Tracing the Trophic Plasticity of the Coral–Dinoflagellate Symbiosis Using Amino Acid Compound-Specific Stable Isotope Analysis
Abstract
1. Introduction
2. Material and Methods
2.1. Experimental Setup
2.2. Compound-Specific Stable Isotope Analysis
2.3. Data Analysis and Corrections
2.4. Statistical Analyses
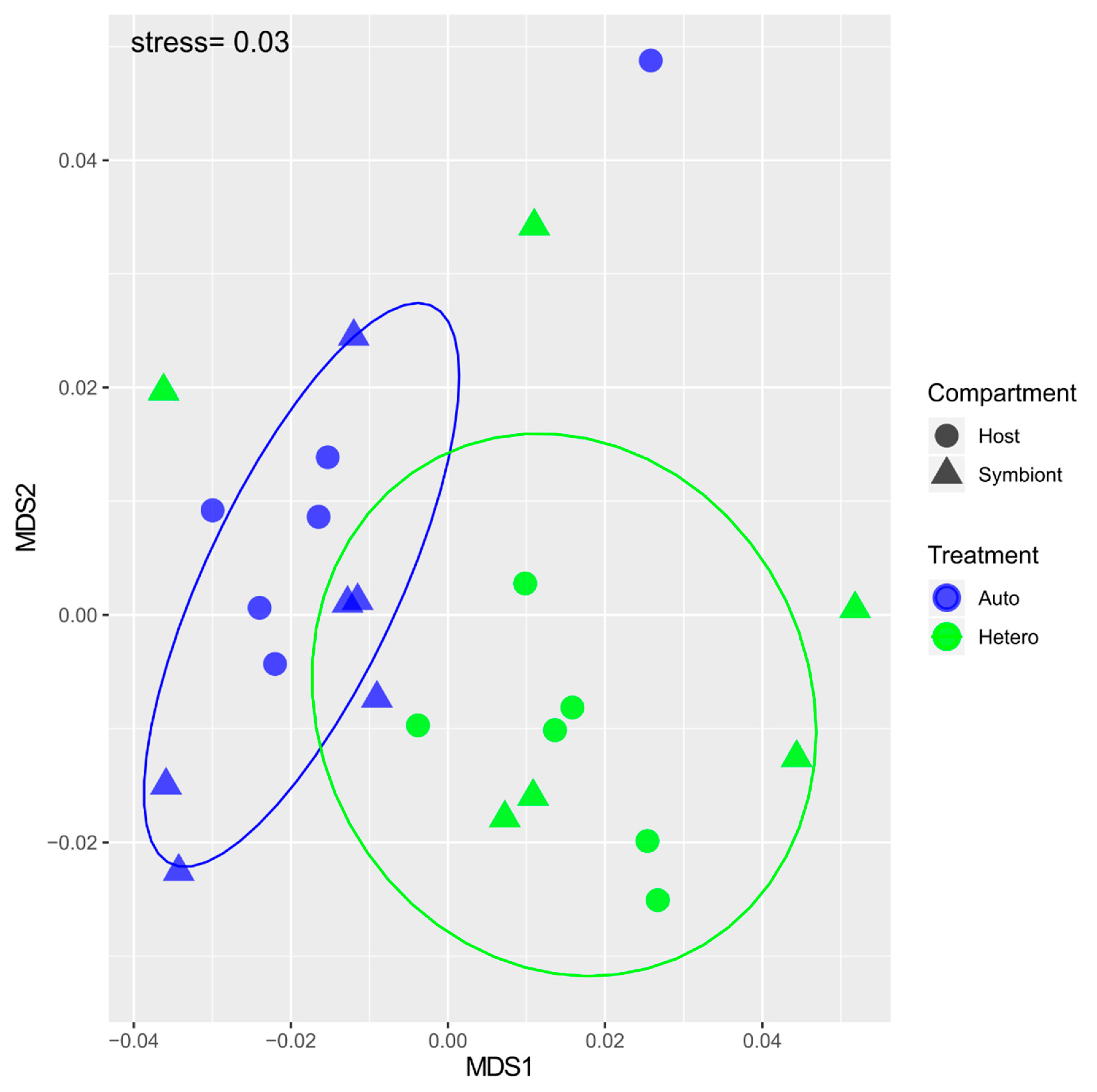
3. Results
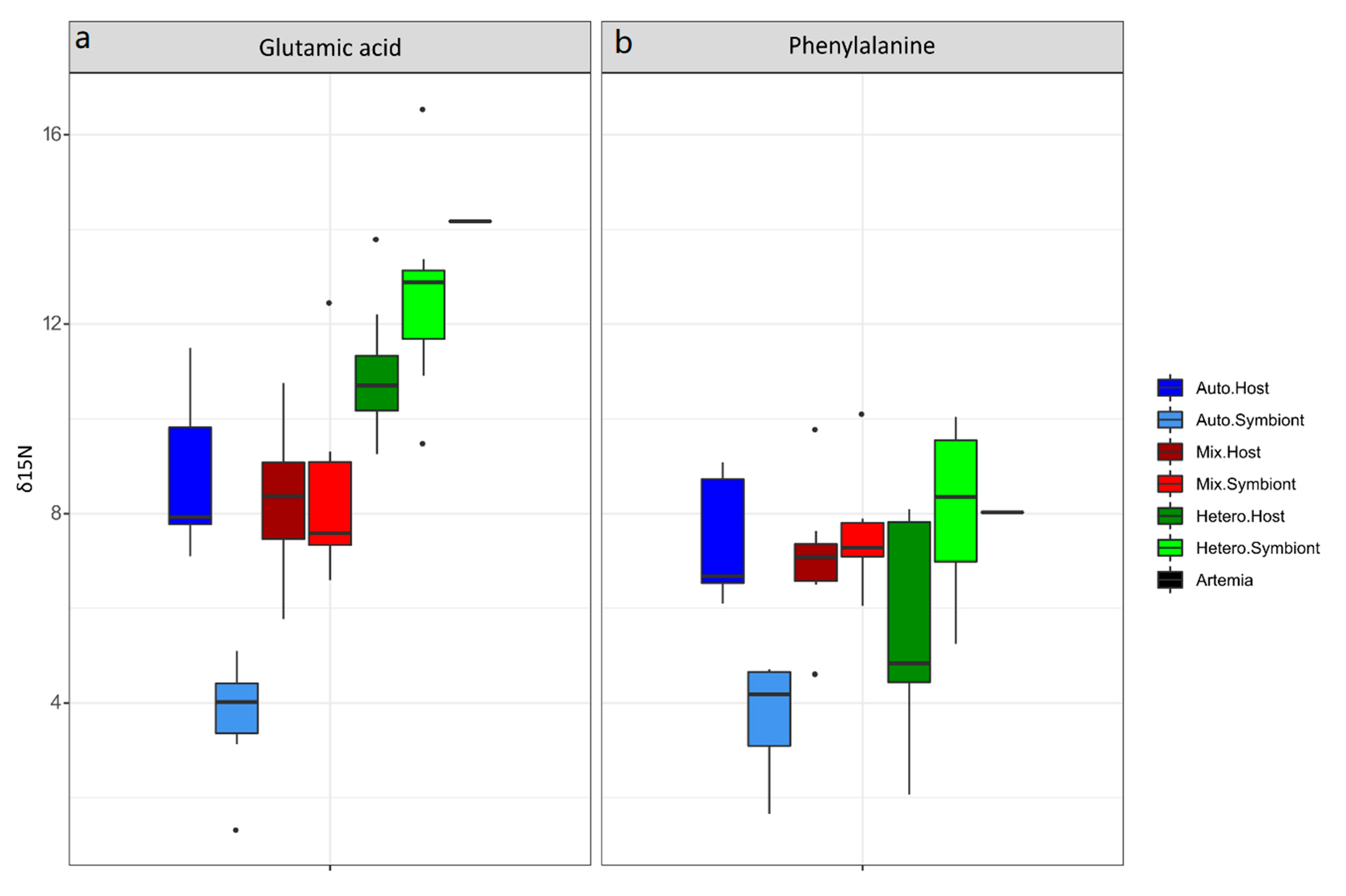
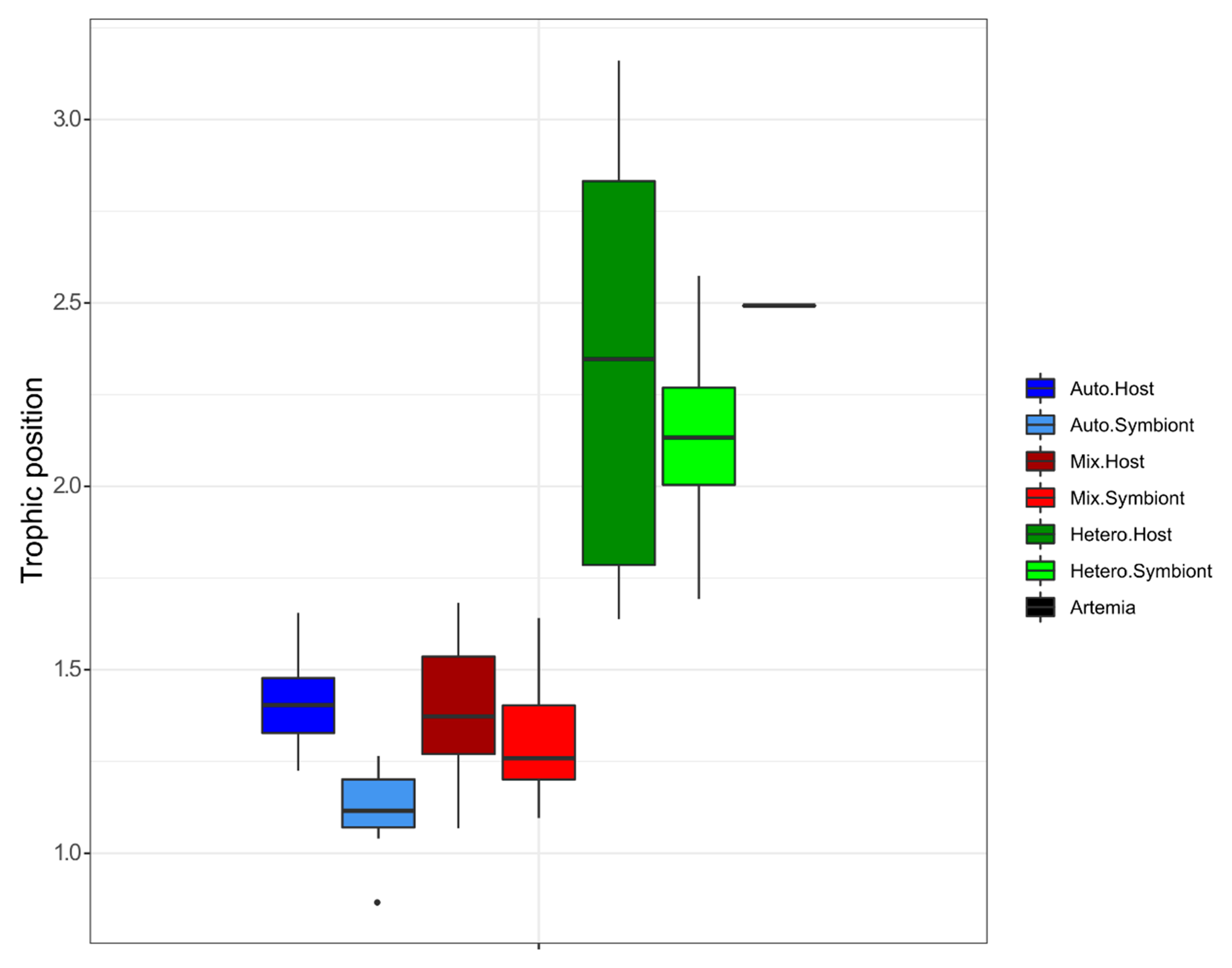
3.1. δ15N-AA
3.2. δ13C-AA
4. Discussion
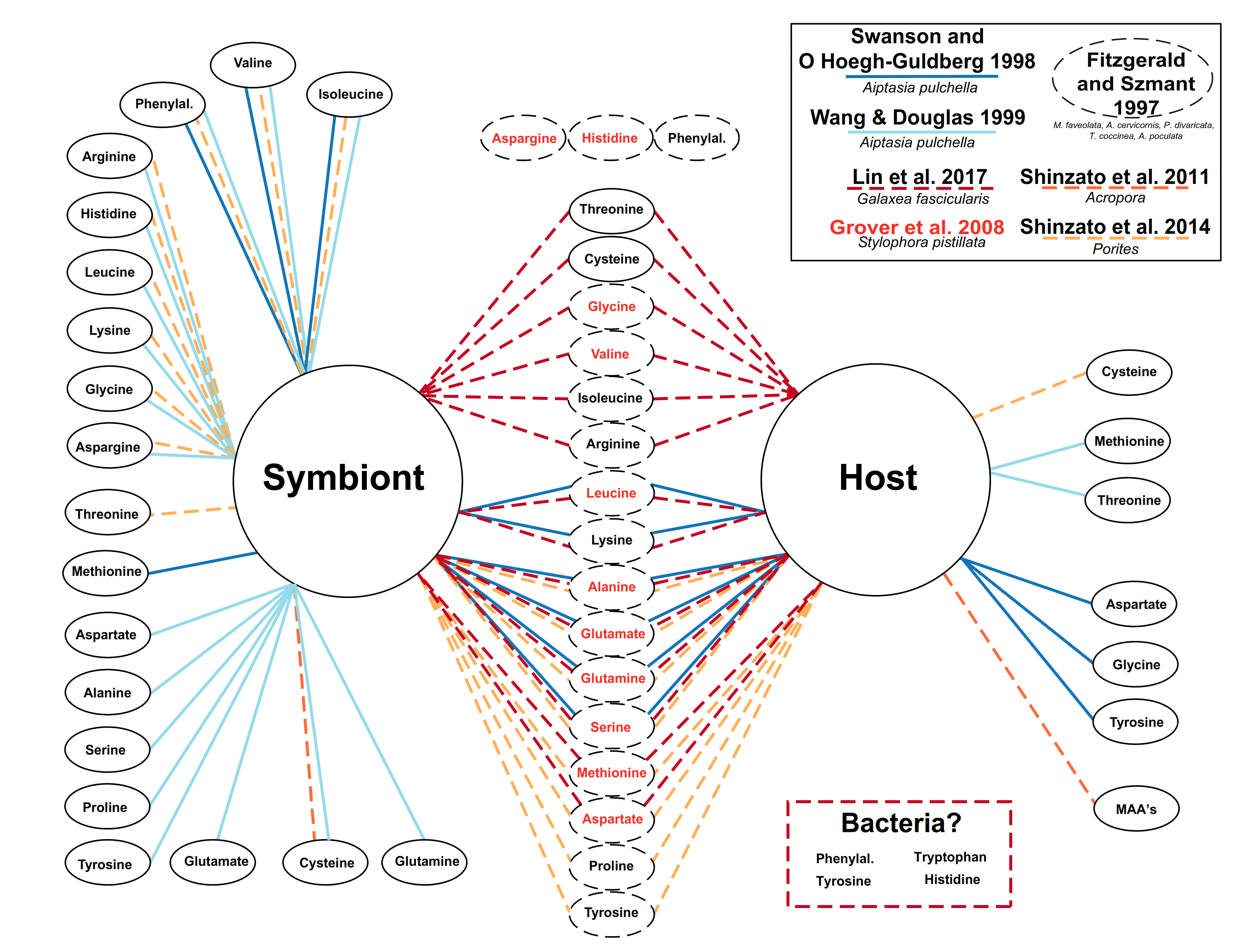
4.1. Lessons from the Amino Acid Profile in Autotrophic Holobionts
4.2. Lessons from the Amino Acid Profile in Mixotrophic and Heterotrophic Holobionts
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corlett, R.T. Restoration, Reintroduction and Rewilding in a Changing World. Trends Ecol. Evol. 2016, 31, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Halpern, B.S.; Frazier, M.; Potapenko, J.; Casey, K.S.; Koenig, K.; Longo, C.; Lowndes, J.S.; Rockwood, R.C.; Selig, E.R.; Selkoe, K.A.; et al. Spatial and temporal changes in cumulative human impacts on the world’s ocean. Nat. Commun. 2015, 6, 7615. [Google Scholar] [CrossRef] [PubMed]
- Fabricius, K.E. Effects of terrestrial runoff on the ecology of corals and coral reefs: Review and synthesis. Mar. Pollut. Bull. 2005, 50, 125–146. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.M.; Freeman, C.J.; Wong, J.C.; Fogel, M.L.; Knowlton, N. Climate change promotes parasitism in a coral symbiosis. ISME J. 2018, 12, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Li, C.; Wang, T.; Zhang, M.; Xu, J. Combined Effects of Experimental Warming and Eutrophication on Phytoplankton Dynamics and Nitrogen Uptake. Water 2018, 10, 1057. [Google Scholar] [CrossRef]
- Urabe, J.; Shimizu, Y.; Yamaguchi, T. Understanding the stoichiometric limitation of herbivore growth: The importance of feeding and assimilation flexibilities. Ecol. Lett. 2018, 21, 197–206. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Fitzgerald, G.F. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Selosse, M.A.; Charpin, M.; Not, F. Mixotrophy everywhere on land and in water: The grand écart hypothesis. Ecol. Lett. 2017, 20, 246–263. [Google Scholar] [CrossRef]
- Knowlton, N.; Jackson, J.B. Shifting Baselines, Local Impacts, and Global Change on Coral Reefs. PLoS Biol. 2008, 6, e54. [Google Scholar] [CrossRef]
- Lajeunesse, T.C.; Parkinson, J.E.; Gabrielson, P.W.; Jeong, H.J.; Reimer, J.D.; Voolstra, C.R.; Santos, S.R. Systematic Revision of Symbiodiniaceae Highlights the Antiquity and Diversity of Coral Endosymbionts. Curr. Biol. 2018, 28, 2570–2580. [Google Scholar] [CrossRef]
- Grover, R.; Maguer, J.F.; Allemand, D.; Ferrier-Pagès, C. Uptake of dissolved free amino acids by the scleractinian coral Stylophora pistillata. J. Exp. Biol. 2008, 211, 860–865. [Google Scholar] [CrossRef] [PubMed]
- Houlbrèque, F.; Ferrier-Pagès, C. Heterotrophy in tropical scleractinian corals. Biol. Rev. 2009, 84, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Benavides, M.; Bednarz, V.N.; Ferrier-Pagès, C. Diazotrophs: Overlooked Key Players within the Coral Symbiosis and Tropical Reef Ecosystems? Front. Mar. Sci. 2017, 4, 10. [Google Scholar] [CrossRef]
- Anthony, K.R.; Hoogenboom, M.O.; Maynard, J.A.; Grottoli, A.G.; Middlebrook, R. Energetics approach to predicting mortality risk from environmental stress: A case study of coral bleaching. Funct. Ecol. 2009, 23, 539–550. [Google Scholar] [CrossRef]
- Grottoli, A.G.; Rodrigues, L.J.; Palardy, J.E. Heterotrophic plasticity and resilience in bleached corals. Nature 2006, 440, 1186–1189. [Google Scholar] [CrossRef]
- Boecklen, W.J.; Yarnes, C.T.; Cook, B.A.; James, A.C. On the Use of Stable Isotopes in Trophic Ecology. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 411–440. [Google Scholar] [CrossRef]
- Rädecker, N.; Pogoreutz, C.; Voolstra, C.R.; Wiedenmann, J.; Wild, C. Nitrogen cycling in corals: The key to understanding holobiont functioning? Trends Microbiol. 2015, 23, 490–497. [Google Scholar] [CrossRef]
- Crandall, J.B.; Teece, M.A.; Estes, B.A.; Manfrino, C.; Ciesla, J.H. Nutrient acquisition strategies in mesophotic hard corals using compound specific stable isotope analysis of sterols. J. Exp. Mar. Biol. Ecol. 2016, 474, 133–141. [Google Scholar] [CrossRef]
- Teece, M.A.; Estes, B.; Gelsleichter, E.; Lirman, D. Heterotrophic and autotrophic assimilation of fatty acids by two scleractinian corals, Montastraea faveolata and Porites astreoides. Limnol. Oceanogr. 2011, 56, 1285–1296. [Google Scholar] [CrossRef]
- Treignier, C.; Tolosa, I.; Grover, R.; Reynaud, S.; Ferrier-Pagès, C. Carbon isotope composition of fatty acids and sterols in the scleractinian coral Turbinaria reniformis: Effect of light and feeding. Limnol. Oceanogr. 2009, 54, 1933–1940. [Google Scholar] [CrossRef]
- Fox, M.D.; Smith, E.A.E.; Smith, J.E.; Newsome, S.D. Trophic plasticity in a common reef-building coral: Insights from δ 13 C analysis of essential amino acids. Funct. Ecol. 2019, 33, 2203–2214. [Google Scholar] [CrossRef]
- Fujii, T.; Tanaka, Y.; Maki, K.; Saotome, N.; Morimoto, N.; Watanabe, A.; Miyajima, T. Organic Carbon and Nitrogen Isoscapes of Reef Corals and Algal Symbionts: Relative Influences of Environmental Gradients and Heterotrophy. Microorganisms 2020, 8, 1221. [Google Scholar] [CrossRef] [PubMed]
- McMahon, K.W.; Thorrold, S.R.; Houghton, L.A.; Berumen, M.L. Tracing carbon flow through coral reef food webs using a compound-specific stable isotope approach. Oecologia 2016, 180, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Schiff, J.T.; Batista, F.C.; Sherwood, O.A.; Guilderson, T.P.; Hill, T.M.; Ravelo, A.C.; McMahon, K.W.; McCarthy, M.D. Compound specific amino acid δ13C patterns in a deep-sea proteinaceous coral: Implications for reconstructing detailed δ13C records of exported primary production. Mar. Chem. 2014, 166, 82–91. [Google Scholar] [CrossRef]
- Chikaraishi, Y.; Ogawa, N.O.; Doi, H.; Ohkouchi, N. 15N/14N ratios of amino acids as a tool for studying terrestrial food webs: A case study of terrestrial insects (bees, wasps, and hornets). Ecol. Res. 2011, 26, 835–844. [Google Scholar] [CrossRef]
- Ohkouchi, N.; Chikaraishi, Y.; Close, H.G.; Fry, B.; Larsen, T.; Madigan, D.J.; McCarthy, M.D.; McMahon, K.W.; Nagata, T.; Naito, Y.I.; et al. Advances in the application of amino acid nitrogen isotopic analysis in ecological and biogeochemical studies. Org. Geochem. 2017, 113, 150–174. [Google Scholar] [CrossRef]
- McClelland, J.W.; Montoya, J.P. Trophic relationships and the nitrogen isotopic composition of amino acids in plankton. Ecology 2002, 83, 2173–2180. [Google Scholar] [CrossRef]
- McMahon, K.W.; McCarthy, M.D. Embracing variability in amino acid δ15N fractionation: Mechanisms, implications, and applications for trophic ecology. Ecosphere 2016, 7. [Google Scholar] [CrossRef]
- Honch, N.V.; McCullagh, J.S.; Hedges, R.E. Variation of bone collagen amino acid δ13C values in archaeological humans and fauna with different dietary regimes: Developing frameworks of dietary discrimination. Am. J. Physic. Anthropol. 2012, 148, 495–511. [Google Scholar] [CrossRef]
- Shinzato, C.; Shoguchi, E.; Kawashima, T.; Hamada, M.; Hisata, K.; Tanaka, M.; Fujie, M.; Fujiwara, M.; Koyanagi, R.; Ikuta, T.; et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 2011, 476, 320–323. [Google Scholar] [CrossRef]
- Cui, G.; Liew, Y.J.; Li, Y.; Kharbatia, N.; Zahran, N.I.; Emwas, A.H.; Eguiluz, V.M.; Aranda, M. Host-dependent nitrogen recycling as a mechanism of symbiont control in Aiptasia. PLoS Genet. 2019, 15, e1008189. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, L.M.; Szmant, A.M. Biosynthesis of ’essential’ amino acids by scleractinian corals. Biochem. J. 1997, 322, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Shinzato, C.; Inoue, M.; Kusakabe, M. A Snapshot of a Coral “Holobiont”: A Transcriptome Assembly of the Scleractinian Coral, Porites, Captures a Wide Variety of Genes from Both the Host and Symbiotic Zooxanthellae. PLoS ONE 2014, 9, e85182. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.T.; Douglas, A.E. Essential amino acid synthesis and nitrogen recycling in an alga-invertebrate symbiosis. Mar. Biol. 1999, 135, 219–222. [Google Scholar] [CrossRef]
- Ying, H.; Cooke, I.; Sprungala, S.; Wang, W.; Hayward, D.C.; Tang, Y.; Huttley, G.; Ball, E.E.; Forêt, S.; Miller, D.J. Comparative genomics reveals the distinct evolutionary trajectories of the robust and complex coral lineages. Genome Biol. 2018, 19, 175. [Google Scholar] [CrossRef]
- Tremblay, P.; Grover, R.; Maguer, J.-F.; Legendre, L.; Ferrier-Pagès, C. Autotrophic carbon budget in coral tissue: A new 13C-based model of photosynthate translocation. J. Exp. Biol. 2012, 215, 1384–1393. [Google Scholar] [CrossRef]
- Cowie, G.L.; Hedges, J.I. Improved amino acid quantification in environmental samples: Charge-matched recovery standards and reduced analysis time. Mar. Chem. 1992, 37, 223–238. [Google Scholar] [CrossRef]
- Martinez, S.; Lalzer, M.; Shemesh, E.; Einbinder, S.; Goodman, B.; Tchernov, D. Effect of different derivatization protocols on the calculation of trophic position using amino acids compound-specific stable isotopes. Front. Mar. Sci. 2020, 7, 1–7. [Google Scholar] [CrossRef]
- Docherty, G.; Jones, V.; Evershed, R.P. Practical and theoretical considerations in the gas chromatography/combustion/isotope ratio mass spectrometry? 13C analysis of small polyfunctional compounds. Rapid Commun. Mass Spectrom. 2001, 15, 730–738. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Anderson, M.J. Distance-Based Tests for Homogeneity of Multivariate Dispersions. Biometrics 2006, 62, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Martinez Arbizu, P. PairwiseAdonis: Pairwise Multilevel Comparison Using Adonis R Package Version 0.3. 2019. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 1 November 2020).
- Chikaraishi, Y.; Ogawa, N.O.; Kashiyama, Y.; Takano, Y.; Suga, H.; Tomitani, A.; Miyashita, H.; Kitazato, H.; Ohkouchi, N. Determination of aquatic food-web structure based on compound-specific nitrogen isotopic composition of amino acids. Limnol. Oceanogr. Methods 2009, 7, 740–750. [Google Scholar] [CrossRef]
- Grover, R.; Maguer, J.F.; Reynaud-Vaganay, S.; Ferrier-Pages, C. Uptake of ammonium by the scleractinian coral Stylophora pistillata: Effect of feeding, light, and ammonium concentrations. Limnol. Oceanogr. 2002, 47, 782–790. [Google Scholar] [CrossRef]
- Dubinsky, Z.; Falkowski, P.G.; Porter, J.W.; Muscatine, L. Absorption and utilization of radiant energy by light- and shade-adapted colonies of the hermatypic coral Stylophora pistillata. Proc. R. Soc. 1984, 222, 203–214. [Google Scholar] [CrossRef]
- Lin, Z.; Chen, M.; Dong, X.; Zheng, X.; Huang, H.; Xu, X.; Chen, J. Transcriptome profiling of Galaxea fascicularis and its endosymbiont Symbiodinium reveals chronic eutrophication tolerance pathways and metabolic mutualism between partners. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Macko, S.A.; Estep, M.L.F.; Engel, M.H.; Hare, P.E. Kinetic fractionation of stable nitrogen isotopes during amino acid transamination. Geochim. Cosmochim. Acta 1986, 50, 2143–2146. [Google Scholar] [CrossRef]
- Blau, N.; van Spronsen, F.J.; Levy, H.L. Phe-nylketonuria. Lancet 2010, 376, 1417–1427. [Google Scholar] [CrossRef]
- Grover, R.; Maguer, J.F.; Allemand, D.; Ferrier-Pagés, C. Nitrate uptake in the scleractinian coral Stylophora pistillata. Limnol. Oceanogr. 2003, 48, 2266–2274. [Google Scholar] [CrossRef]
- Yellowlees, D.; Rees, T.A.V.; Leggat, W. Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 2008, 31, 679–694. [Google Scholar] [CrossRef]
- Pernice, M.; Meibom, A.; Van Den Heuvel, A.; Kopp, C.; Domart-Coulon, I.; Hoegh-Guldberg, O.; Dove, S. A single-cell view of ammonium assimilation in coral–dinoflagellate symbiosis. ISME J. 2012, 6, 1314–1324. [Google Scholar] [CrossRef]
- Tanaka, Y.; Suzuki, A.; Sakai, K. The stoichiometry of coral-dinoflagellate symbiosis: Carbon and nitrogen cycles are balanced in the recycling and double translocation system. ISME J. 2018, 12, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Ferrier-Pagès, C.; Godinot, C.; D’Angelo, C.; Wiedenmann, J.; Grover, R. Phosphorus metabolism of reef organisms with algal symbionts. Ecol. Monogr. 2016, 86, 262–277. [Google Scholar] [CrossRef]
- Davy, S.K.; Allemand, D.; Weis, V.M. Cell Biology of Cnidarian-Dinoflagellate Symbiosis. Microbiol. Mol. Biol. Rev. 2012, 76, 229–261. [Google Scholar] [CrossRef] [PubMed]
- Wangpraseurt, D.; Jacques, S.; Lyndby, N.; Holm, J.B.; Ferrier-Pages, C.; Kühl, M. Microscale light management and inherent optical properties of intact corals studied with optical coherence tomography. J. R. Soc. Interface 2019, 16, 20180567. [Google Scholar] [CrossRef] [PubMed]
- Chikaraishi, Y.; Kashiyama, Y.; Ogawa, N.O.; Kitazato, H.; Ohkouchi, N. Metabolic control of nitrogen isotope composition of amino acids in macroalgae and gastropods: Implications for aquatic food web studies. Mar. Ecol. Prog. Ser. 2007, 342, 85–90. [Google Scholar] [CrossRef]
- Smith, G.J.; Muscatine, L. Cell cycle of symbiotic dinoflagellates: Variation in G1 phase-duration with anemone nutritional status and macronutrient supply in the Aiptasia pulchella—Symbiodinium pulchrorum symbiosis. Mar. Biol. 1999, 134, 405–418. [Google Scholar] [CrossRef]
- Tremblay, P.; Maguer, J.-F.; Grover, R.; Ferrier-Pagès, C. Trophic dynamics of scleractinian corals: Stable isotope evidence. J. Exp. Biol. 2015, 218, 1223–1234. [Google Scholar] [CrossRef]
- Deane, E.M.; O’Brien, R.W. Uptake of sulphate, taurine, cysteine and methionine by symbiotic and free-living dinoflagellates. Arch. Microbiol. 1981, 128, 311–319. [Google Scholar] [CrossRef]
- John, E.H.; Flynn, K.J. Amino acid uptake by the toxic dinoflagellate Alexandrium fundyense. Mar. Biol. 1999, 133, 11–19. [Google Scholar] [CrossRef]
- Douglas, A.E. Molecular dissection of nutrient exchange at the insect-microbial interface. Curr. Opin. Insect Sci. 2014, 4, 23–28. [Google Scholar] [CrossRef]
- Shigenobu, S.; Watanabe, H.; Hattori, M.; Sakaki, Y.; Ishikawa, H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 2000, 407, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Rangel, M.S.; Erler, D.; Tagliafico, A.; Cowden, K.; Scheffers, S.; Christidis, L. Quantifying the transfer of prey δ15N signatures into coral holobiont nitrogen pools. Mar. Ecol. Prog. Ser. 2019, 610, 33–49. [Google Scholar] [CrossRef]
- Wegley, L.W.; Edwards, R.; Rodriguez-Brito, B.; Liu, H.; Rohwer, F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 2007, 9, 2707–2719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N. Role of methionine on epigenetic modification of DNA methylation and gene expression in animals. Anim. Nutr. 2018, 4, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Loram, J.E.; Trapido-Rosenthal, H.G.; Douglas, A.E. Functional significance of genetically different symbiotic algae Symbiodinium in a coral reef symbiosis. Mol. Ecol. 2007, 16, 4849–4857. [Google Scholar] [CrossRef] [PubMed]
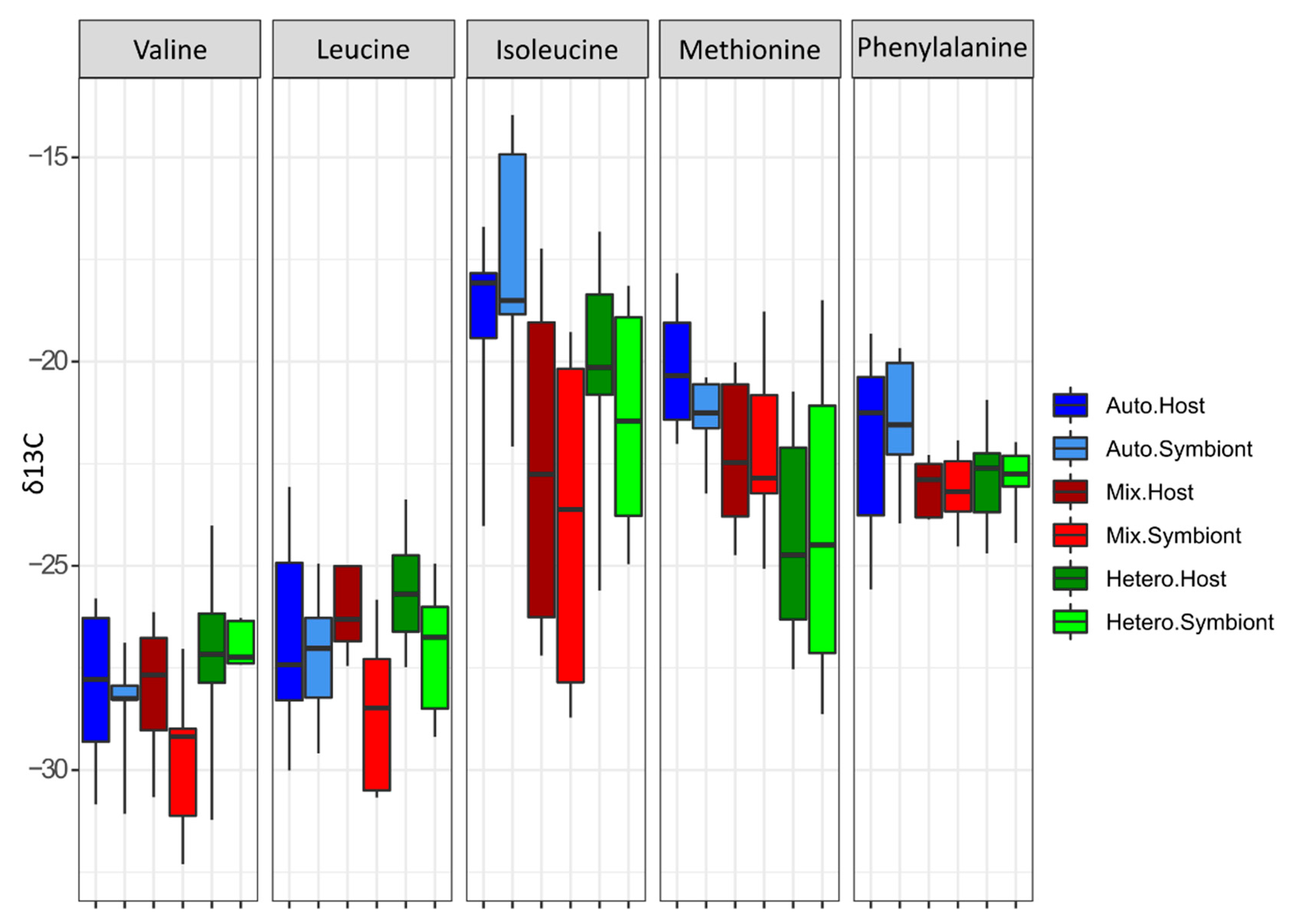
| Amino Acid | Mean δ13C | Standard Deviation |
|---|---|---|
| Valine | −30.527 | 1.393 |
| Leucine | −27.465 | 1.064 |
| Isoleucine | −21.086 | 1.046 |
| Methionine | −19.899 | 1.021 |
| Phenylalanine | −25.540 | 1.083 |
| Valine | Df | Sum Sqs | Mean Sqs | F. Model | p |
|---|---|---|---|---|---|
| Feeding | 1 | 7.748 | 7.7482 | 2.84605 | 0.1081 |
| Host.symbiont | 1 | 0.074 | 0.074 | 0.02718 | 0.8694 |
| Residuals | 23 | 62.616 | 2.7224 | 0.88895 | |
| Total | 25 | 70.438 | 1 | ||
| Leucine | |||||
| Feeding | 1 | 4.111 | 4.1106 | 1.2997 | 0.2648 |
| Host.symbiont | 1 | 5.182 | 5.1824 | 1.6386 | 0.215 |
| Residuals | 23 | 72.744 | 3.1628 | 0.88672 | |
| Total | 25 | 82.037 | 1 | ||
| Isoleucine | |||||
| Feeding | 1 | 33.704 | 33.704 | 4.2303 | 0.050 |
| Host.symbiont | 1 | 0.118 | 0.118 | 0.0148 | 0.9034 |
| Residuals | 23 | 183.244 | 7.967 | 0.84419 | |
| Total | 25 | 217.065 | 1 | ||
| Methionine | |||||
| Feeding | 1 | 76.003 | 76.003 | 12.9126 | 0.0014 |
| Host.symbiont | 1 | 1.164 | 1.164 | 0.1978 | 0.6554 |
| Residuals | 23 | 135.377 | 5.886 | 0.63694 | |
| Total | 25 | 212.545 | 1 | ||
| Phenylalanine | |||||
| Feeding | 1 | 8.45 | 8.4502 | 3.6157 | 0.0705 |
| Host.symbiont | 1 | 0.194 | 0.1939 | 0.083 | 0.768 |
| Residuals | 23 | 53.752 | 2.3371 | 0.86146 | |
| Total | 25 | 62.397 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrier-Pagès, C.; Martinez, S.; Grover, R.; Cybulski, J.; Shemesh, E.; Tchernov, D. Tracing the Trophic Plasticity of the Coral–Dinoflagellate Symbiosis Using Amino Acid Compound-Specific Stable Isotope Analysis. Microorganisms 2021, 9, 182. https://doi.org/10.3390/microorganisms9010182
Ferrier-Pagès C, Martinez S, Grover R, Cybulski J, Shemesh E, Tchernov D. Tracing the Trophic Plasticity of the Coral–Dinoflagellate Symbiosis Using Amino Acid Compound-Specific Stable Isotope Analysis. Microorganisms. 2021; 9(1):182. https://doi.org/10.3390/microorganisms9010182
Chicago/Turabian StyleFerrier-Pagès, Christine, Stephane Martinez, Renaud Grover, Jonathan Cybulski, Eli Shemesh, and Dan Tchernov. 2021. "Tracing the Trophic Plasticity of the Coral–Dinoflagellate Symbiosis Using Amino Acid Compound-Specific Stable Isotope Analysis" Microorganisms 9, no. 1: 182. https://doi.org/10.3390/microorganisms9010182
APA StyleFerrier-Pagès, C., Martinez, S., Grover, R., Cybulski, J., Shemesh, E., & Tchernov, D. (2021). Tracing the Trophic Plasticity of the Coral–Dinoflagellate Symbiosis Using Amino Acid Compound-Specific Stable Isotope Analysis. Microorganisms, 9(1), 182. https://doi.org/10.3390/microorganisms9010182







