Loss of Motility as a Non-Lethal Mechanism for Intercolony Inhibition (“Sibling Rivalry”) in Marinobacter
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Growth Media, and Conditions
2.2. AHL Bioassays
2.3. AHL Extraction from Marinobacter Supernatants
2.4. In Silico Search for Putative AHL Biosynthetic Genes
2.5. Bacteriocin-Like Inhibitory Substance (BLIS) Assay
2.6. Metabolic and Cell Viability Assays
2.7. Isolation of Active Compounds Secreted by DG893
2.8. Protein Identification
2.9. Microscopic Sample Preparation and Imaging
3. Results
3.1. Quorum Sensing (QS)
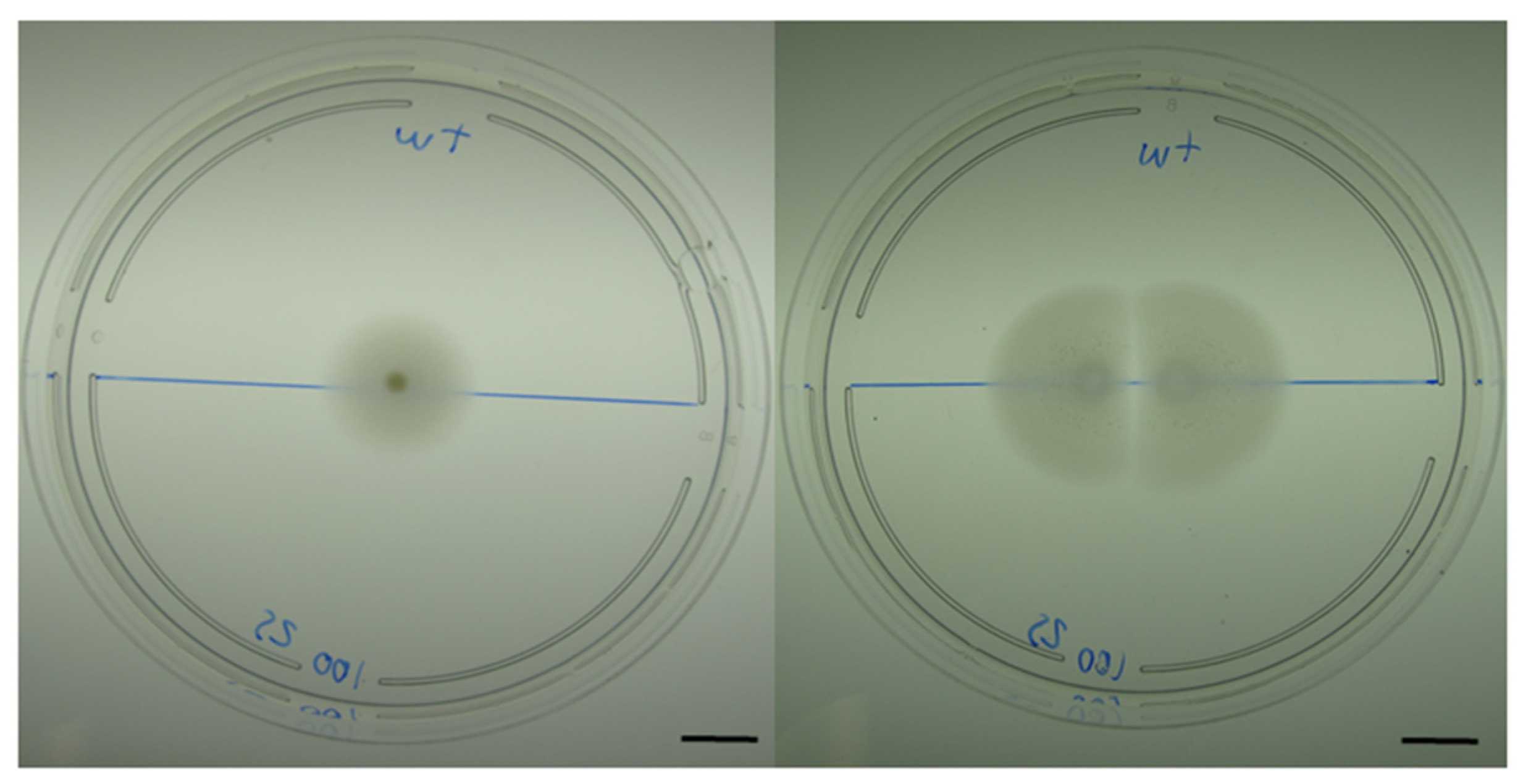
3.2. Intraspecies Interactions
3.3. Identification of Potential Bioactive Material
3.4. GDPD Network Analysis
4. Discussion
5. Conclusions
- (1)
- Most if not all of the bacteria in the genus Marinobacter neither produce nor utilize either of the two main QS systems (i.e., those based on acylhomoserine lactones or alternatively AI-2) commonly found in Gram-negative bacteria.
- (2)
- Many bacteria of the Marinobacter genus display clear and unambiguous sibling intercolony inhibition.
- (3)
- The intercolony inhibition is not lethal.
- (4)
- The inhibition between colonies is likely due to a loss in motility of the cells at the intercolony interface, and such motility is not permanent and is restored when cells are placed on fresh media.
- (5)
- A secreted protein related to GDPD may be part of this process.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Duran, R. Marinobacter. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Glatz, R.E.; Lepp, P.W.; Ward, B.B.; Francis, C.A. Planktonic microbial community composition across steep physical/chemical gradients in permanently ice-covered Lake Bonney, Antarctica. Geobiology 2006, 4, 53–67. [Google Scholar] [CrossRef]
- Shivaji, S.; Gupta, P.; Chaturvedi, P.; Suresh, K.; Delille, D. Marinobacter maritimus sp. nov., a psychrotolerant strain isolated from sea water off the sub Antarctic Kerguelen islands. Int. J. Syst. Evol. Microbiol. 2005, 55, 1453–1456. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.C.; Li, H.R.; Xin, Y.H.; Chi, Z.M.; Zhou, P.J.; Yu, Y. Marinobacter psychrophilus sp.nov. a psychrophilic bacterium isolated from the Arctic. Int. J. Syst. Evol. Microbiol. 2008, 58, 1463–1466. [Google Scholar] [CrossRef] [PubMed]
- Abed, R.M.; Dobretsov, S.; Al-Fori, M.; Gunasekera, S.P.; Sudesh, K.; Paul, V.J. Quorum-sensing inhibitory compounds from extremophilic microorganisms isolated from a hypersaline cyanobacterial mat. J. Ind. Microbiol. Biotech. 2013, 40, 759–772. [Google Scholar] [CrossRef]
- Bagheri, M.; Amoozegar, M.A.; Didari, M.; Makhdoumi-Kakhki, A.; Schumann, P.; Sproer, C.; Sanchez-Porro, C.; Ventosa, A. Marinobacter persicus sp. nov., a moderately halophilic bacterium from a saline lake in Iran. Antonie Van Leeuwenhoek 2013, 104, 47–54. [Google Scholar] [CrossRef]
- Huu, N.B.; Denner, E.B.M.; Ha, D.T.C.; Wanner, G.; Stan-Lotter, H. Marinobacter aquaeolei sp. nov., a halophilic bacterium isolated form a Vietnamese oil-producing well. Int. J. Syst. Evol. Microbiol. 1999, 49, 367–375. [Google Scholar] [CrossRef]
- Wan, X.; Hou, S.; Burns, S.L.; Saito, J.A.; Donachie, S.P. Draft Genome Sequence of a Novel Marinobacter sp. Strain from Honolulu Harbor, Hawai’i. Genome Announc. 2016, 4, e01354-16. [Google Scholar] [CrossRef]
- De França, P.; Camilo, E.; Fantinatti-Garboginni, F. Draft Genome Sequence of Marinobacter sp. strain ANT_B65, isolated from Antarctic marine sponge. Genome Announc. 2018, 6, e01404-17. [Google Scholar] [CrossRef]
- Green, D.H.; Llewellyn, L.E.; Negri, A.P.; Blackburn, S.I.; Bolch, C.J.S. Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol. Ecol. 2004, 47, 345–357. [Google Scholar] [CrossRef]
- Green, D.H.; Bowman, J.P.; Smith, E.A.; Gutierrez, T.; Bolch, C.J.S. Marinobacter algicola sp. nov., isolated from laboratory cultures of paralytic shellfish toxin producing dinoflagellates. Int. J. Syst. Evol. Microbiol. 2006, 56, 523–527. [Google Scholar] [CrossRef]
- Kaeppel, E.C.; Gardes, A.; Seebah, S.; Grossart, H.P.; Ullrich, M.S. Marinobacter adhaerens sp.nov. isolated from marine aggregates formed with the diatom Thalassiosira weissflogii. Int. J. Syst. Evol. Microbiol. 2012, 62, 124–128. [Google Scholar] [PubMed]
- Orata, F.D.; Rosana, A.R.; Xu, Y.; Simkus, D.N.; Bramucci, A.R.; Boucher, Y.; Case, R.J. Draft genome sequences of four bacterial strains isolated from a polymicrobial culture of naked (N-type) Emiliania huxleyi CCMP. Genome Announc. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Rosana, A.R.R.; Orata, F.D.; Xu, Y.; Simkus, D.N.; Bramucci, A.R.; Boucher, Y.; Case, R.J. Draft genome sequences of seven bacterial strains isolated from a polymicrobial culture of coccolith-bearing (C-type) Emiliana huxleyi M217. Genome Announc. 2016, 4, e00673-16. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.S.; Yung, C.M.; Davis, K.M.; Rynearson, T.; Hunt, D.E. Draft genome sequences of three bacterial isolates from cultures of the marine diatom Thalassiosira rotula. Genome Announc. 2017, 5, e00316-17. [Google Scholar] [CrossRef] [PubMed]
- Singer, E.; Webb, E.A.; Nelson, W.C.; Heidelberg, J.F.; Ivanova, N.; Pati, A.; Edwards, K.J. Genomic potential of Marinobacter aquaeolei, a biogeochemical “opportunitroph”. Appl. Environ. Microbiol. 2011, 77, 2763–2771. [Google Scholar] [CrossRef] [PubMed]
- Green, D.H.; Echavarri-Bravo, V.; Brennan, D.; Hart, M.C. Bacterial diversity associated with the coccolithophorid algae Emiliania huxleyi and Coccolithus pelagicus f. braarudii. Biomed. Res. Int. 2015, 2015, 194540. [Google Scholar] [CrossRef]
- Cruz-López, R.; Maske, H. The vitamin B1 and B12 are required by the marine dinoflagellate Lingulodinium polyedrum and can be provided by its associated bacterial community in culture. Front. Microbiol. 2016, 7, 560. [Google Scholar] [CrossRef]
- Hattenrath-Lehmann, T.K.; Gobler, C.J. Identification of unique microbiomes associated with harmful algal blooms caused by Alexandrium fundyense and Dinophysis acuminata. Harmful Algae 2017, 68, 17–30. [Google Scholar] [CrossRef]
- Shin, H.; Lee, E.; Shin, J.; Ko, S.-R.; Oh, H.S.; Ahn, C.Y.; Oh, H.-M.; Cho, B.K.; Cho, S. Elucidation of the bacterial communities associated with the harmful microalgae Alexandrium tamarense and Cochlodinium polykrikoides using nanopore sequencing. Sci. Rep. 2018, 8, 5323. [Google Scholar] [CrossRef]
- Zhou, J.; Richlen, M.L.; Sehein, T.R.; Kulis, D.M.; Anderson, D.M.; Cai, Z. Microbial Community Structure and Associations during a Marine Dinoflagellate Bloom. Front. Microbiol. 2018, 9, 1201. [Google Scholar] [CrossRef]
- Amin, S.A.; Green, D.H.; Hart, M.C.; Küpper, F.C.; Sunda, W.G.; Carrano, C.J. Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc. Natl. Acad. Sci. USA 2009, 106, 17071–17076. [Google Scholar] [CrossRef] [PubMed]
- Gärdes, A.A.; Iversen, M.H.; Grossart, H.; Passow, U.; Ullrich, M.S. Diatom-associated bacteria are required for aggregation of Thalassiosira weissflogii. ISME J. 2011, 5, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Gärdes, A.A.; Ramaye, Y.; Grossart, H.; Passow, U.; Ullrich, M.S. Effects of Marinobacter adhaerens HP15 on polymer exudation by Thalassiosira weissflogii at different N: P ratios. Mar. Ecol. Prog. Ser. 2012, 461, 1–14. [Google Scholar] [CrossRef]
- Bolch, C.J.; Subramanian, T.A.; Green, D.H. The toxic dinoflagellate Gymnodinium catenatum (Dinophyceae) requires marine bacteria for growth. J. Phycol. 2011, 47, 1009–1022. [Google Scholar] [CrossRef] [PubMed]
- Bolch, C.J.; Bejoy, T.A.; Green, D.H. Bacterial Associates Modify Growth Dynamics of the Dinoflagellate Gymnodinium catenatum. Front. Microbiol. 2017, 8, 670. [Google Scholar] [CrossRef] [PubMed]
- Mayali, X.; Franks, P.S.; Burton, R.S. Temporal attachment dynamics by distinct bacterial taxa during a dinoflagellate bloom. Aquat. Microb. Ecol. 2011, 63, 111–122. [Google Scholar] [CrossRef]
- Romero, M.; Martin-Cuadrado, A.B.; Roca-Rivada, A.; Cabello, A.M.; Otero, A. Quorum quenching in cultivable bacteria from dense marine coastal microbial communities. FEMS Microbiol. Ecol. 2011, 75, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Hmelo, L.R. Quorum sensing in marine microbial environments. Annu. Rev. Mar. Sci. 2017, 9, 257–281. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Ann. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Williams, P.; Winzer, K.; Chan, W.C.; Cámara, M. Look who’s talking: Communication and quorum sensing in the bacterial world. Philos. Trans. R Soc. B Biol. Sci. 2007, 362, 1119–1134. [Google Scholar] [CrossRef]
- Parsek, M.R.; Greenberg, E.P. Acyl-homoserine lactone quorum sensing in gram negative bacteria: A signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 2000, 97, 8789–8793. [Google Scholar] [CrossRef] [PubMed]
- Kostka, J.E.; Prakash, O.; Overholt, W.A.; Green, S.J.; Freyer, G.; Canion, A.; Delgardio, J.; Norton, N.; Hazen, T.C.; Huettel, M. Hydrocarbon-Degrading Bacteria and the Bacterial Community Response in Gulf of Mexico Beach Sands Impacted by the Deepwater Horizon Oil Spill. Appl. Environ. Microbiol. 2011, 77, 7962–7974. [Google Scholar] [CrossRef] [PubMed]
- Tait, K.; Williamson, H.; Atkinson, S.; Williams, P.; Cámar, M.; Joint, I. Turnover of quorum sensing signal molecules modulates cross-kingdom signaling. Environ. Microbial. 2009, 11, 1792–1802. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Grossart, H.P.; Schlingloff, A.; Kiørboe, T. Possible quorum sensing in marine snow bacteria: Production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 2002, 68, 4111–4116. [Google Scholar] [CrossRef]
- Paul, R.; Ghosh, T.; Tang, T.; Kumar, A.; Riley, M.A. Molecular mechanisms of bacteriocin evolution. Annu. Rev. Genet. 1998, 32, 255–278. [Google Scholar]
- Todorov, S.D. Bacteriocins from Lactobacillus plantarum—production, genetic organization and mode of action. Brazilian J. Microbiol. 2009, 40, 209–221. [Google Scholar] [CrossRef]
- Riley, M.A.; Wertz, J.E. Bacteriocins: Evolution, ecology, and application. Annu. Rev. Microbiol. 2002, 56, 117–137. [Google Scholar] [CrossRef]
- Maldonado-Barragán, A.; Ruiz-Barba, J.L.; Jiménez-Díaz, R. Knockout of three-component regulatory systems reveals that the apparently constitutive plantaricin-production phenotype shown by Lactobacillus plantarum on solid medium is regulated via quorum sensing. Int. J. Food. Microbiol. 2009, 130, 35–42. [Google Scholar] [CrossRef]
- Majeed, H.A.; Gillor, O.; Kerr, B.; Riley, M.A. Competitive interactions in Escherichia coli populations: The role of bacteriocins. ISME J. 2011, 5, 71–81. [Google Scholar] [CrossRef]
- Freilich, S.; Zarecki, R.; Eilam, O.; Segal, E.S.; Henry, C.S.; Kupiec, M.; Gophna, U.; Sharan, R.; Ruppin, E. Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2011, 2, 589. [Google Scholar] [CrossRef]
- Bruce, J.B.; West, S.A.; Griffin, A.S. Bacteriocins and the assembly of natural Pseudomonas fluorescens populations. J. Evol. Biol. 2017, 30, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Bakkal, S.; Robinson, S.M.; Riley, M.A. Bacteriocins of Aquatic Microorganisms and Their Potential Applications in the Seafood Industry, Health and Environment in Aquaculture; Carvalho, D., David, G.S., Reinaldo, J.S., Eds.; Intech Open: London, UK, 2012. [Google Scholar]
- McLean, R.J.C.; Pierson, L.C., III; Fuqua, C. A simple screening protocol for the identification of quorum signal antagonists. J. Microbiol. Meth. 2004, 58, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Twigg, M. The Signal Based Relationship between the Green Seaweed Ulva and its Indigenous Bacterial Community. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2013. [Google Scholar]
- Markowitz, V.M.; Chen, I.M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Pillay, M.; Ratner, A.; Huang, J.; Woyke, T.; Huntemann, M.; et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014, 42, D560–D567. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.R.; Nobel, T.; Steele, J.A.; Schwalbach, M.S.; Hewson, I.; Fuhrman, J.A. Virus and prokaryote enumeration from planktonic marine environments. Epifluorescence microscopy with SYBR GreenI. Nat. Prot. 2007, 2, 269–276. [Google Scholar] [CrossRef]
- Be’er, A.; Ariel, G.; Kalisman, O.; Helman, Y.; Sirota-Madi, A.; Zhang, H.P.; Florin, E.; Payne, S.M.; Ben-Jacob, E.; Swinney, H.L. Lethal protein produced in response to competition between sibling bacterial colonies. Proc. Natl. Acad. Sci. USA 2010, 107, 6258–6263. [Google Scholar] [CrossRef]
- Be’er, A.; Zhang, H.G.; Florin, E.; Payne, S.M.; Ben-Jacob, E.; Swinney, H.L. Deadly competition between sibling bacterial colonies. Proc. Natl. Acad. Sci. USA 2009, 106, 428–433. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Owji, H.; Nezafat, N.; Negahdaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018, 97, 422–441. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Jensen, L.J. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Seper, A.; Fengler, V.H.; Roier, S.; Wolinski, H.; Kohlwein, S.D.; Bishop, A.L.; Camilli, A.; Reidl, J.; Schild, S. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol. Microbiol. 2011, 82, 1015–1037. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, H.; Matsushita, M. Bacterial fractal growth in the concentration field of nutrient. J. Phys. Soc. Jpn. 1991, 60, 88–94. [Google Scholar] [CrossRef]
- Wakita, J.I.; Komatsu, K.; Nakahara, A.; Matsuyama, T.; Matsushita, M. Experimental investigation on the validity of population dynamics approach to bacterial colony formation. J. Phys. Soc. Jpn. 1994, 63, 1205–1211. [Google Scholar] [CrossRef]
- Paul, R.; Ghosh, T.; Tang, T.; Kumar, A. Rivalry in Bacillus subtilis colonies: Enemy or family? Soft Matter 2019, 15, 5400–5411. [Google Scholar] [CrossRef] [PubMed]
- Sekowska, A.; Masson, J.B.; Celani, A.; Danchin, A.; Vergassola, M. Repulsion and Metabolic Switches in the Collective Behavior of Bacterial Colonies. Biophys. J. 2009, 97, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, S.; Nakayama, M.; Shoji, W. Self-organization of bacterial communities against environmental pH variation: Controlled chemotactic motility arranges cell population structures in biofilms. PLoS ONE 2017, 12, e0173195. [Google Scholar] [CrossRef]
- Troselj, V.; Treuner-Lange, A.; Søgaard-Andersen, L.; Wall, D. Physiological Heterogeneity Triggers Sibling Conflict Mediated by the Type VI Secretion System in an Aggregative Multicellular Bacterium. mBio 2018, 9, e01645-17. [Google Scholar] [CrossRef]
- Tipping, M.J.; Gibbs, K.A. Peer pressure from a Proteus mirabilis selfrecognition system controls participation in cooperative swarm motility. PLoS Pathog. 2019, 15. [Google Scholar] [CrossRef]
- Antelmann, H.; Scharf, C.; Hecker, M. Phosphate Starvation-Inducible Proteins of Bacillus subtilis: Proteomics and Transcriptional Analysis. J. Bacterial. 2000, 182, 4478–4490. [Google Scholar] [CrossRef]
- Roy, A.B.; Petrova, O.E.; Sauer, K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacterial. 2012, 194, 2904–2915. [Google Scholar] [CrossRef]
- Calvez, S.; Rincé, A.; Auffray, Y.; Prévost, H.; Drider, D. Identification of new genes associated with intermediate resistance of Enterococcus faecalis to divercin V41, a pediocin-like bacteriocin. Microbiology 2007, 153, 1609–1618. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pesavento, C.; Hengge, R. Bacterial nucleotide-based second messengers. Curr. Opin. Microbiol. 2009, 12, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Jenal, U.A.; Reinders, C. Lori Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 2017, 15, 271–284. [Google Scholar] [CrossRef] [PubMed]



| Node | Annotation | Acc. Number | Score |
|---|---|---|---|
| EDM46611.1 | Glycerophosphoryl diester phosphodiesterase | MDG893_19429 | Query |
| EDM49251.1 | FAD dependent oxidoreductase | MDG893_07635 | 0.890 |
| EDM47432.1 | Glycerophosphoryl diester phosphodiesterase | MDG893_00180 | 0.877 |
| gpsA | Glycerol-3-phosphate dehydrogenase (GPDH) | MDG893_06880 | 0.807 |
| EDM47089.1 | Glycerol kinase | MDG893_11889 | 0.756 |
| EDM45841.1 | Glycerol kinase | MDG893_05159 | 0.756 |
| EDM46512.1 | Phosphodiesterase/alkaline phosphatase D | MDG893_20089 | 0.754 |
| EDM47357.1 | Flagellar secretion chaperone protein FliS | MDG893_01080 | 0.724 |
| EDM46845.1 | Extracellular nuclease | MDG893_17732 | 0.721 |
| EDM47042.1 | Predicted phosphatase | MDG893_02110 | 0.712 |
| EDM47259.1 | Chemotaxis protein histidine kinase | MDG893_18974 | 0.681 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-López, R.; Kolesinski, P.; De Boever, F.; Green, D.H.; Carrano, M.W.; Carrano, C.J. Loss of Motility as a Non-Lethal Mechanism for Intercolony Inhibition (“Sibling Rivalry”) in Marinobacter. Microorganisms 2021, 9, 103. https://doi.org/10.3390/microorganisms9010103
Cruz-López R, Kolesinski P, De Boever F, Green DH, Carrano MW, Carrano CJ. Loss of Motility as a Non-Lethal Mechanism for Intercolony Inhibition (“Sibling Rivalry”) in Marinobacter. Microorganisms. 2021; 9(1):103. https://doi.org/10.3390/microorganisms9010103
Chicago/Turabian StyleCruz-López, Ricardo, Piotr Kolesinski, Frederik De Boever, David H. Green, Mary W. Carrano, and Carl J. Carrano. 2021. "Loss of Motility as a Non-Lethal Mechanism for Intercolony Inhibition (“Sibling Rivalry”) in Marinobacter" Microorganisms 9, no. 1: 103. https://doi.org/10.3390/microorganisms9010103
APA StyleCruz-López, R., Kolesinski, P., De Boever, F., Green, D. H., Carrano, M. W., & Carrano, C. J. (2021). Loss of Motility as a Non-Lethal Mechanism for Intercolony Inhibition (“Sibling Rivalry”) in Marinobacter. Microorganisms, 9(1), 103. https://doi.org/10.3390/microorganisms9010103






