Identification of an Acidic Amino Acid Permease Involved in d-Aspartate Uptake in the Yeast Cryptococcus humicola
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Strains, Media, and Growth Conditions
2.3. Identification of Aap Homologs
2.4. DNA and RNA Preparation
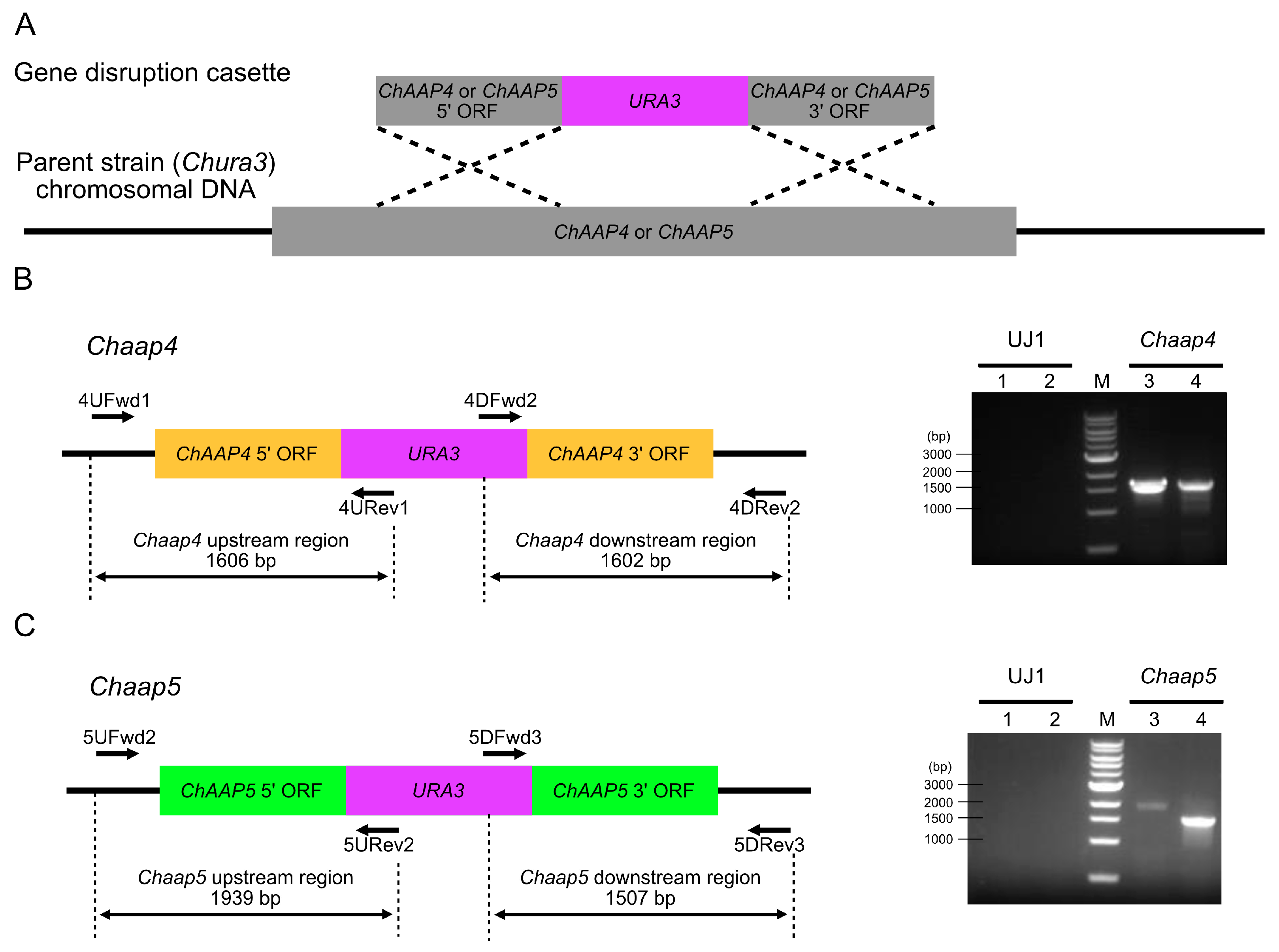
2.5. Disruption of ChAAP Genes
2.6. ChDDO Induction Experiment
2.7. DDO Assay
2.8. Quantitative Real-Time RT-PCR (qRT-PCR)
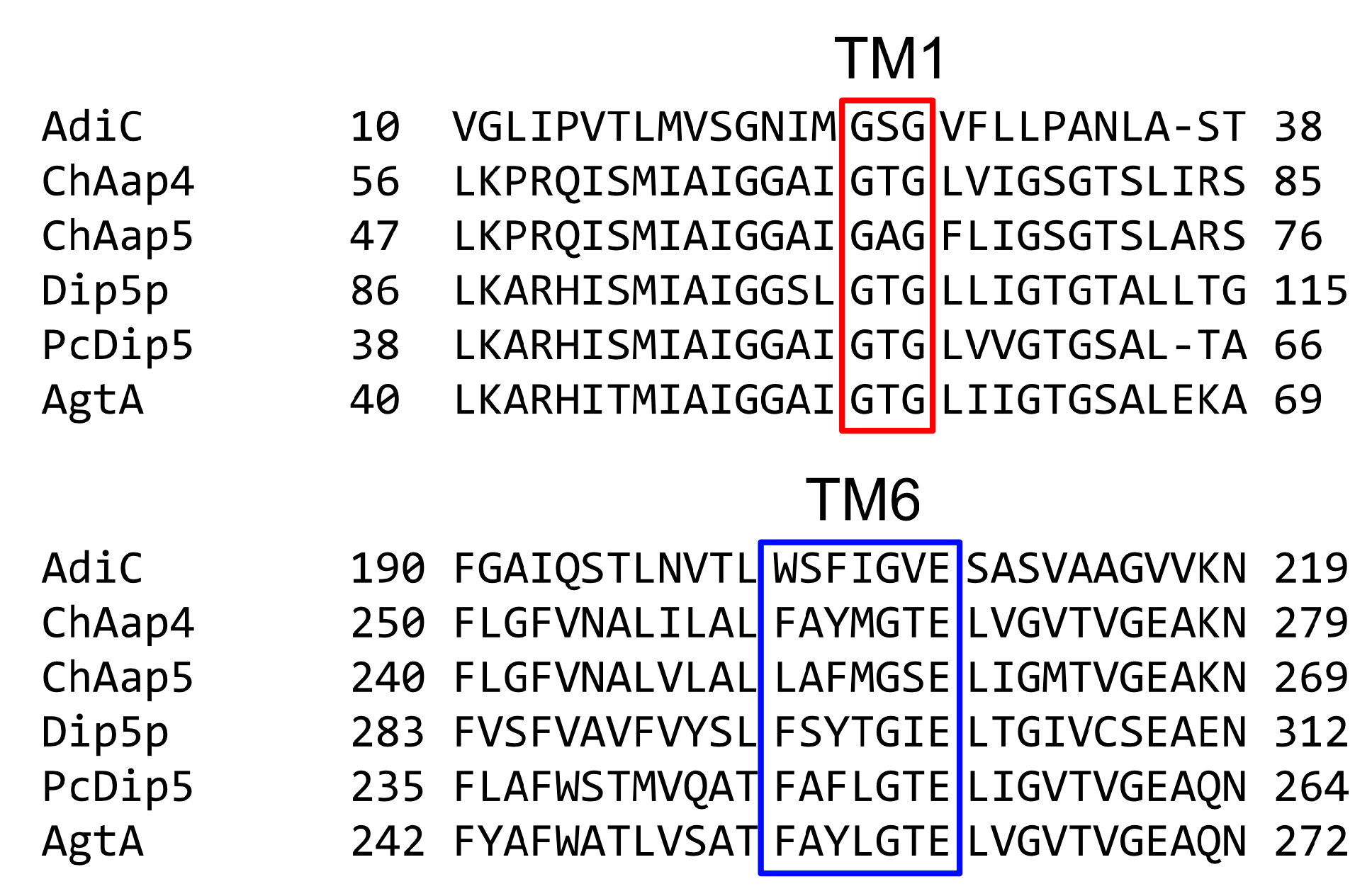
2.9. Sequence Analyses and Structural Modeling
3. Results
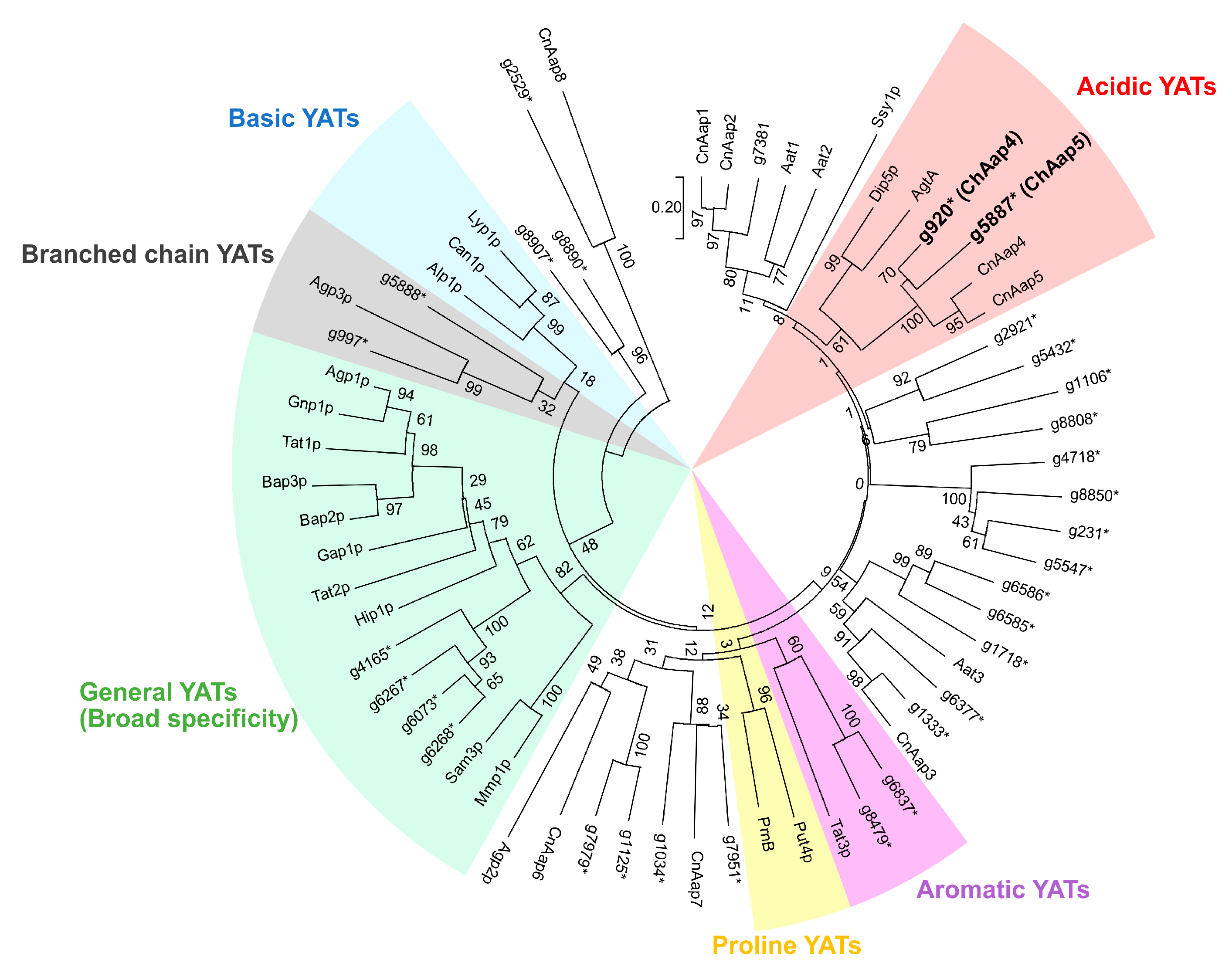
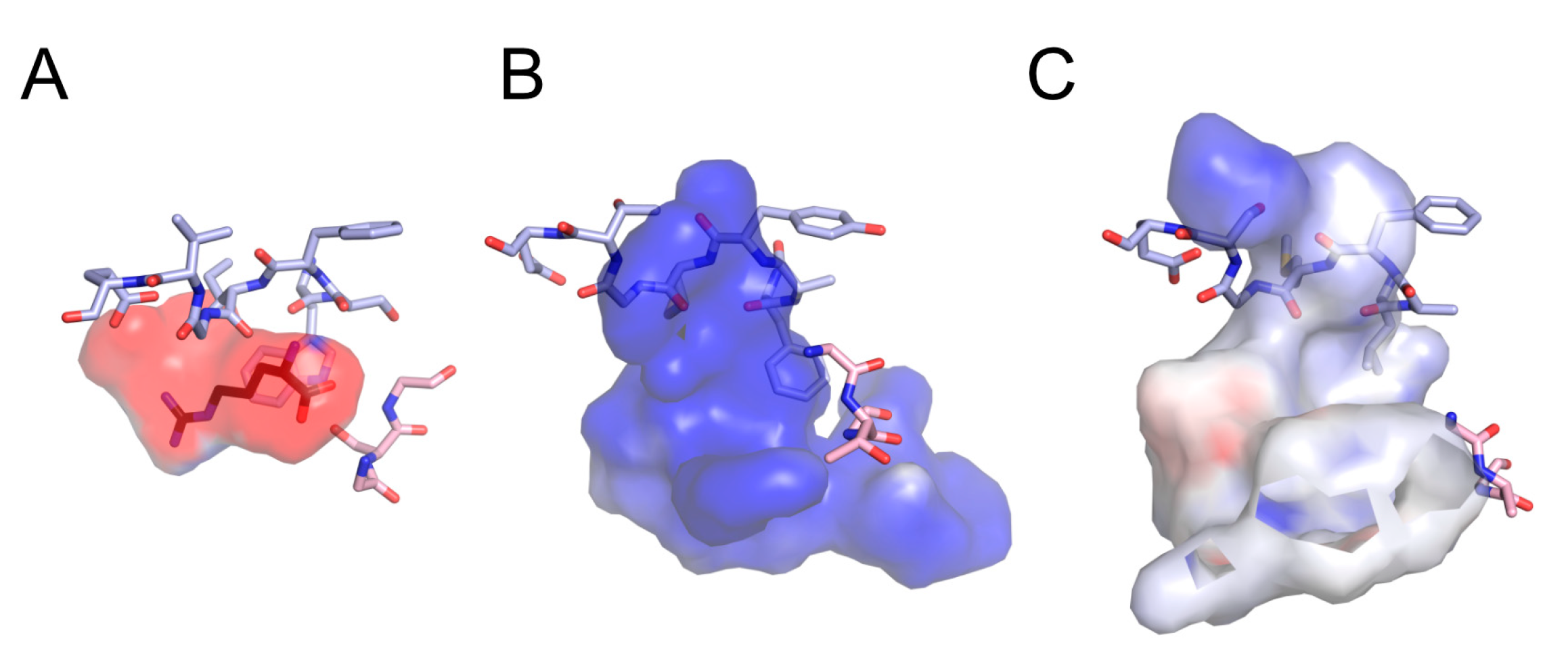
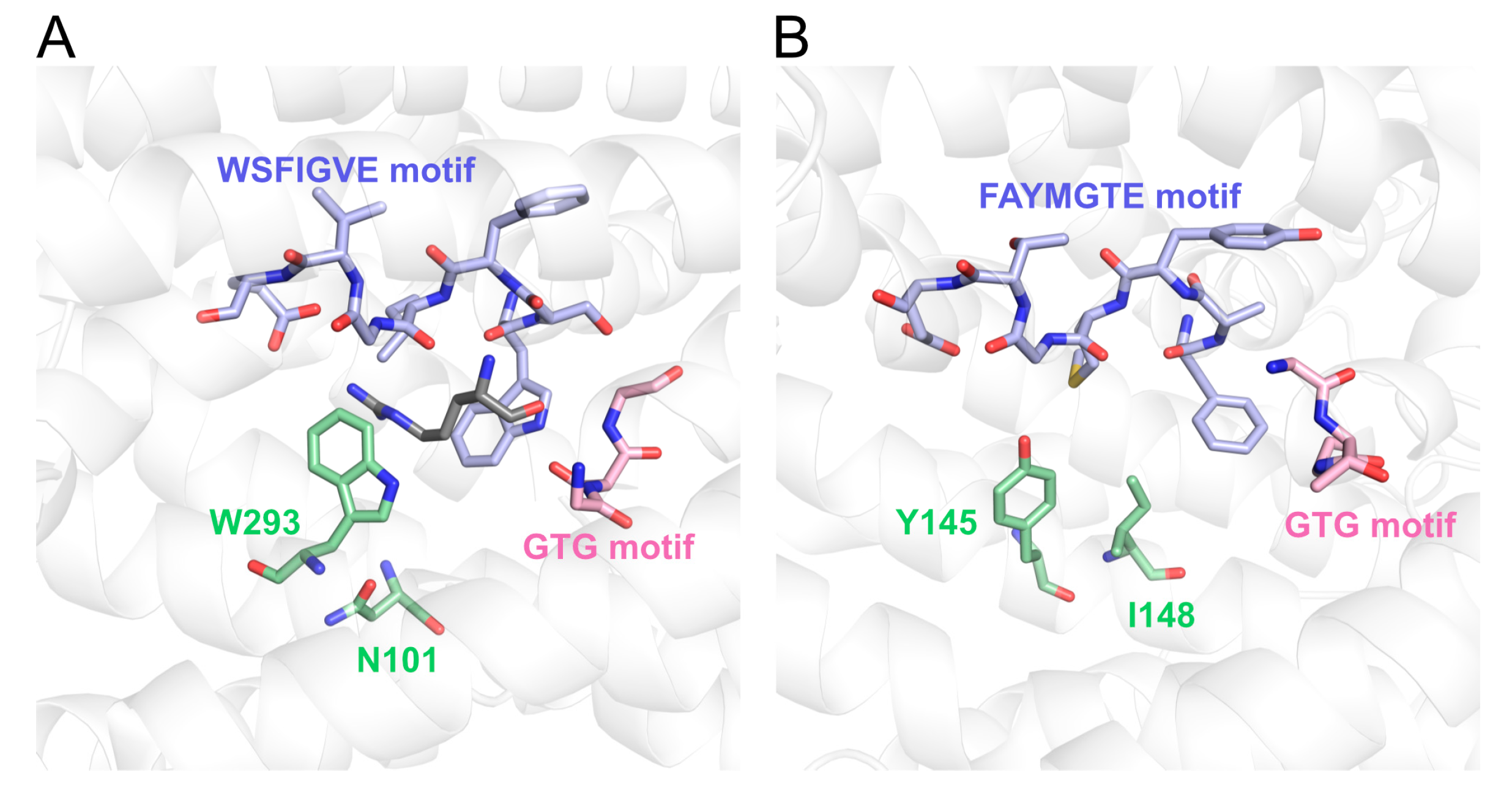
3.1. Identification of Acidic Aap Homologs of C. humicola Strain UJ1
3.2. Growth Characteristics of Chaap4 and Chaap5 Strains on Amino Acids
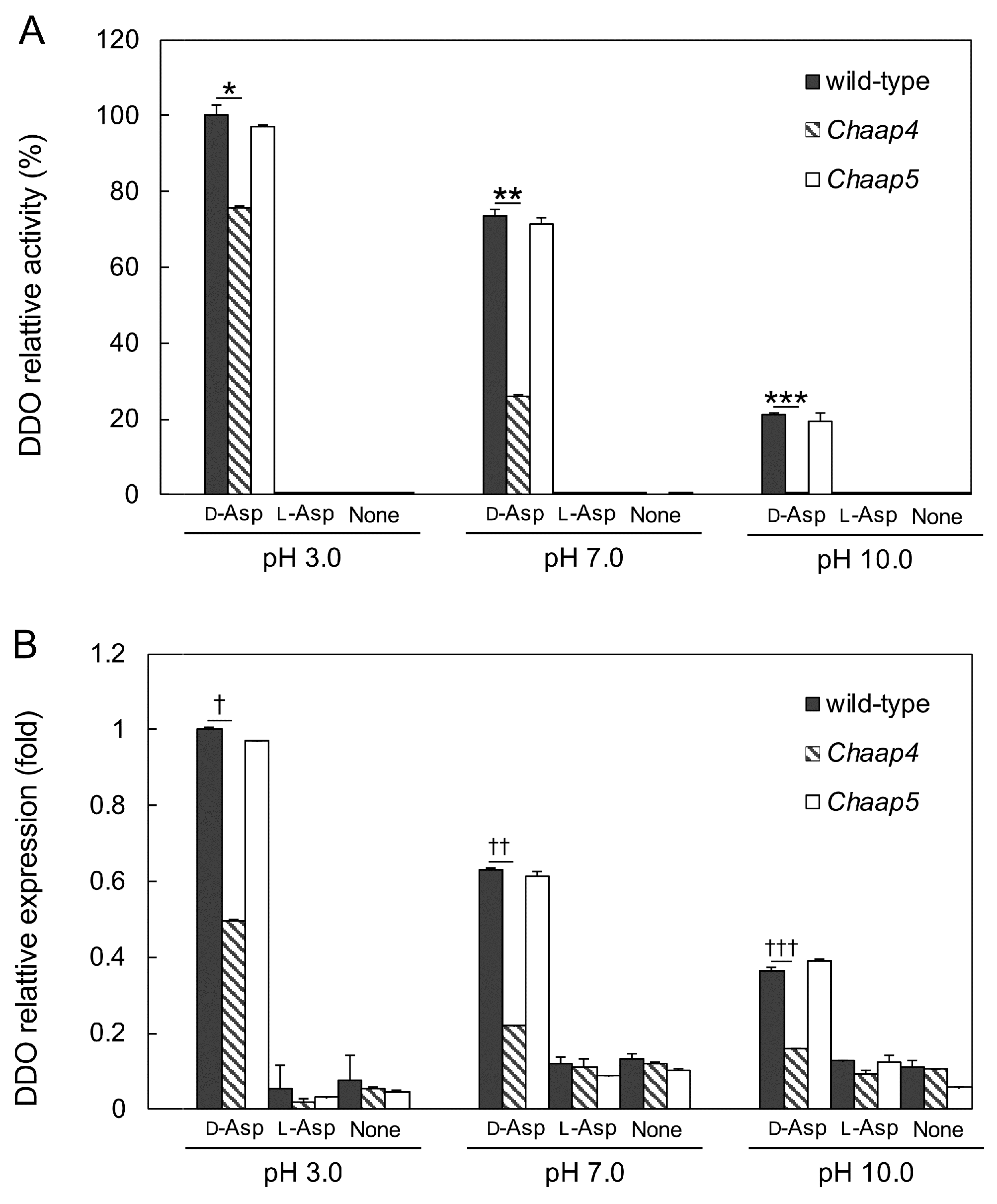
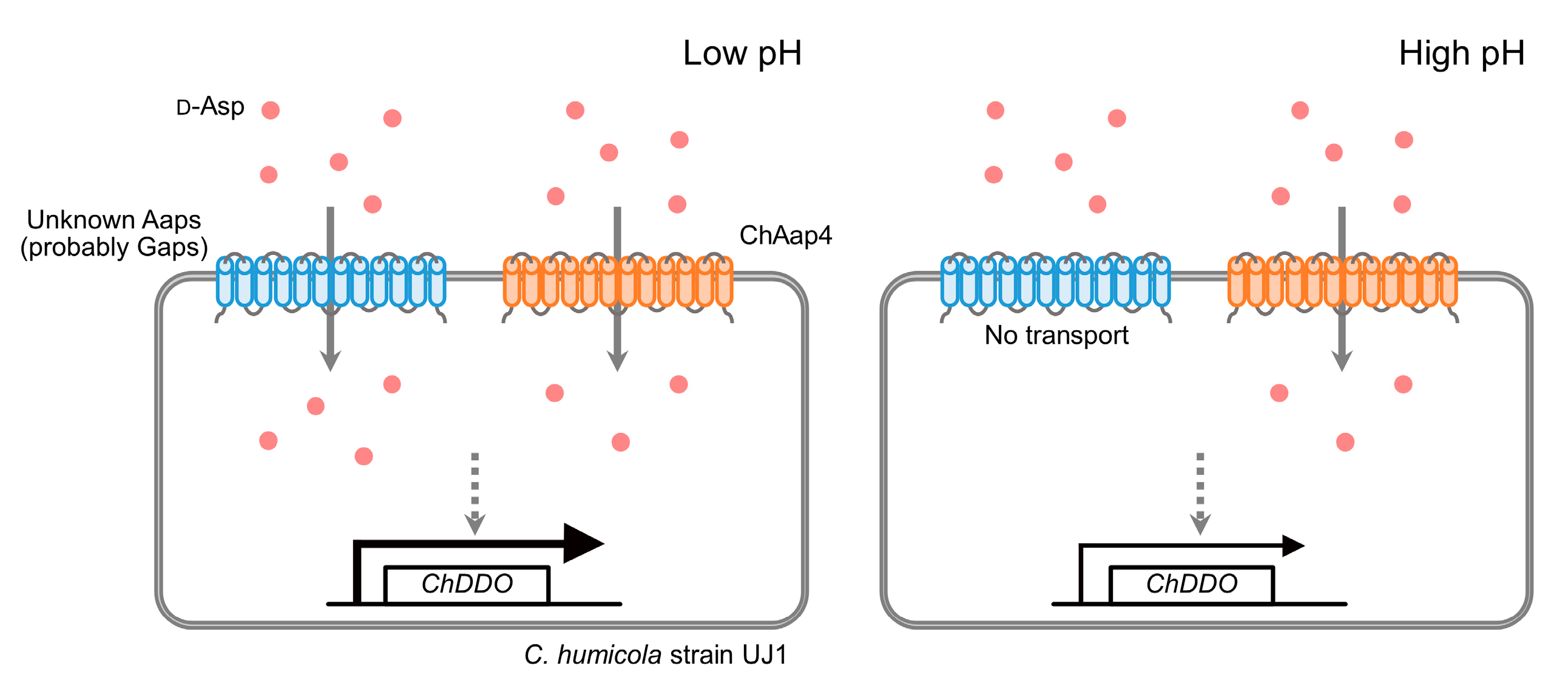
3.3. Effect of Medium pH on the Growth of Chaap4 and Chaap5 Strains on Amino Acids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- D’Aniello, A.; Palescandolo, R.; Scardi, B. The distribution of the d-aspartate oxidase activity in Cephalopoda. Comp. Biochem. Physiol. B 1975, 50, 209–210. [Google Scholar] [CrossRef]
- Yamada, R.; Nagasaki, H.; Wakabayashi, Y.; Iwashima, A. Presence of d-aspartate oxidase in rat liver and mouse tissues. Biochim. Biophys. Acta 1988, 965, 202–205. [Google Scholar] [CrossRef]
- Kera, Y.; Nagasaki, H.; Iwashima, A.; Yamada, R. Presence of d-aspartate oxidase and free d-aspartate in amphibian (Xenopus laevis, Cynops pyrrhogaster) tissues. Comp. Biochem. Physiol. B 1992, 103, 345–348. [Google Scholar] [CrossRef]
- Kera, Y.; Aoyama, H.; Watanabe, N.; Yamada, R.H. Distribution of d-aspartate oxidase and free d-glutamate and d-aspartate in chicken and pigeon tissues. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1996, 115, 121–126. [Google Scholar] [CrossRef]
- Yamada, R.; Ujiie, H.; Kera, Y.; Nakase, T.; Kitagawa, K.; Imasaka, T.; Arimoto, K.; Takahashi, M.; Matsumura, Y. Purification and properties of d-aspartate oxidase from Cryptococcus humicolus UJ1. Biochim. Biophys. Acta 1996, 1294, 153–158. [Google Scholar] [CrossRef]
- Takahashi, S. d-Aspartate Oxidase: Distribution, Functions, Properties, and Biotechnological Applications. Appl. Microbiol. Biotechnol. 2020, 104, 2883–2895. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Takahashi, T.; Kera, Y.; Matsunaga, R.; Shibuya, H.; Yamada, R.H. Cloning and expression in Escherichia coli of the d-aspartate oxidase gene from the yeast Cryptococcus humicola and characterization of the recombinant enzyme. J. Biochem. 2004, 135, 533–540. [Google Scholar] [CrossRef]
- Takahashi, S.; Kakuichi, T.; Fujii, K.; Kera, Y.; Yamada, R.H. Physiological role of d-aspartate oxidase in the assimilation and detoxification of d-aspartate in the yeast Cryptococcus humicola. Yeast 2005, 22, 1203–1212. [Google Scholar] [CrossRef]
- D’Aniello, A.; D’Onofrio, G.; Pischetola, M.; D’Aniello, G.; Vetere, A.; Petrucelli, L.; Fisher, G.H. Biological role of d-amino acid oxidase and d-aspartate oxidase. Effects of d-amino acids. J. Biol. Chem. 1993, 268, 26941–26949. [Google Scholar] [CrossRef]
- Huang, A.S.; Beigneux, A.; Weil, Z.M.; Kim, P.M.; Molliver, M.E.; Blackshaw, S.; Nelson, R.J.; Young, S.G.; Snyder, S.H. d-Aspartate regulates melanocortin formation and function: Behavioral alterations in d-aspartate oxidase-deficient mice. J. Neurosci. 2006, 26, 2814–2819. [Google Scholar] [CrossRef]
- Errico, F.; Nuzzo, T.; Carella, M.; Bertolino, A.; Usiello, A. The Emerging Role of Altered d-Aspartate Metabolism in Schizophrenia: New Insights from Preclinical Models and Human Studies. Front. Psychiatry 2018, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Simonetta, M.P.; Verga, R.; Fretta, A.; Hanozet, G.M. Induction of d-amino-acid oxidase by d-alanine in Rhodotorula gracilis grown in defined medium. Microbiology 1989, 135, 593–600. [Google Scholar] [CrossRef][Green Version]
- Godard, P.; Urrestarazu, A.; Vissers, S.; Kontos, K.; Bontempi, G.; van Helden, J.; Andre, B. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2007, 27, 3065–3086. [Google Scholar] [CrossRef] [PubMed]
- Wakayama, M.; Nakashima, S.; Sakai, K.; Moriguchi, M. Isolation, Enzyme-Production and Characterization of d-Aspartate Oxidase from Fusarium-Sacchari var. Elongatum Y-105. J. Ferment. Bioeng. 1994, 78, 377–379. [Google Scholar] [CrossRef]
- Fukunaga, S.; Yuno, S.; Takahashi, M.; Taguchi, S.; Kera, Y.; Odani, S.; Yamada, R.H. Purification and properties of d-glutamate oxidase from Candida boidinii 2201. J. Ferment. Bioeng. 1998, 85, 579–583. [Google Scholar] [CrossRef]
- Yamada, R.; Nagasaki, H.; Nagata, Y.; Wakabayashi, Y.; Iwashima, A. Administration of d-aspartate increases d-aspartate oxidase activity in mouse liver. Biochim. Biophys. Acta 1989, 990, 325–328. [Google Scholar] [CrossRef]
- Gournas, C.; Athanasopoulos, A.; Sophianopoulou, V. On the Evolution of Specificity in Members of the Yeast Amino Acid Transporter Family as Parts of Specific Metabolic Pathways. Int. J. Mol. Sci. 2018, 19, 1398. [Google Scholar] [CrossRef]
- Reizer, J.; Finley, K.; Kakuda, D.; MacLeod, C.L.; Reizer, A.; Saier, M.H., Jr. Mammalian integral membrane receptors are homologous to facilitators and antiporters of yeast, fungi, and eubacteria. Protein Sci. 1993, 2, 20–30. [Google Scholar] [CrossRef]
- Horak, J. Amino acid transport in eucaryotic microorganisms. Biochim. Biophys. Acta 1986, 864, 223–256. [Google Scholar] [CrossRef]
- Horak, J. Yeast nutrient transporters. Biochim. Biophys. Acta 1997, 1331, 41–79. [Google Scholar] [CrossRef]
- Grenson, M.; Hou, C.; Crabeel, M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J. Bacteriol. 1970, 103, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Jauniaux, J.C.; Grenson, M. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 1990, 190, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Andre, B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 1995, 11, 1575–1611. [Google Scholar] [CrossRef] [PubMed]
- Regenberg, B.; Holmberg, S.; Olsen, L.D.; Kielland-Brandt, M.C. Dip5p mediates high-affinity and high-capacity transport of l-glutamate and l-aspartate in Saccharomyces cerevisiae. Curr. Genet. 1998, 33, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Regenberg, B.; During-Olsen, L.; Kielland-Brandt, M.C.; Holmberg, S. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 1999, 36, 317–328. [Google Scholar] [CrossRef]
- Trip, H.; Evers, M.E.; Kiel, J.A.; Driessen, A.J. Uptake of the beta-lactam precursor alpha-aminoadipic acid in Penicillium chrysogenum is mediated by the acidic and the general amino acid permease. Appl. Environ. Microbiol. 2004, 70, 4775–4783. [Google Scholar] [CrossRef]
- Apostolaki, A.; Erpapazoglou, Z.; Harispe, L.; Billini, M.; Kafasla, P.; Kizis, D.; Penalva, M.A.; Scazzocchio, C.; Sophianopoulou, V. AgtA, the dicarboxylic amino acid transporter of Aspergillus nidulans, is concertedly down-regulated by exquisite sensitivity to nitrogen metabolite repression and ammonium-elicited endocytosis. Eukaryot. Cell 2009, 8, 339–352. [Google Scholar] [CrossRef]
- Takahashi, S.; Matsunaga, R.; Kera, Y.; Yamada, R.H. Isolation of the Cryptococcus humicolus URA3 gene encoding orotidine-5’-phosphate decarboxylase and its use as a selective marker for transformation. J. Biosci. Bioeng. 2003, 96, 23–31. [Google Scholar] [CrossRef]
- Imanishi, D.; Abe, K.; Kera, Y.; Takahashi, S. Draft Genome Sequence of the Yeast Vanrija humicola (Formerly Cryptococcus humicola) Strain UJ1, a Producer of d-Aspartate Oxidase. Genome Announc. 2018, 6, e00068-18. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Fernandes, J.D.; Martho, K.; Tofik, V.; Vallim, M.A.; Pascon, R.C. The Role of Amino Acid Permeases and Tryptophan Biosynthesis in Cryptococcus neoformans Survival. PLoS ONE 2015, 10, e0132369. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Molla, G.; Piubelli, L.; Volonte, F.; Pilone, M.S. Enzymatic detection of d-amino acids. Methods Mol. Biol. 2012, 794, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhou, L.; Jiao, X.; Lu, F.; Yan, C.; Zeng, X.; Wang, J.; Shi, Y. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 2010, 463, 828–832. [Google Scholar] [CrossRef]
- Pall, M.L. Amino acid transport in Neurospora crassa. 3. Acidic amino acid transport. Biochim. Biophys. Acta 1970, 211, 513–520. [Google Scholar] [CrossRef]
- Hunter, D.R.; Segel, I.H. Acidic and basic amino acid transport systems of Penicillium chrysogenum. Arch. Biochem. Biophys. 1971, 144, 168–183. [Google Scholar] [CrossRef]
- Robinson, J.H.; Anthony, C.; Drabble, W.T. The acidic amino-acid permease of Aspergillus nidulans. J. Gen. Microbiol. 1973, 79, 53–63. [Google Scholar] [CrossRef][Green Version]
- Ma, D.; Lu, P.; Yan, C.; Fan, C.; Yin, P.; Wang, J.; Shi, Y. Structure and mechanism of a glutamate-GABA antiporter. Nature 2012, 483, 632–636. [Google Scholar] [CrossRef]
- De Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef]
- Martho, K.F.; de Melo, A.T.; Takahashi, J.P.; Guerra, J.M.; Santos, D.C.; Purisco, S.U.; Melhem, M.S.; Fazioli, R.D.; Phanord, C.; Sartorelli, P.; et al. Amino Acid Permeases and Virulence in Cryptococcus neoformans. PLoS ONE 2016, 11, e0163919. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, L.; Ratera, M.; Paladino, A.; Bartoccioni, P.; Errasti-Murugarren, E.; Valencia, E.; Portella, G.; Bial, S.; Zorzano, A.; Fita, I.; et al. Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc. Natl. Acad. Sci. USA 2011, 108, 3935–3940. [Google Scholar] [CrossRef] [PubMed]
- Gournas, C.; Prevost, M.; Krammer, E.M.; Andre, B. Function and Regulation of Fungal Amino Acid Transporters: Insights from Predicted Structure. Adv. Exp. Med. Biol. 2016, 892, 69–106. [Google Scholar] [CrossRef] [PubMed]
- Olivera, H.; Gonzalez, A.; Pena, A. Regulation of the amino acid permeases in nitrogen-limited continuous cultures of the yeast Saccharomyces cerevisiae. Yeast 1993, 9, 1065–1073. [Google Scholar] [CrossRef]
- Casal, M.; Paiva, S.; Queiros, O.; Soares-Silva, I. Transport of carboxylic acids in yeasts. FEMS Microbiol. Rev. 2008, 32, 974–994. [Google Scholar] [CrossRef]
- Rytka, J. Positive selection of general amino acid permease mutants in Saccharomyces cerevisiae. J. Bacteriol. 1975, 121, 562–570. [Google Scholar] [CrossRef]
- Hofman-Bang, J. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotechnol. 1999, 12, 35–73. [Google Scholar] [CrossRef]
- Forsberg, H.; Ljungdahl, P.O. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 2001, 21, 814–826. [Google Scholar] [CrossRef]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005, 59, 407–450. [Google Scholar] [CrossRef]
- Sellick, C.A.; Reece, R.J. Modulation of transcription factor function by an amino acid: Activation of Put3p by proline. EMBO J. 2003, 22, 5147–5153. [Google Scholar] [CrossRef] [PubMed]

| N Source | Chaap4 | Chaap5 | |
|---|---|---|---|
| None | Below Detection | Below Detection | |
| NH4Cl | 95% | 98% | |
| Aliphatic | l-Ala | 106% | 115% |
| Gly | 108% | 115% | |
| l-Ile | 95% | 107% | |
| l-Val | 113% | 104% | |
| Aromatic | l-Phe | 60% * | 79% * |
| Sulfur | l-Met | 102% | 95% |
| Hydroxylated | l-Ser | 106% | 106% |
| Acidic | l-Asp | 36% *** | 94% |
| d-Asp | 67% ** | 102% | |
| l-Glu | 84% ** | 103% | |
| Amide | l-Asn | 96% | 107% |
| l-Gln | 102% | 108% | |
| Basic | l-Lys | 96% | 92% * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imanishi, D.; Kera, Y.; Takahashi, S. Identification of an Acidic Amino Acid Permease Involved in d-Aspartate Uptake in the Yeast Cryptococcus humicola. Microorganisms 2021, 9, 192. https://doi.org/10.3390/microorganisms9010192
Imanishi D, Kera Y, Takahashi S. Identification of an Acidic Amino Acid Permease Involved in d-Aspartate Uptake in the Yeast Cryptococcus humicola. Microorganisms. 2021; 9(1):192. https://doi.org/10.3390/microorganisms9010192
Chicago/Turabian StyleImanishi, Daiki, Yoshio Kera, and Shouji Takahashi. 2021. "Identification of an Acidic Amino Acid Permease Involved in d-Aspartate Uptake in the Yeast Cryptococcus humicola" Microorganisms 9, no. 1: 192. https://doi.org/10.3390/microorganisms9010192
APA StyleImanishi, D., Kera, Y., & Takahashi, S. (2021). Identification of an Acidic Amino Acid Permease Involved in d-Aspartate Uptake in the Yeast Cryptococcus humicola. Microorganisms, 9(1), 192. https://doi.org/10.3390/microorganisms9010192





