oligosaccharides on macrophage cells. Molecules 2018, 23, 2118. [Google Scholar] [CrossRef] [PubMed]
- Teitelbaum, J.E.; Walker, W.A. Nutritional impact of pre- and probiotics as protective gastrointestinal organisms. Ann. Rev. Nutr. 2002, 22, 107–138. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Pijning, T.; Tietema, M.; Dobruchowska, J.M.; Yin, H.; Gerwig, G.J.; Kralj, S.; Dijkhuizen, L. Characterization of the glucansucrase GTF180 W1065 mutant enzymes producing polysaccharides and oligosaccharides with altered linkage composition. Food Chem. 2017, 217, 81–90. [Google Scholar] [CrossRef]
- Lee, S.; Song, I.H.; Park, Y.S. In vivo and In vitro study of immunostimulation by Leuconostoc lactis-produced gluco-oligosaccharides. Molecules 2019, 24, 3994. [Google Scholar] [CrossRef]
- Lee, S.; Park, J.; Jang, J.K.; Lee, B.H.; Park, Y.S. Structural analysis of gluco-oligosaccharides produced by Leuconostoc lactis and their prebiotic effect. Molecules 2019, 24, 3998. [Google Scholar] [CrossRef]
- Cicotello, J.; Wolf, I.V.; D’Angelo, L.; Guglielmotti, D.M.; Quiberoni, A.; Suárez, V.B. Response of Leuconostoc strains against technological stress factors: Growth performance and volatile profiles. Food Microbiol. 2018, 73, 362–370. [Google Scholar] [CrossRef]
- D’Angelo, L.; Cicotello, J.; Zago, M.; Guglielmotti, D.; Quiberoni, A.; Suárez, V. Leuconostoc strains isolated from dairy products: Response against food stress conditions. Food Microbiol. 2017, 66, 28–39. [Google Scholar] [CrossRef]
- Chung, C.H.; Day, D.F. Glucooligosaccharides from Leuconostoc mesenteroides B-742 (ATCC 13146): A potential prebiotic. J. Ind. Microbiol. Biotechol. 2002, 29, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Kothari, D.; Baruah, R.; Goyal, A. Immobilization of glucansucrase for the production of gluco-oligosaccharides from Leuconostoc mesenteroides. Biotechnol. Lett. 2012, 34, 2101–2106. [Google Scholar] [CrossRef]
- Kothari, D.; Goyal, A. Gentio-oligosaccharides from Leuconostoc mesenteroides NRRL B-1426 dextransucrase as prebiotics and as a supplement for functional foods with anti-cancer properties. Food Funct. 2015, 6, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, S.; Park, Y.-S. Prebiotic and Anti-Inflammatory Effects of Oligosaccharides Produced from Leuconostoc lactis SBC001. In Proceedings of the 2019 KoSFoST International Symposium and Annual Meeting, SONGDO Convensia, Incheon, Korea, 26–28 June 2019; Korean Society of Food Science and Technology: Seoul, Korea, 2019; p. 456. [Google Scholar]
- Zou, P.; Yuan, S.; Yang, X.; Guo, Y.; Li, L.; Xu, C.; Zhai, X.; Wang, J. Structural characterization and antitumor effects of chitosan oligosaccharides against orthotopic liver tumor via NF-κB signaling pathway. J. Funct. Foods 2019, 57, 157–165. [Google Scholar] [CrossRef]
- Li, P.-j.; Xia, J.-I.; Nie, Z.-y.; Shan, Y. Pectic oligosaccharides hydrolyzed from orange peel by fungal multi-enzyme complexes and their prebiotic and antibacterial potentials. LWT-Food Sci. Technol. 2016, 69, 203–210. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, Y.; Zhou, W. In vitro and in silico studies of the inhibition activity of anthocyanins against porcine pancreatic α-amylase. J. Funct. Foods 2016, 21, 50–57. [Google Scholar] [CrossRef]
- Lordan, S.; Smyth, T.J.; Soler-Vila, A.; Stanton, C.; Ross, R.P. The α-amylase and α-glucosidase inhibitory effects of Irish seaweed extracts. Food Chem. 2013, 141, 2170–2176. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.A.; Morais, M.A.B.; Terrett, O.M.; Lyczakowski, J.J.; Zanphorlin, L.M.; Ferreira-Filho, J.A.; Tonoli, C.C.C.; Murakami, M.T.; Dupree, P.; Souza, A.P. An engineered GH1 β-glucosidase displays enhanced glucose tolerance and increased sugar release from lignocellulosic materials. Sci. Rep. 2019, 9, 4903. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Chen, W.; Wang, W.; Tan, H.; Li, S.; Yin, H. Effective degradation of curdlan powder by a novel endo-β-1→ 3-glucanase. Carbohydr. Polym. 2018, 201, 122–130. [Google Scholar] [CrossRef]
- González-Alfonso, J.L.; Rodrigo-Frutos, D.; Belmonte-Reche, E.; Peñalver, P.; Poveda, A.; Jiménez-Barbero, J.; Ballesteros, A.O.; Hirose, Y.; Polaina, J.; Morales, J.C.; et al. Enzymatic synthesis of a novel pterostilbene α-glucoside by the combination of cyclodextrin glucanotransferase and amyloglucosidase. Molecules 2018, 23, 1271. [Google Scholar] [CrossRef]
- Aslan, Y.; Sharif, Y.M.; Şahin, Ö. Covalent immobilization of Aspergillus niger amyloglucosidase (ANAG) with ethylenediamine-functionalized and glutaraldehyde-activated active carbon (EFGAAC) obtained from sesame seed shell. Int. J. Biol. Macromol. 2020, 142, 222–231. [Google Scholar] [CrossRef]
- Kahar, U.M.; Chan, K.G.; Sani, M.H.; Noh, N.I.M.; Goh, K.M. Effects of single and co-immobilization on the product specificity of type I pullulanase from Anoxybacillus sp. SK3-4. Int. J. Biol. Macromol. 2017, 104, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Mao, W.; Liu, X.; Wang, S.; Xia, Z.; Cao, S.; Li, L.; Zhang, Q.; Liu, S. Sequence analysis of the pyruvylated galactan sulfate-derived oligosaccharides by negative-ion electrospray tandem mass spectrometry. Carbohydr. Res. 2016, 433, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Wen, J.Y.; Wang, Y.Y.; Bian, J.; Li, M.F.; Peng, F.; Sun, R.C. NMR and ESI–MS spectrometry characterization of autohydrolysis xylo-oligosaccharides separated by gel permeation chromatography. Carbohydr. Polym. 2018, 195, 303–310. [Google Scholar] [CrossRef] [PubMed]
- İspirli, H.; Şimşek, Ö.; Skory, C.; Sağdıç, O.; Dertli, E. Characterization of a 4, 6-α-glucanotransferase from Lactobacillus reuteri E81 and production of malto-oligosaccharides with immune-modulatory roles. Int. J. Biol. Macromol. 2019, 124, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Mano, M.C.R.; Neri-Numa, I.A.; da Silva, J.B.; Paulino, B.N.; Pessoa, M.G.; Pastore, G.M. Oligosaccharide biotechnology: An approach of prebiotic revolution on the industry. Appl. Microbiol. Biotechnol. 2018, 102, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.J.; Yoon, K.Y. Enzymatic hydrolysis of perilla seed meal yields water-soluble dietary fiber as a potential functional carbohydrate source. Food Sci. Biotechnol. 2020, 29, 987–996. [Google Scholar] [CrossRef]
- Kim, H.; Lee, S.J.; Shin, K.S. Characterization of new oligosaccharide converted from cellobiose by novel strain of Bacillus subtilis. Food Sci. Biotechnol. 2018, 27, 37–45. [Google Scholar] [CrossRef]
- Yoon, S.B.; Lee, Y.J.; Park, S.K.; Kim, H.C.; Bae, H.; Kim, H.M.; Ko, S.G.; Choi, H.Y.; Oh, M.S.; Park, W. Anti-inflammatory effects of Scutellaria baicalensis water extract on LPS-activated RAW 264.7 macrophages. J. Ethnopharmacol. 2009, 125, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Delgado, G.T.C.; Thomé, R.; Gabriel, D.L.; Tamashiro, W.M.S.C.; Pastore, G.M. Yacon (Smallanthus sonchifolius)-derived fructooligosaccharides improves the immune parameters in the mouse. Nutr. Res. 2012, 32, 884–892. [Google Scholar] [CrossRef]
- Lara-Villoslada, F.; de Haro, O.; Camuesco, M.; Velasco, A.; Zarzuelo, A.; Xaus, J.; Galvez, J. Short-chain gructooligosaccharides, in spite of being fermented in the upper part of the large intestine, have anti-inflammatory activity in the TNBS model of colitis. Eur. J. Nutr. 2006, 45, 418–425. [Google Scholar] [CrossRef]
- Chun, S.Y.; Lee, K.S.; Nam, K.S. Refined deep-sea water suppresses inflammatory responses via the MAPK/AP-1 and NF-κB signaling pathway in LPS-treated RAW 264.7 macrophage cells. Int. J. Mol. Sci. 2017, 18, 2282. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.T.; Ignarro, L.J. Lipopolysaccharide-induced expression of interferon-β mediates the timing of inducible nitric-oxide synthase induction in RAW 264.7 macrophages. J. Biol. Chem. 2001, 276, 47950–47957. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, R.; Lahti, A.; Kankaanranta, H.; Moilanen, E. Nitric oxide production and signaling in inflammation. Inflam. Allergy Drug Targets 2005, 4, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Hirano, T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 2002, 13, 357–368. [Google Scholar] [CrossRef]
- Kim, K.N.; Heo, S.J.; Yoon, W.J.; Kang, S.M.; Ahn, G.; Yi, T.H.; Jeon, Y.J. Fucoxanthin inhibits the inflammatory response by suppressing the activation of NF-κB and MAPKs in lipopolysaccharide-induced RAW 264.7 macrophages. Eur. J. Pharmacol. 2010, 649, 369–375. [Google Scholar] [CrossRef]
- Jung, W.K.; Choi, I.; Lee, D.Y.; Yea, S.S.; Choi, Y.H.; Kim, M.M.; Park, S.G.; Seo, S.K.; Lee, S.W.; Lee, C.M. Caffeic acid phenethyl ester protects mice from lethal endotoxin shock and inhibits lipopolysaccharide-induced cyclooxygenase-2 and inducible nitric oxide synthase expression in RAW 264.7 macrophages via the p38/ERK and NF-κB pathways. Int. J. Biochem. Cell Biol. 2008, 40, 2572–2582. [Google Scholar] [CrossRef]
- Zhang, D.K.; Cheng, L.N.; Huang, X.L.; Shi, W.; Xiang, J.Y.; Gan, H.T. Tetrandrine ameliorates dextran-sulfate-sodium-induced colitis in mice through inhibition of nuclear factor-κB activation. Int. J. Colorectal Dis. 2009, 24, 5–12. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, Y.; Ma, L.; Cao, X.; Xiao, J.; Chen, J.; Jiao, S.; Gao, Y.; Liu, C.; Duan, Z. Activation of vascular endothelial growth factor receptor-3 in macrophages restrains TLR4-NF-κB signaling and protects against endotoxin shock. Immunity 2014, 40, 501–514. [Google Scholar] [CrossRef]
- Hwang, D.; Kang, M.J.; Jo, M.J.; Seo, Y.B.; Park, N.G.; Kim, G.-D. Anti-inflammatory activity of β-thymosin peptide derived from pacific oyster (Crassostrea gigas) on NO and PGE2 production by down-regulating NF-κB in LPS-induced RAW264.7 macrophage cells. Mar. Drugs 2019, 17, 129. [Google Scholar] [CrossRef]
- Karin, M.; Lawrence, T.; Nizet, V. Innate immunity gone awry: Linking microbial infections to chronic inflammation and cancer. Cell 2006, 124, 823–835. [Google Scholar] [CrossRef]
- Yoon, H.J.; Moon, M.E.; Park, H.S.; Kim, H.W.; Im, S.Y.; Lee, J.H.; Kim, Y.H. Effects of chitosan oligosaccharide (COS) on the glycerol-induced acute renal failure in vitro and in vivo. Food Chem. Toxicol. 2008, 46, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Pangestuti, R.; Bak, S.S.; Kim, S.K. Attenuation of pro-inflammatory mediators in LPS-stimulated BV2 microglia by chitooligosaccharides via the MAPK signaling pathway. Int. J. Biol. Macromol. 2011, 49, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.S.F.; Meijerink, M.; Zeuner, B.; Holck, J.; Louis, P.; Meyer, A.S.; Wells, J.M.; Flint, H.J.; Duncan, S.H. Prebiotic potential of pectin and pectic oligosaccharides to promote anti-inflammatory commensal bacteria in the human colon. FEMS Microbiol. Ecol. 2017, 93, 127. [Google Scholar] [CrossRef] [PubMed]

| Gene | Primer Sequence | |
|---|---|---|
| iNOS | Forward Reverse | 5′-AATGGCAACATCAGGTCGGCCATCACT-3 ′5′-GCTGTGTGTCACAGAAGTCTCGAACTC-3′ |
| IL-6 | Forward Reverse | 5′-CAAGAGACTTCCATCCAGTTGC-3 ′5′-TTGCCGAGTTCTCAAAGTGAC-3′ |
| IL-1β | Forward Reverse | 5′-ATGGCAACTGTTCCTGAACTCAACT-3 ′5′-CAGGACAGGTATAGATTCTTTCCTT-3′ |
| TNF-α | Forward Reverse | 5′-ATGAGCACAGAAAGCATGATCCG-3 ′5′-CCAAAGTAGACCTGCCCGGACTC-3′ |
| GAPDH | Forward Reverse | 5′-ATCCCATCACCATCTTCCAG-3 ′5′-CCTGCTTCACCACCTTCTTG-3′ |
| Run | Factor Value | SBC-Oligosaccharide Production (Relative Peak Area) | ||
|---|---|---|---|---|
| Initial pH | Sucrose (M) | Temperature (°C) | ||
| 1 | 5.0 (−1) | 0.2 (−1) | 29 (−1) | 117.43 ± 2.71 g |
| 2 | 7.0 (1) | 0.2 (−1) | 29 (−1) | 154.47 ± 5.60 i |
| 3 | 5.0 (−1) | 0.4 (1) | 29 (−1) | 134.52 ± 2.92 h |
| 4 | 7.0 (1) | 0.4 (1) | 29 (−1) | 209.44 ± 0.60 k |
| 5 | 5.0 (−1) | 0.2 (−1) | 37 (1) | 28.98 ± 1.97 b |
| 6 | 7.0 (1) | 0.2 (−1) | 37 (1) | 5.02 ± 0.67 a |
| 7 | 5.0 (1) | 0.4 (1) | 37 (1) | 33.81 ± 0.15 bc |
| 8 | 4.0 (−2) | 0.3 (0) | 33 (0) | 5.47 ± 2.09 a |
| 9 | 8.0 (2) | 0.3 (0) | 33 (0) | 1.28 ± 0.34 a |
| 10 | 6.0(0) | 0.1 (−2) | 33 (0) | 57.62 ± 2.66 e |
| 11 | 6.0 (0) | 0.5 (2) | 33 (0) | 37.76 ± 8.23 bc |
| 12 | 6.0 (0) | 0.3 (0) | 41 (2) | 53.50 ± 2.23 de |
| 13 | 6.0 (0) | 0.3 (0) | 25 (−2) | 185.60 ± 1.14 j |
| 14 | 6.0 (0) | 0.3 (0) | 41 (2) | 76.03 ± 0.25 f |
| 15 | 6.0 (0) | 0.3 (0) | 33 (0) | 57.67 ± 21.04 e |
| 16 | 6.0 (0) | 0.3 (0) | 33 (0) | 44.46 ± 2.84 cde |
| 17 | 6.0 (0) | 0.3 (0) | 33 (0) | 53.30 ± 10.6 de |
| 18 | 6.0 (0) | 0.3 (0) | 33 (0) | 56.38 ± 10.61 de |
| 19 | 6.0 (0) | 0.3 (0) | 33 (0) | 42.88 ± 3.68 bcd |
| 20 | 6.0 (0) | 0.3 (0) | 33 (0) | 49.78 ± 1.71 de |
| Variables | DF | Adj SS | Adj MS | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 9 | 114,385 | 12,709.4 | 19.59 | 0.000 |
| Linear | 3 | 32,181 | 107,26.9 | 16.54 | 0.000 |
| Quadratic | 3 | 28,351 | 9450.3 | 14.57 | 0.000 |
| Cross-product | 3 | 8162 | 2720.7 | 4.19 | 0.014 |
| X1 | 1 | 7033 | 7032.5 | 10.84 | 0.003 |
| X2 | 1 | 827 | 826.8 | 1.27 | 0.268 |
| X3 | 1 | 18,490 | 18,490.3 | 28.5 | 0.000 |
| X12 | 1 | 549 | 549.4 | 0.85 | 0.365 |
| X22 | 1 | 5 | 4.8 | 0.01 | 0.932 |
| X32 | 1 | 24,461 | 24,461 | 37.71 | 0.000 |
| X1X2 | 1 | 286 | 285.9 | 0.44 | 0.512 |
| X1X3 | 1 | 6772 | 6771.7 | 10.44 | 0.003 |
| X2X3 | 1 | 1104 | 1104.5 | 1.7 | 0.202 |
| Residual | 50 | 19,461 | 648.7 | - | - |
| Lack of fit | 5 | 18,323 | 3664.6 | 80.5 | 0.000 |
| Prue error | 45 | 1138 | 45.5 | - | - |
| Cor Total | 59 | 133,846 | - | - | - |
| α-1,4 Glycosidic Linkages (%) | α-1,6 Glycosidic Linkages (%) | Ratio ofα-1,4 to α-1,6 | |
|---|---|---|---|
| Waxy corn starch | 96.9 ± 0.11 | 3.11 ± 0.11 | 31.22 ± 1.10 |
| Oligosaccharides produced by Leu. lactis SBC001 | 23.63 ± 0.91 | 76.37 ± 0.91 | 0.31 ± 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Jang, J.-K.; Park, Y.-S. Production Optimization, Structural Analysis, and Prebiotic- and Anti-Inflammatory Effects of Gluco-Oligosaccharides Produced by Leuconostoc lactis SBC001. Microorganisms 2021, 9, 200. https://doi.org/10.3390/microorganisms9010200
Kim M, Jang J-K, Park Y-S. Production Optimization, Structural Analysis, and Prebiotic- and Anti-Inflammatory Effects of Gluco-Oligosaccharides Produced by Leuconostoc lactis SBC001. Microorganisms. 2021; 9(1):200. https://doi.org/10.3390/microorganisms9010200
Chicago/Turabian StyleKim, Minhui, Jae-Kweon Jang, and Young-Seo Park. 2021. "Production Optimization, Structural Analysis, and Prebiotic- and Anti-Inflammatory Effects of Gluco-Oligosaccharides Produced by Leuconostoc lactis SBC001" Microorganisms 9, no. 1: 200. https://doi.org/10.3390/microorganisms9010200
APA StyleKim, M., Jang, J.-K., & Park, Y.-S. (2021). Production Optimization, Structural Analysis, and Prebiotic- and Anti-Inflammatory Effects of Gluco-Oligosaccharides Produced by Leuconostoc lactis SBC001. Microorganisms, 9(1), 200. https://doi.org/10.3390/microorganisms9010200





