Gold Nanoparticles and Graphene Oxide Flakes Synergistic Partaking in Cytosolic Bactericidal Augmentation: Role of ROS and NOX2 Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bone Marrow-Derived Macrophages (BMDMs)
2.3. Blood Sampling and Preparation
2.3.1. Blood Samples
2.3.2. Human Neutrophils Isolation
2.4. Intracellular Bacterial Killing Assay
2.5. Determination of pH of Phagosomes
2.6. Assay of Phagosome/Lysosome Fusion
2.7. Phagocytosis of pHrodo E. coli BioParticles by BMDMs
2.8. NADPH Oxidase Assay
2.9. Statistical Analysis
3. Results and Discussion
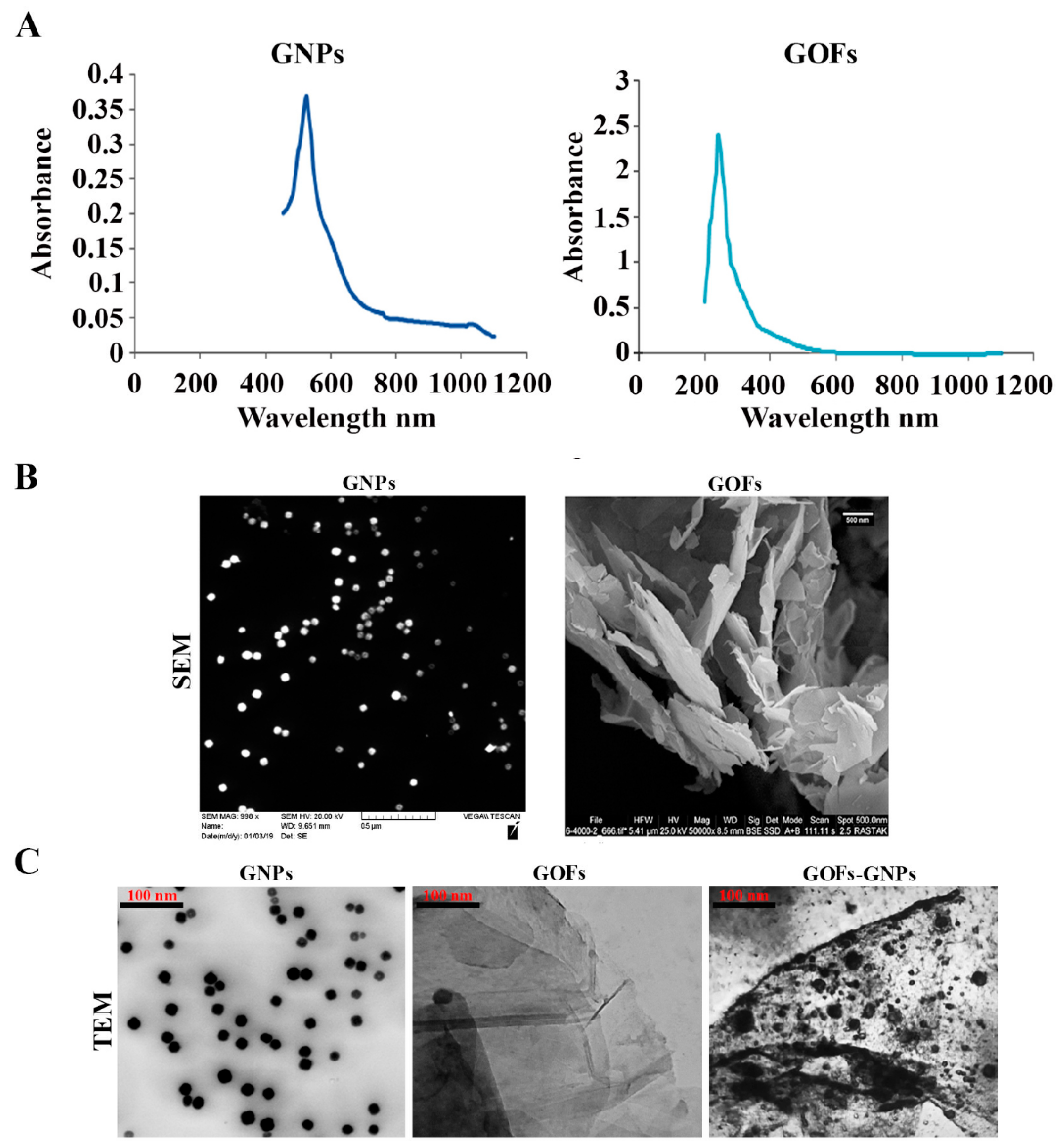
3.1. Characterization of the GNPs and GOFs
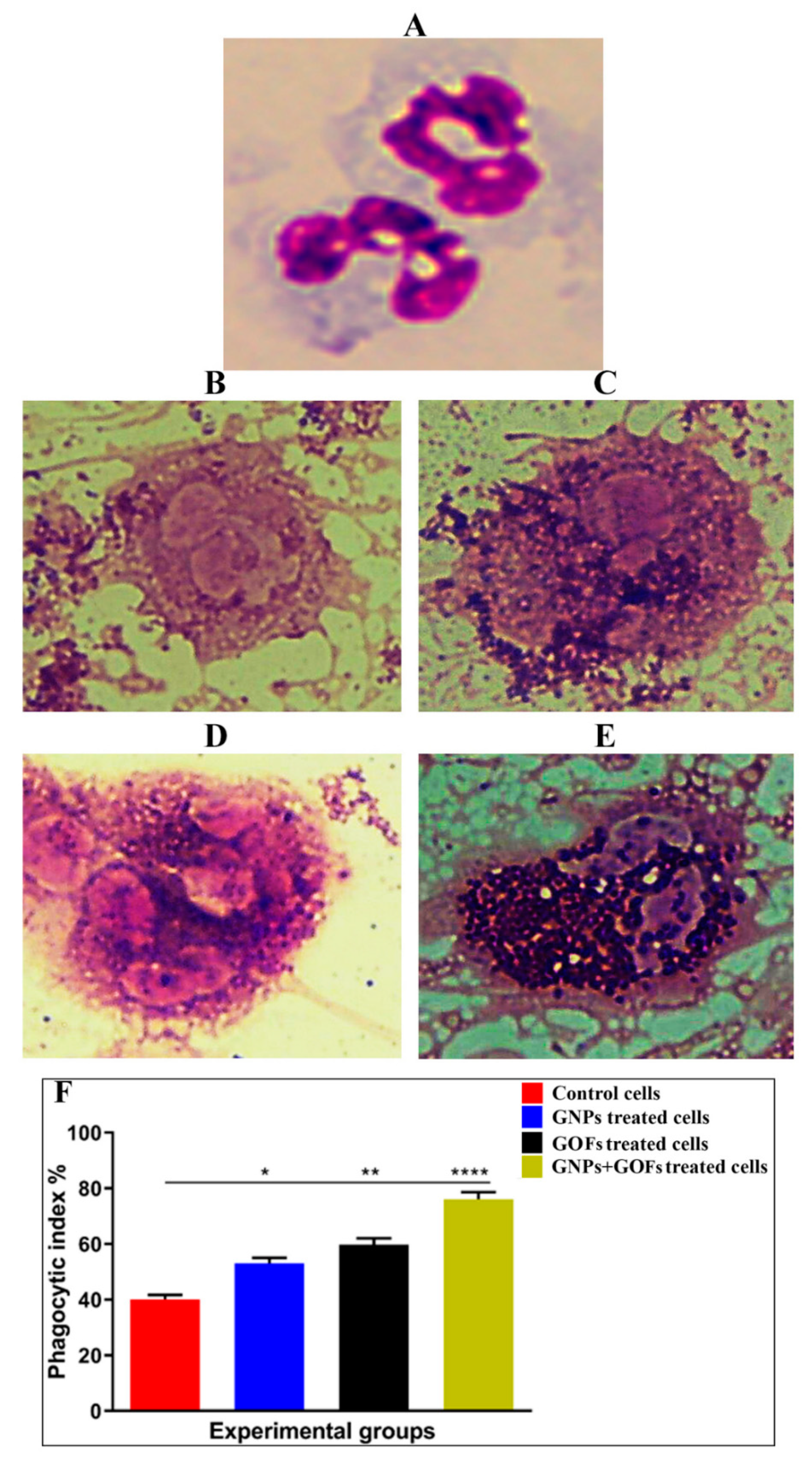
3.2. GNPs and GOFs Increase the Phagocytic Index of Human Neutrophils
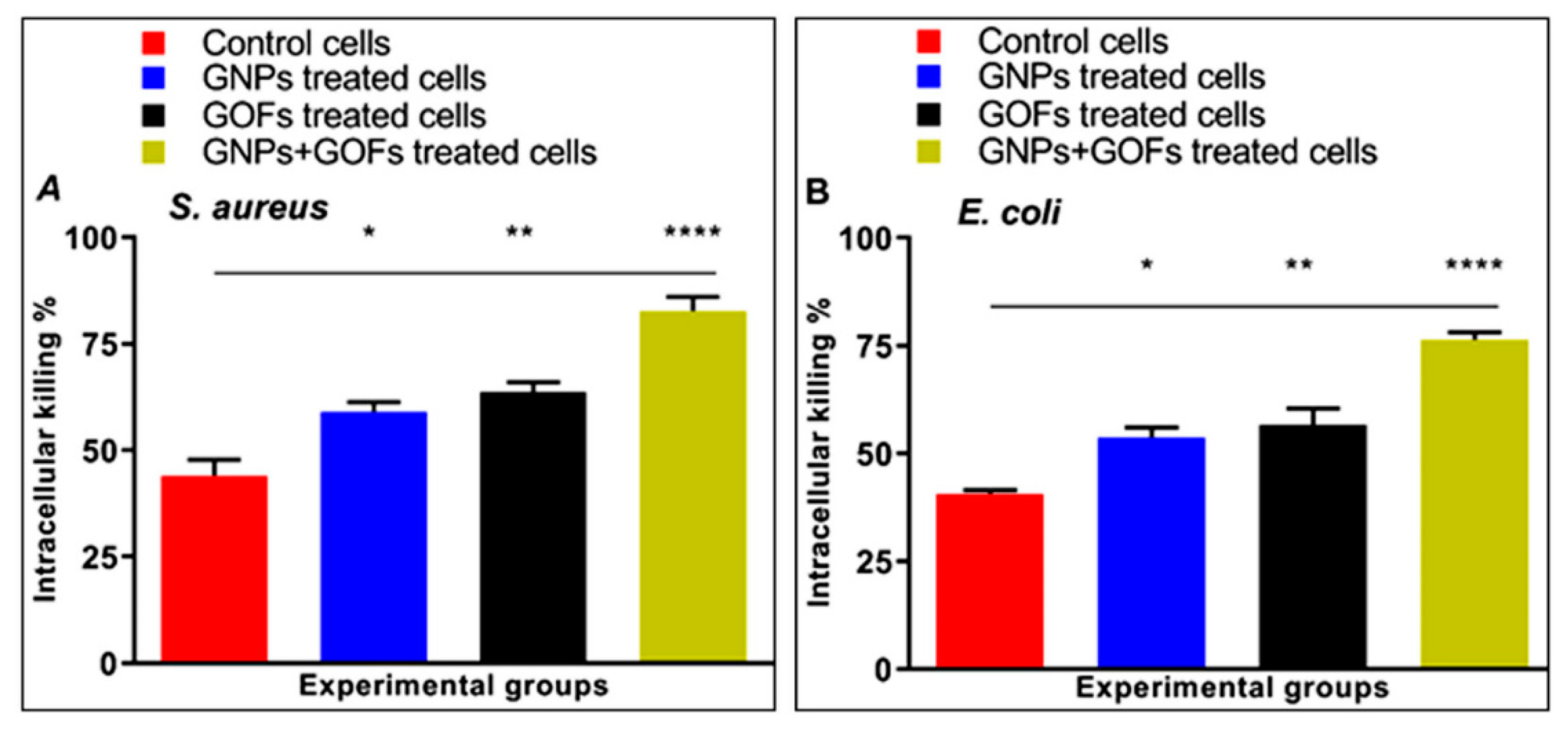
3.3. GNPs and GOFs Increase Bacterial Intracellular Killings
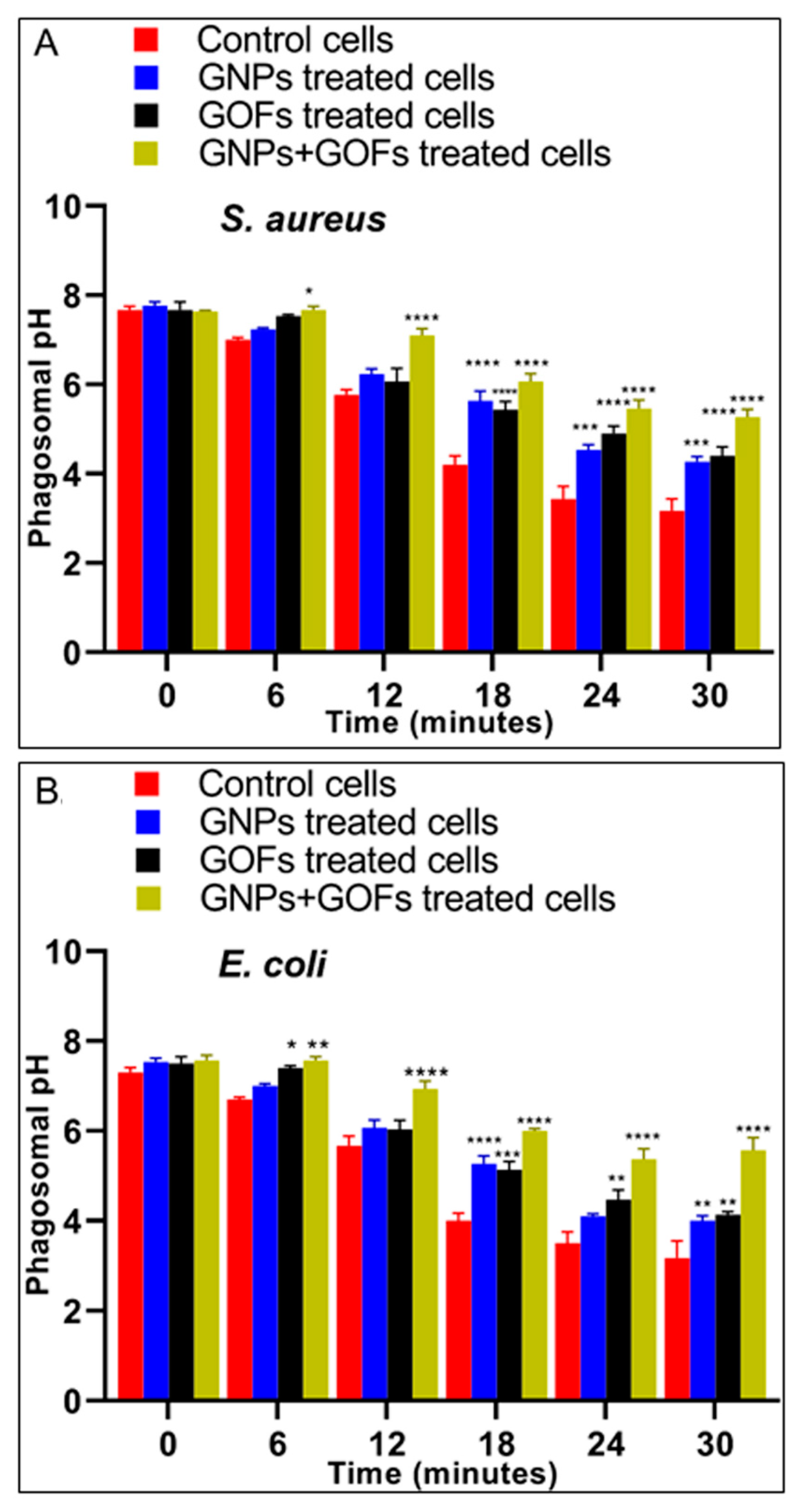
3.4. The GNPs and GOFs in Phagosome Maturation
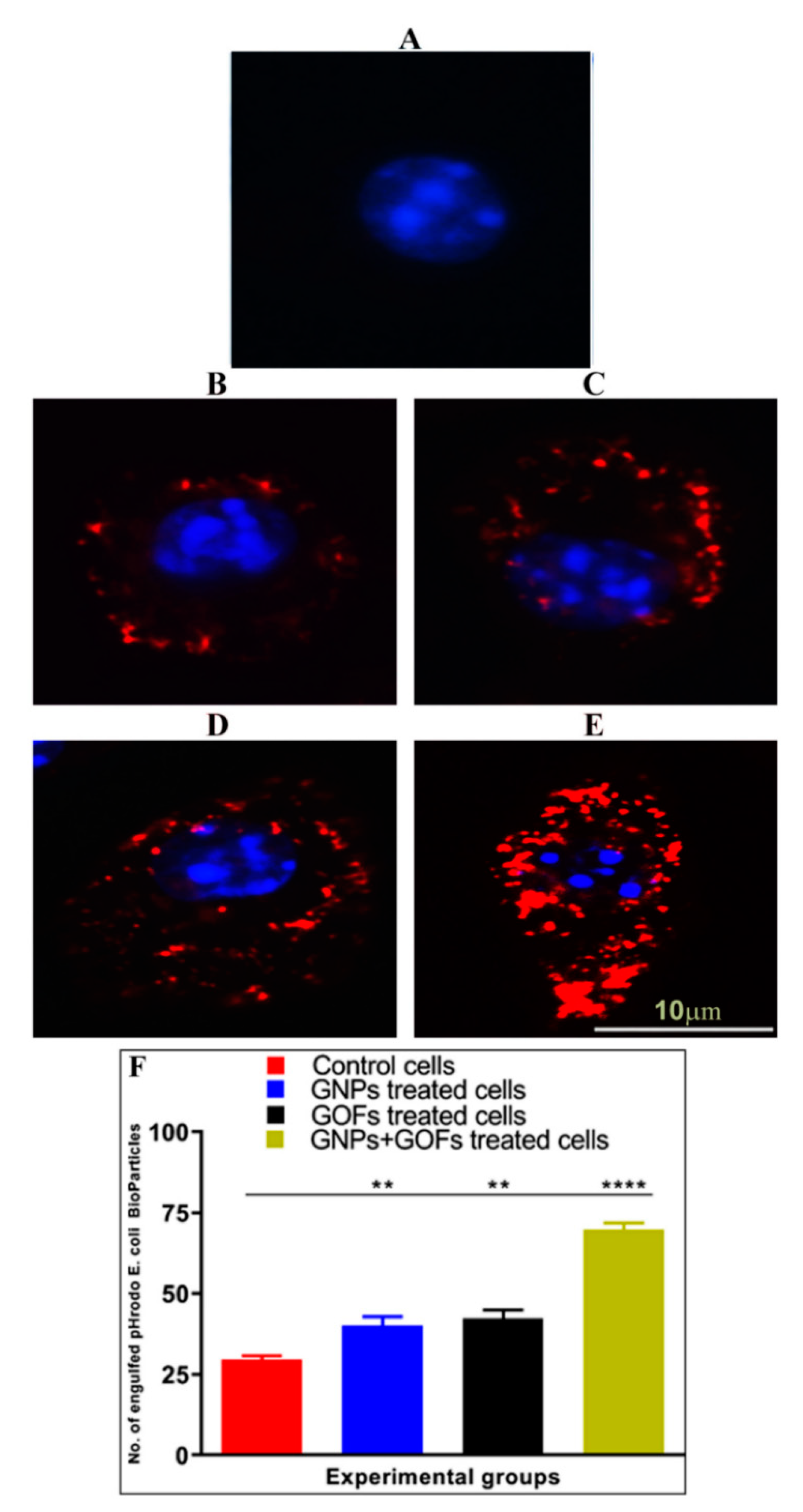
3.5. GNPs and GOFs Increase Phagocytosis of pHrodo E. coli Bioparticles by BMDMs
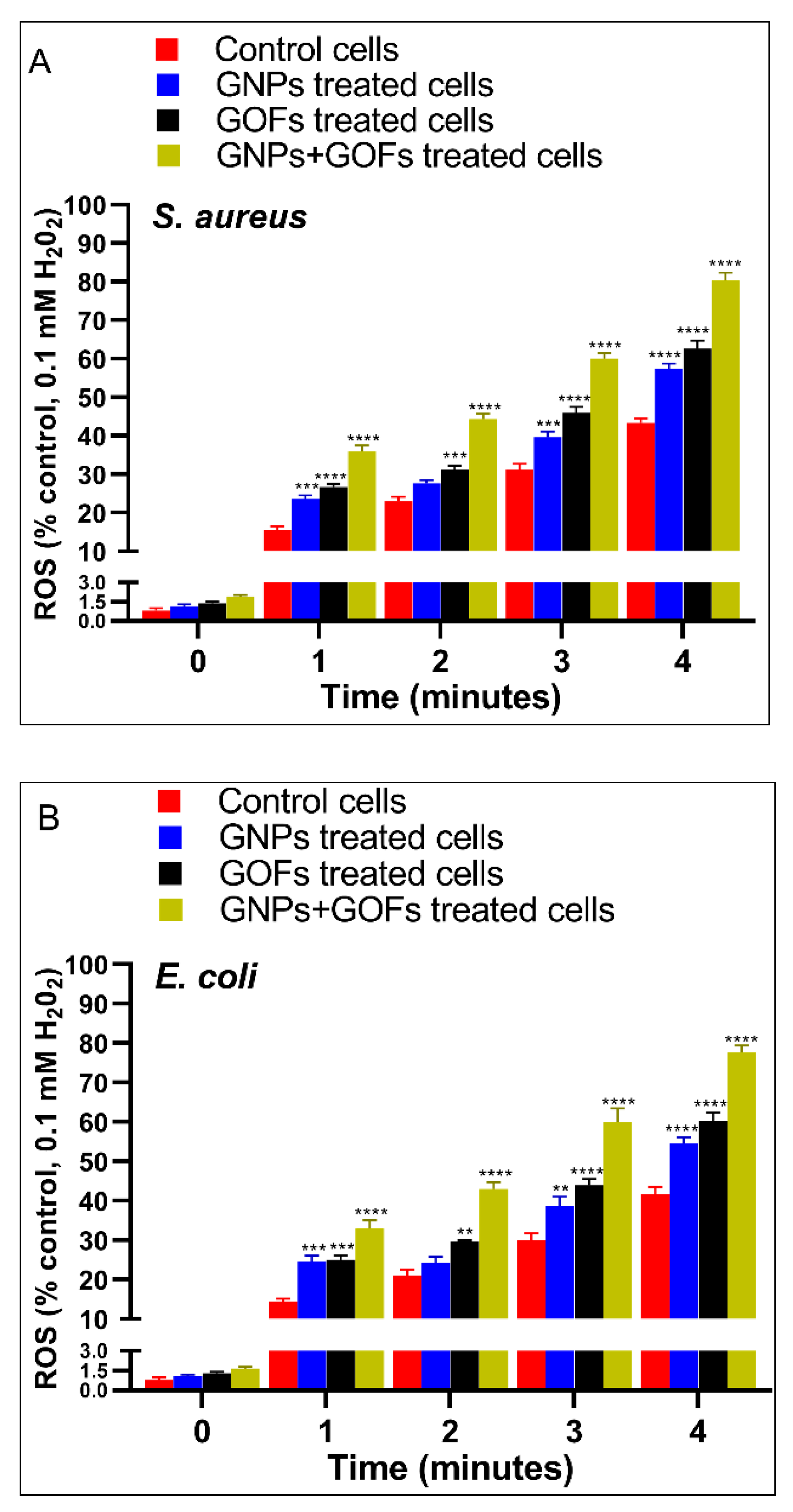
3.6. GNPs and GOFs Induce NOX2 Function in BMDMs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Taki, A.; Smooker, P. Small Wonders: The Use of Nanoparticles for Delivering Antigen. Vaccines 2015, 3, 638–661. [Google Scholar] [CrossRef] [PubMed]
- Kaittanis, C.; Shaffer, T.M.; Thorek, D.L.J.; Grimm, J. Dawn of Advanced Molecular Medicine: Nanotechnological Advancements in Cancer Imaging and Therapy. Crit. Rev. Oncog. 2014, 19, 143–176. [Google Scholar] [CrossRef]
- Raza, A.; Sime, F.B.; Cabot, P.J.; Maqbool, F.; Roberts, J.A.; Falconer, J.R. Solid nanoparticles for oral antimicrobial drug delivery: A review. Drug Discov. Today 2019, 24, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Canaparo, R.; Foglietta, F.; Giuntini, F.; Della Pepa, C.; Dosio, F.; Serpe, L. Recent developments in antibacterial therapy: Focus on stimuli-responsive drug-delivery systems and therapeutic nanoparticles. Molecules 2019, 24, 1991. [Google Scholar] [CrossRef]
- Getts, D.R.; Shea, L.D.; Miller, S.D.; King, N.J.C. Harnessing nanoparticles for immune modulation. Trends Immunol. 2015, 36, 419–427. [Google Scholar] [CrossRef]
- Assali, A.; Razzazan, S.; Akhavan, O.; Mottaghitalab, F.; Adeli, M.; Atyabi, F. The bio-interface between functionalized Au NR@GO nanoplatforms with protein corona and their impact on delivery and release system. Colloids Surf. B Biointerfaces 2019, 173, 891–898. [Google Scholar] [CrossRef]
- Assali, A.; Akhavan, O.; Adeli, M.; Razzazan, S.; Dinarvand, R.; Zanganeh, S.; Soleimani, M.; Dinarvand, M.; Atyabi, F. Multifunctional core-shell nanoplatforms (gold@graphene oxide) with mediated NIR thermal therapy to promote miRNA delivery. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1891–1903. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Singh, V.K.; Hunter, R.L.; Jagannath, C. Macrophage heterogeneity and plasticity in tuberculosis. J. Leukoc. Biol. 2019, 106, 275–282. [Google Scholar] [CrossRef]
- Zhang, Y.; Mortimer, M.; Guo, L.-H. Interplay between engineered nanomaterials and microbiota. Environ. Sci. Nano 2020, 7, 2454–2485. [Google Scholar] [CrossRef]
- Giovanni, M.; Yue, J.; Zhang, L.; Xie, J.; Ong, C.N.; Leong, D.T. Pro-inflammatory responses of RAW264.7 macrophages when treated with ultralow concentrations of silver, titanium dioxide, and zinc oxide nanoparticles. J. Hazard. Mater. 2015, 297, 146–152. [Google Scholar] [CrossRef]
- Zanganeh, S.; Hutter, G.; Spitler, R.; Lenkov, O.; Mahmoudi, M.; Shaw, A.; Pajarinen, J.S.; Nejadnik, H.; Goodman, S.; Moseley, M.; et al. Iron oxide nanoparticles inhibit tumour growth by inducing pro-inflammatory macrophage polarization in tumour tissues. Nat. Nanotechnol. 2016, 11, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.A.; Bolus, W.R.; Hasty, A.H. A decade of progress in adipose tissue macrophage biology. Immunol. Rev. 2014, 262, 134–152. [Google Scholar] [CrossRef] [PubMed]
- Roszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef] [PubMed]
- MacParland, S.A.; Tsoi, K.M.; Ouyang, B.; Ma, X.-Z.; Manuel, J.; Fawaz, A.; Ostrowski, M.A.; Alman, B.A.; Zilman, A.; Chan, W.C.W.; et al. Phenotype Determines Nanoparticle Uptake by Human Macrophages from Liver and Blood. ACS Nano 2017, 11, 2428–2443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Cao, S.; Liang, S.; Tan, C.H.; Luo, B.; Xu, X.; Saw, P.E. Differently Charged Super-Paramagnetic Iron Oxide Nanoparticles Preferentially Induced M1-Like Phenotype of Macrophages. Front. Bioeng. Biotechnol. 2020, 8, 537. [Google Scholar] [CrossRef]
- Yao, Y.; Zang, Y.; Qu, J.; Tang, M.; Zhang, T. The Toxicity Of Metallic Nanoparticles On Liver: The Subcellular Damages, Mechanisms, And Outcomes. Int. J. Nanomed. 2019, 14, 8787. [Google Scholar] [CrossRef]
- Dukhinova, M.S.; Prilepskii, A.; Shtil, A.A.; Vinogradov, V.V. Metal oxide nanoparticles in therapeutic regulation of macrophage functions. Nanomaterials 2019, 9, 1631. [Google Scholar] [CrossRef]
- Alphandéry, E. Iron oxide nanoparticles for therapeutic applications. Drug Discov. Today 2020, 25, 141–149. [Google Scholar] [CrossRef]
- Fuchs, A.-K.; Syrovets, T.; Haas, K.A.; Loos, C.; Musyanovych, A.; Mailänder, V.; Landfester, K.; Simmet, T. Carboxyl- and amino-functionalized polystyrene nanoparticles differentially affect the polarization profile of M1 and M2 macrophage subsets. Biomaterials 2016, 85, 78–87. [Google Scholar] [CrossRef]
- Talekar, M.; Tran, T.-H.; Amiji, M. Translational Nano-Medicines: Targeted Therapeutic Delivery for Cancer and Inflammatory Diseases. AAPS J. 2015, 17, 813–827. [Google Scholar] [CrossRef]
- Oh, N.; Park, J.H. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int. J. Nanomed. 2014, 9, 51–63. [Google Scholar]
- Peng, J.; Liang, X. Progress in research on gold nanoparticles in cancer management. Medicine 2019, 98, e15311. [Google Scholar] [CrossRef] [PubMed]
- Nicol, J.R.; Dixon, D.; Coulter, J.A. Gold nanoparticle surface functionalization: A necessary requirement in the development of novel nanotherapeutics. Nanomedicine 2015, 10, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Dellacherie, M.O.; Seo, B.R.; Mooney, D.J. Macroscale biomaterials strategies for local immunomodulation. Nat. Rev. Mater. 2019, 4, 379–397. [Google Scholar] [CrossRef]
- Hegab, H.M.; Elmekawy, A.; Zou, L.; Mulcahy, D.; Saint, C.P.; Ginic-Markovic, M. The controversial antibacterial activity of graphene-based materials. Carbon N. Y. 2016, 105, 362–376. [Google Scholar] [CrossRef]
- Kumar, P.; Huo, P.; Zhang, R.; Liu, B. Antibacterial Properties of Graphene-Based Nanomaterials. Nanomaterials 2019, 9, 737. [Google Scholar] [CrossRef] [PubMed]
- Sima, L.E.; Chiritoiu, G.; Negut, I.; Grumezescu, V.; Orobeti, S.; Munteanu, C.V.A.; Sima, F.; Axente, E. Functionalized Graphene Oxide Thin Films for Anti-tumor Drug Delivery to Melanoma Cells. Front. Chem. 2020, 8, 184. [Google Scholar] [CrossRef]
- Orecchioni, M.; Cabizza, R.; Bianco, A.; Delogu, L.G. Graphene as cancer theranostic tool: Progress and future challenges. Theranostics 2015, 5, 710–723. [Google Scholar] [CrossRef]
- Liu, J.; Cui, L.; Losic, D. Graphene and graphene oxide as new nanocarriers for drug delivery applications. Acta Biomater. 2013, 9, 9243–9257. [Google Scholar] [CrossRef]
- Bai, R.G.; Husseini, G.A. Graphene-based drug delivery systems. In Biomimetic Nanoengineered Materials for Advanced Drug Delivery; Elsevier: Amsterdam, The Netherlands, 2019; pp. 149–168. [Google Scholar]
- Henna, T.K.; Nivitha, K.P.; Raphey, V.R.; Sabu, C.; Pramod, K. Functionalized Graphene for Drug Delivery Applications. In Carbon Nanostructures; Springer International Publishing: Singapore, 2019; pp. 247–278. [Google Scholar]
- Bai, Y.; Xu, T.; Zhang, X. Graphene-based biosensors for detection of biomarkers. Micromachines 2020, 11, 60. [Google Scholar] [CrossRef]
- Sreejith, S.; Joshi, H.; Zhao, Y. Graphene-based materials in biosensing, bioimaging, and therapeutics. In Carbon Nanostructures; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 35–61. [Google Scholar]
- Zhu, X.; Liu, Y.; Li, P.; Nie, Z.; Li, J. Applications of graphene and its derivatives in intracellular biosensing and bioimaging. Analyst 2016, 141, 4541–4553. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Huang, Y.; Huang, P. Graphene-Based Nanomaterials in Bioimaging. In Biomedical Applications of Functionalized Nanomaterials: Concepts, Development and Clinical Translation; Elsevier: Amsterdam, The Netherlands, 2018; pp. 247–287. ISBN 9780323508797. [Google Scholar]
- Campbell, E.; Hasan, M.T.; Pho, C.; Callaghan, K.; Akkaraju, G.R.; Naumov, A.V. Graphene Oxide as a Multifunctional Platform for Intracellular Delivery, Imaging, and Cancer Sensing. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Bitounis, D.; Ali-Boucetta, H.; Hong, B.H.; Min, D.-H.; Kostarelos, K. Prospects and Challenges of Graphene in Biomedical Applications. Adv. Mater. 2013, 25, 2258–2268. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, M. Polymer Nanocomposites-A Comparison between Carbon Nanotubes, Graphene, and Clay as Nanofillers. Materials 2016, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kim, J.H. Synthesis, toxicity, biocompatibility, and biomedical applications of graphene and graphene-related materials. Int. J. Nanomed. 2016, 1927. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.P.; Bottini, M.; Fadeel, B. Graphene and the Immune System: A Romance of Many Dimensions. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Yue, H.; Wei, W.; Yue, Z.; Wang, B.; Luo, N.; Gao, Y.; Ma, D.; Ma, G.; Su, Z. The role of the lateral dimension of graphene oxide in the regulation of cellular responses. Biomaterials 2012, 33, 4013–4021. [Google Scholar] [CrossRef]
- Mendes, R.G.; Koch, B.; Bachmatiuk, A.; Ma, X.; Sanchez, S.; Damm, C.; Schmidt, O.G.; Gemming, T.; Eckert, J.; Rümmeli, M.H. A size dependent evaluation of the cytotoxicity and uptake of nanographene oxide. J. Mater. Chem. B 2015, 3, 2522–2529. [Google Scholar] [CrossRef]
- Zhang, W.; Yan, L.; Li, M.; Zhao, R.; Yang, X.; Ji, T.; Gu, Z.; Yin, J.-J.; Gao, X.; Nie, G. Deciphering the underlying mechanisms of oxidation-state dependent cytotoxicity of graphene oxide on mammalian cells. Toxicol. Lett. 2015, 237, 61–71. [Google Scholar] [CrossRef]
- Gurunathan, S.; Woong Han, J.; Abdal Daye, A.; Eppakayala, V.; Kim, J. Oxidative stress-mediated antibacterial activity of graphene oxide and reduced graphene oxide in Pseudomonas aeruginosa. Int. J. Nanomed. 2012, 7, 5901. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.W.; Eppakayala, V.; Dayem, A.A.; Kwon, D.-N.; Kim, J.-H. Biocompatibility effects of biologically synthesized graphene in primary mouse embryonic fibroblast cells. Nanoscale Res. Lett. 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Park, J.H.; Kim, J.-H. An in vitro evaluation of graphene oxide reduced by Ganoderma spp. in human breast cancer cells (MDA-MB-231). Int. J. Nanomed. 2014, 1783. [Google Scholar] [CrossRef] [PubMed]
- Jabir, M.S.; Sulaiman, G.M.; Taqi, Z.J.; Li, D. Iraqi propolis increases degradation of IL-1beta and NLRC4 by autophagy following Pseudomonas aeruginosa infection. Microbes Infect. 2018, 20, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, G.M.; Waheeb, H.M.; Jabir, M.S.; Khazaal, S.H.; Dewir, Y.H.; Naidoo, Y. Hesperidin Loaded on Gold Nanoparticles as a Drug Delivery System for a Successful Biocompatible, Anti-Cancer, Anti-Inflammatory and Phagocytosis Inducer Model. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.; Siano, B.; Diamond, S. Neutrophil isolation protocol. J. Vis. Exp. 2008, 17. [Google Scholar] [CrossRef]
- Berger, S.B.; Romero, X.; Ma, C.; Wang, G.; Faubion, W.A.; Liao, G.; Compeer, E.; Keszei, M.; Rameh, L.; Wang, N.; et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat. Immunol. 2010, 11, 920–927. [Google Scholar] [CrossRef]
- Gunia-Krzyżak, A.; Bareyre, F.M.; Marona, H.; Waszkielewicz, A.M. Four N-(E)-cinnamoyl (cinnamamide) derivatives of aminoalkanols with promising anticonvulsant and analgesic activity. Bioorg. Med. Chem. Lett. 2019. [Google Scholar] [CrossRef]
- Akhavan, O. Bacteriorhodopsin as a superior substitute for hydrazine in chemical reduction of single-layer graphene oxide sheets. Carbon N. Y. 2015, 81, 158–166. [Google Scholar] [CrossRef]
- Kudin, K.N.; Ozbas, B.; Schniepp, H.C.; Prud’homme, R.K.; Aksay, I.A.; Car, R. Raman spectra of graphite oxide and functionalized graphene sheets. Nano Lett. 2008, 8, 36–41. [Google Scholar] [CrossRef]
- Graf, D.; Molitor, F.; Ensslin, K.; Stampfer, C.; Jungen, A.; Hierold, C.; Wirtz, L. Spatially resolved raman spectroscopy of single- and few-layer graphene. Nano Lett. 2007, 7, 238–242. [Google Scholar] [CrossRef]
- Beste, M.T.; Lomakina, E.B.; Hammer, D.A.; Waugh, R.E. Immobilized IL-8 Triggers Phagocytosis and Dynamic Changes in Membrane Microtopology in Human Neutrophils. Ann. Biomed. Eng. 2015, 43, 2207–2219. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.A.; Kemp, S.; Young, L.; Ross, M.; Prach, M.; Hutchison, G.R.; Malone, E. Silver nanoparticles promote the emergence of heterogeneic human neutrophil sub-populations. Sci. Rep. 2018, 8, 7506. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, O.; Ghaderi, E. Escherichia coli bacteria reduce graphene oxide to bactericidal graphene in a self-limiting manner. Carbon N. Y. 2012, 50, 1853–1860. [Google Scholar] [CrossRef]
- Lee, H.-J.; Woo, Y.; Hahn, T.-W.; Jung, Y.M.; Jung, Y.-J. Formation and Maturation of the Phagosome: A Key Mechanism in Innate Immunity against Intracellular Bacterial Infection. Microorganisms 2020, 8, 1298. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Biological chemistry of superoxide radicals. Chemtexts 2020, 6, 7. [Google Scholar] [CrossRef]
- Nauseef, W.M. The phagocyte NOX2 NADPH oxidase in microbial killing and cell signaling. Curr. Opin. Immunol. 2019, 60, 130–140. [Google Scholar] [CrossRef]
- Liu, S.; Zeng, T.H.; Hofmann, M.; Burcombe, E.; Wei, J.; Jiang, R.; Kong, J.; Chen, Y. Antibacterial activity of graphite, graphite oxide, graphene oxide, and reduced graphene oxide: Membrane and oxidative stress. ACS Nano 2011, 5, 6971–6980. [Google Scholar] [CrossRef]
- Koyyada, A.; Orsu, P. Safety and toxicity concerns of graphene and its composites. Anal. Appl. Graphene Compr. Anal. Chem. 2020, 91, 327. [Google Scholar]
- Akhavan, O.; Ghaderi, E.; Akhavan, A. Size-dependent genotoxicity of graphene nanoplatelets in human stem cells. Biomaterials 2012, 33, 8017–8025. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E.; Esfandiar, A. Wrapping bacteria by graphene nanosheets for isolation from environment, reactivation by sonication, and inactivation by near-infrared irradiation. J. Phys. Chem. B 2011, 115, 6279–6288. [Google Scholar] [CrossRef]
- Hashemi, E.; Akhavan, O.; Shamsara, M.; Rahighi, R.; Esfandiar, A.; Tayefeh, A.R. Cyto and genotoxicities of graphene oxide and reduced graphene oxide sheets on spermatozoa. RSC Adv. 2014, 4, 27213–27223. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Rugaie, O.; Jabir, M.; Kadhim, R.; Karsh, E.; Sulaiman, G.M.; Mohammed, S.A.A.; Khan, R.A.; Mohammed, H.A. Gold Nanoparticles and Graphene Oxide Flakes Synergistic Partaking in Cytosolic Bactericidal Augmentation: Role of ROS and NOX2 Activity. Microorganisms 2021, 9, 101. https://doi.org/10.3390/microorganisms9010101
Al Rugaie O, Jabir M, Kadhim R, Karsh E, Sulaiman GM, Mohammed SAA, Khan RA, Mohammed HA. Gold Nanoparticles and Graphene Oxide Flakes Synergistic Partaking in Cytosolic Bactericidal Augmentation: Role of ROS and NOX2 Activity. Microorganisms. 2021; 9(1):101. https://doi.org/10.3390/microorganisms9010101
Chicago/Turabian StyleAl Rugaie, Osamah, Majid Jabir, Rua Kadhim, Esraa Karsh, Ghassan M. Sulaiman, Salman A. A. Mohammed, Riaz A. Khan, and Hamdoon A. Mohammed. 2021. "Gold Nanoparticles and Graphene Oxide Flakes Synergistic Partaking in Cytosolic Bactericidal Augmentation: Role of ROS and NOX2 Activity" Microorganisms 9, no. 1: 101. https://doi.org/10.3390/microorganisms9010101
APA StyleAl Rugaie, O., Jabir, M., Kadhim, R., Karsh, E., Sulaiman, G. M., Mohammed, S. A. A., Khan, R. A., & Mohammed, H. A. (2021). Gold Nanoparticles and Graphene Oxide Flakes Synergistic Partaking in Cytosolic Bactericidal Augmentation: Role of ROS and NOX2 Activity. Microorganisms, 9(1), 101. https://doi.org/10.3390/microorganisms9010101







