Enhancing the Rice Seedlings Growth Promotion Abilities of Azoarcus sp. CIB by Heterologous Expression of ACC Deaminase to Improve Performance of Plants Exposed to Cadmium Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Seeds and Plasmids Used
2.2. Culture Media and Growth Conditions
2.3. Molecular Biology Techniques
2.4. Construction of Plasmids pIZacds and pSEVA237acdS
2.5. Inoculation of Rice Seedlings with Bacteria
2.6. Examination of the Growth of Inoculated Rice Plants in Climate Chambers
2.7. Examination of the Growth of Inoculated Rice Plants under Greenhouse Conditions
2.8. ACC Deaminase Enzymatic Assay
2.9. Cadmium Concentration in Plants Tissues
2.10. Superoxide Dismutase (SOD) Enzymatic Assay
2.11. Statistical Analysis
3. Results and Discussion
3.1. Azoarcus sp. CIB Is Able to Promote Plant Growth
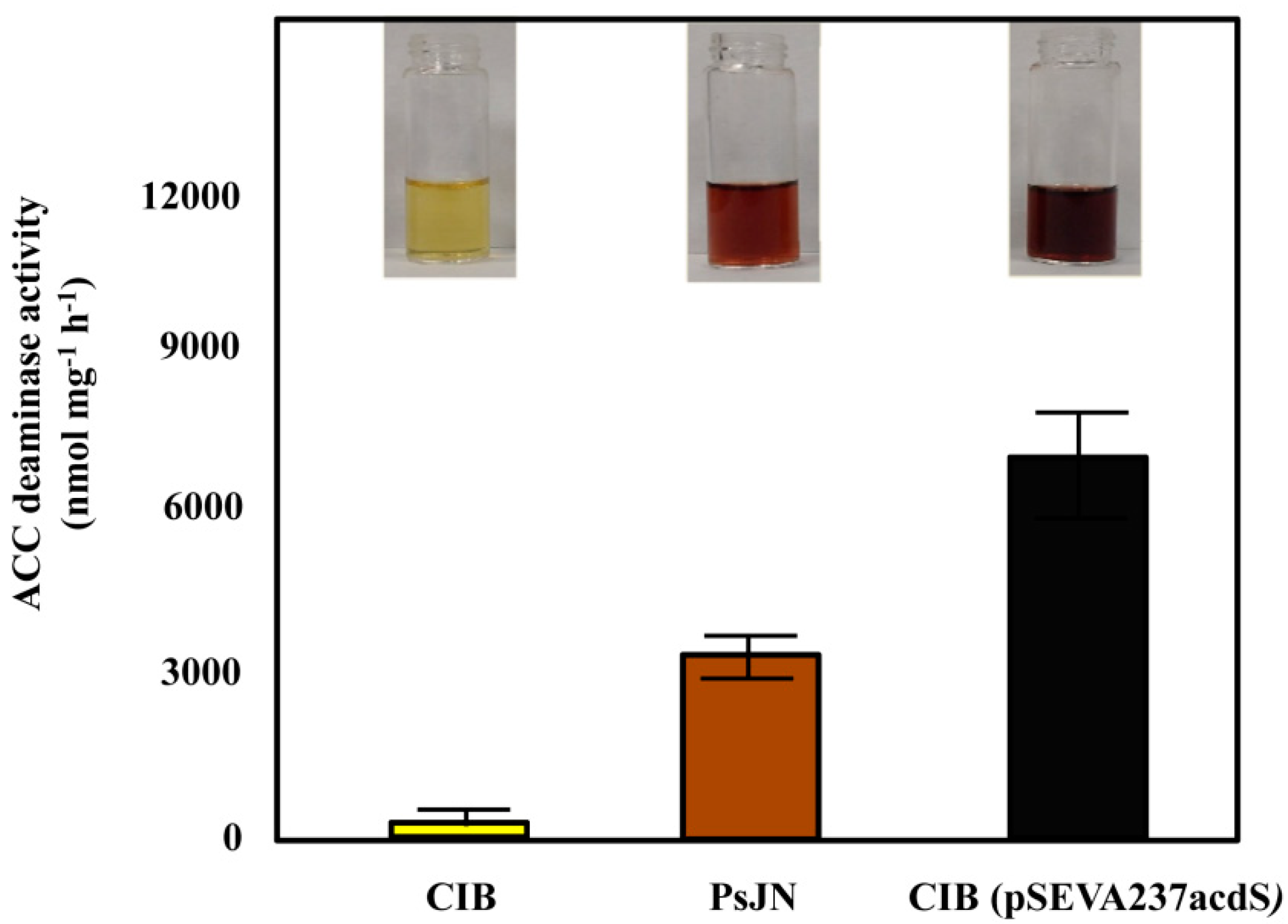
3.2. Engineering an ACC Deaminase-Producing Azoarcus sp. CIB Strain
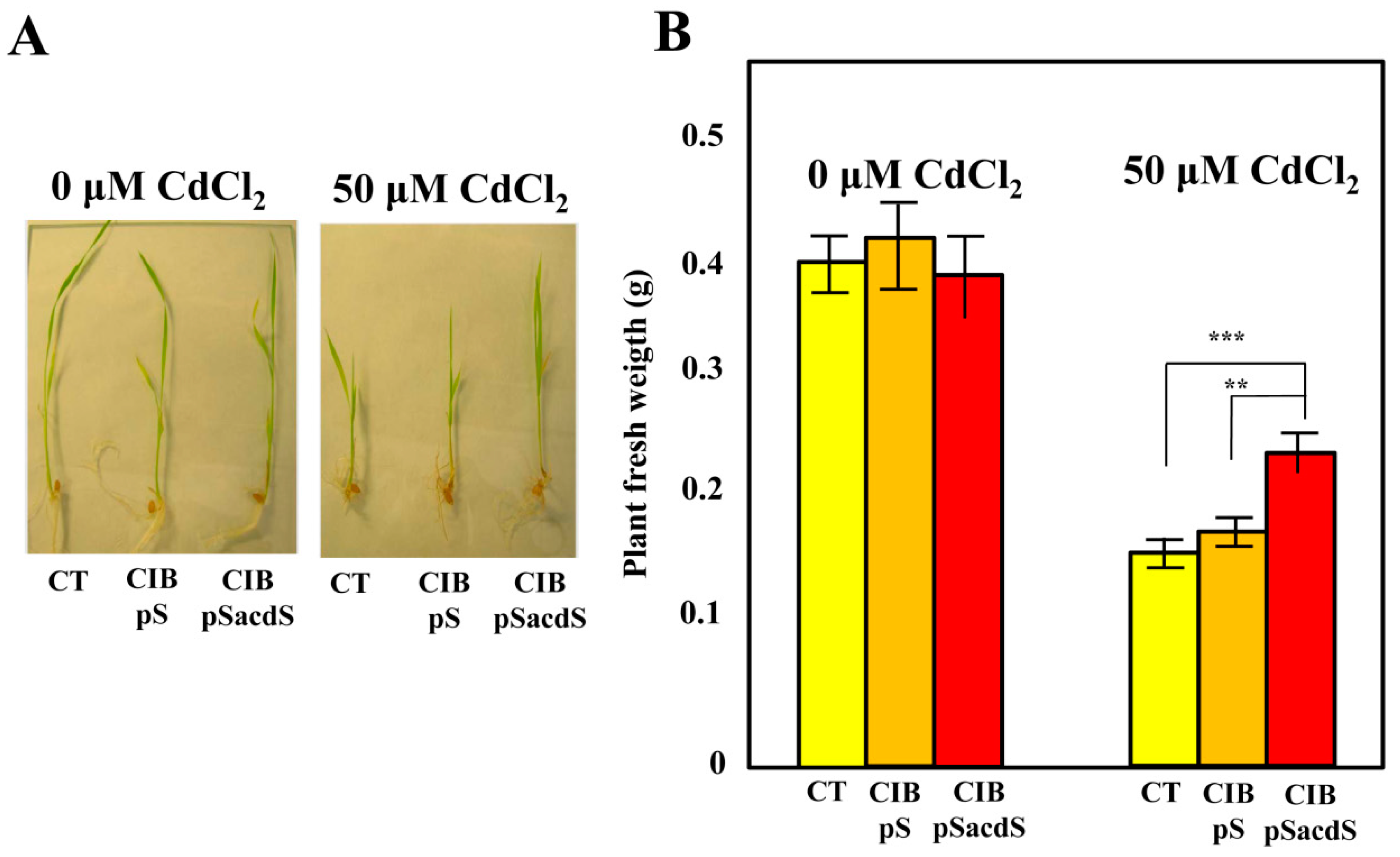
3.3. Azoarcus sp. CIB-Expressing acdS Gene Promotes Rice Growth under Stress Conditions
3.4. Enhanced Cd Concentration in the Shoots by Strain CIB Expressing the acdS Gene
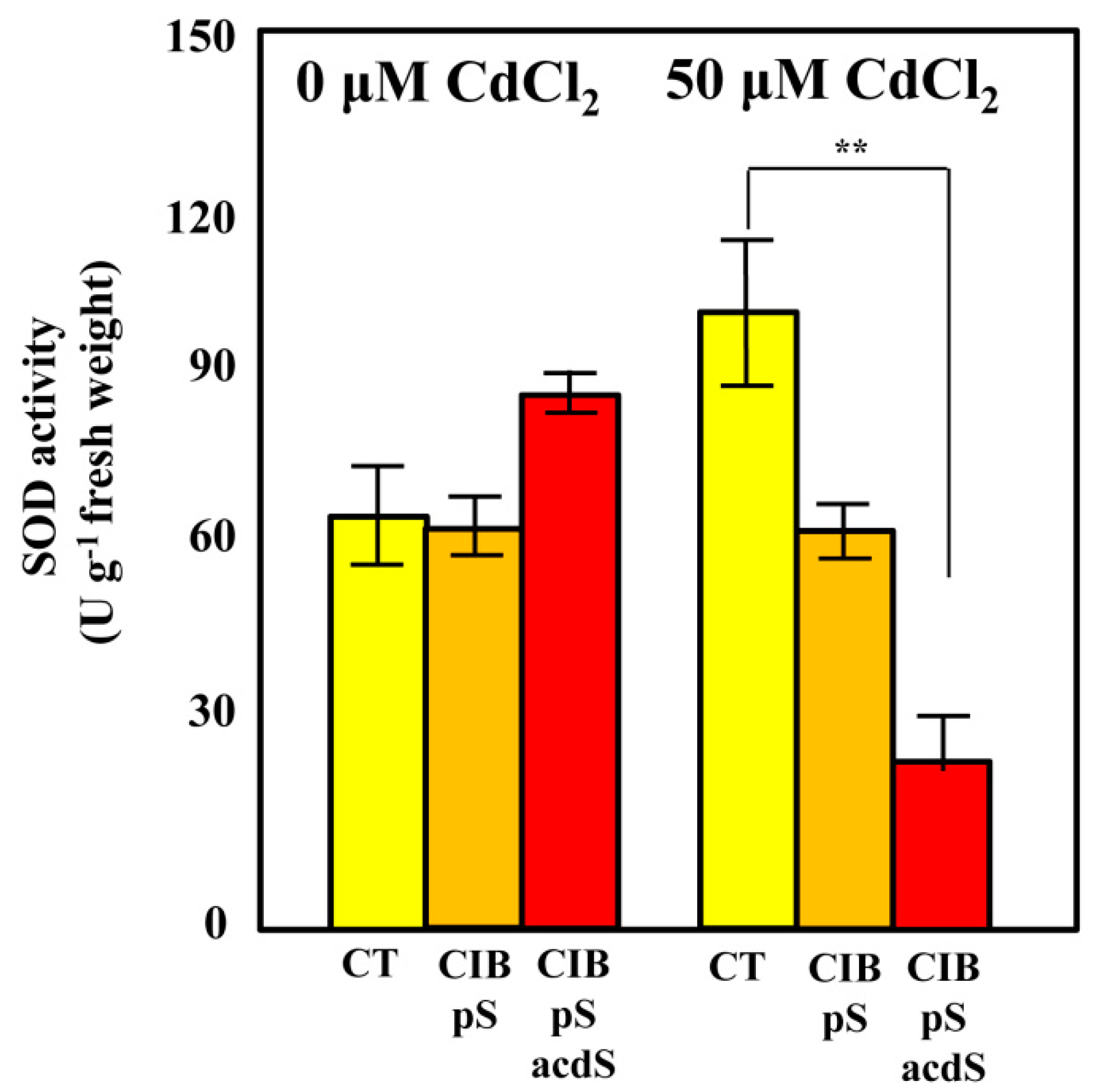
3.5. Azoarcus sp. CIB Expressing acdS Gene May Increase the ROS Quenching Capacity in Rice Roots
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vangronsveld, J.; Clijsters, H.; Van Poucke, M. Phytochrome-controlled ethylene biosynthesis of intact etiolated bean seedlings. Planta 1988, 174, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Frankenberger, W.T. Ethylene accumulation in soil in response to organic amendments. Soil Sci. Soc. Am. J. 1990, 54, 1026–1031. [Google Scholar] [CrossRef]
- Jackson, M.B. Ethylene in root growth and development. In The Plant Hormone Ethylene; Matoo, A.K., Suttle, J.C., Eds.; Springer: Boston, MA, USA, 1991; pp. 159–181. [Google Scholar]
- Abeles, F.B.; Morgan, P.W.; Saltveit, M.E. Ethylene in Plant Biology; Science Direct: Amsterdam, The Netherlands, 1992. [Google Scholar]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Shelke, G.M.; Kumar, A.; Jha, P.N. Biochemistry and genetics of ACC deaminase: A weapon to “stress ethylene” produced in plants. Front. Microbiol. 2015, 6, 937. [Google Scholar]
- Schellingen, K.; Van der Straeten, D.; Remans, T.; Vangronsveld, J.; Keunen, E.; Cuypers, A. Ethylene biosynthesis is involved in the early oxidative challenge induced by moderate Cd exposure in Arabidopsis thaliana. Environ. Exp. Bot. 2015, 117, 1–11. [Google Scholar] [CrossRef]
- Schellingen, K.; Van der Straeten, D.; Remans, T.; Vangronsveld, J.; Keunen, E.; Cuypers, A. Ethylene signalling is mediating the early cadmium-induced oxidative challenge in Arabidopsis thaliana. Plant Sci. 2015, 239, 137–146. [Google Scholar] [CrossRef]
- Keunen, E.; Schellingen, K.; Vangronsveld, J.; Cuypers, A. Ethylene and metal stress: Small molecule, big impact. Front. Plant Sci. 2016, 7, 23. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Geraats, B.P.J.J.; Linthorst, H.J.M.M. Ethylene as a modulator of disease resistance in plants. Trends Plant. Sci. 2006, 11, 184–191. [Google Scholar] [CrossRef]
- Glick, B.R. Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 2007, 26, 227–242. [Google Scholar] [CrossRef]
- Cuypers, A.; Plusquin, M.; Remans, T.; Jozefczak, M.; Keunen, E.; Gielen, H.; Opdenakker, K.; Nair, A.R.; Munters, E.; Artois, T.J.; et al. Cadmium stress: An oxidative challenge. Biometals 2010, 23, 927–940. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; Adrees, M.; Rizvi, H.; Rehman, M.Z.; Hannan, F.; Qayyum, M.F.; Hafeez, F.; Ok, Y.S. Cadmium stress in rice: Toxic effects, tolerance mechanisms and management: A critical review. Environ. Sci. Pollut. Res. 2016, 23, 17859–17879. [Google Scholar] [CrossRef]
- Ma, Q.; Cao, X.; Tan, X.; Si, L.; Wu, L. Effects of cadmium stress on pakchoi (Brassica chinensis L.) growth and uptake of inorganic and organic nitrogenous compounds. Environ. Exp. Bot. 2017, 137, 49–57. [Google Scholar] [CrossRef]
- Mitra, S.; Pramanik, K.; Sarkar, A.; Ghosh, P.K.; Soren, T.; Maiti, T.K. Bioaccumulation of cadmium by Enterobacter sp. and enhancement of rice seedling growth under cadmium stress. Ecotox. Environ. Saf. 2018, 156, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.A. Popova, L.P. Functions and toxicity of cadmium in plants: Recent advances and future prospects. Tur. J. Bot. 2013, 37, 1–13. [Google Scholar]
- Glick, B.R.; Cheng, Z.; Czarny, J.; Duan, J. Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur. J. Plant Pathol. 2007, 119, 329–339. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Nascimento, F.; Brígido, C.; Alho, L.; Glick, B.R.; Oliveira, S. Enhanced chickpea growth-promotion ability of a Mesorhizobium strain expressing an exogenous ACC deaminase gene. Plant Soil 2012, 353, 221–230. [Google Scholar] [CrossRef]
- Kong, Z.; Glick, B.R.; Duan, J.; Ding, S.; Tian, J.; McConkey, B.J.; Wei, G. Effects of 1-aminocyclopropane-1-carboxylate (ACC) deaminase-overproducing Sinorhizobium meliloti on plant growth and copper tolerance of Medicago lupulina. Plant Soil 2015, 391, 383–398. [Google Scholar] [CrossRef]
- Brígido, C.; Nascimento, F.X.; Duan, J.; Glick, B.R.; Oliveira, S. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Mesorhizobium spp. reduces the negative effects of salt stress in chickpea. FEMS Microbiol. Lett. 2013, 349, 46–53. [Google Scholar]
- Krause, A.; Ramakumar, A.; Bartels, D.; Battistoni, F.; Bekel, T.; Boch, J.; Böhm, M.; Friedrich, F.; Hurek, T.; Krause, L.; et al. Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 2006, 24, 1385–1391. [Google Scholar] [CrossRef]
- Faoro, H.; Rene Menegazzo, R.; Battistoni, F.; Gyaneshwar, P.; do Amaral, F.P.; Taulé, C.; Rausch, S.; Gonçalves Galvãom, P.; de Los Santos, C.; Miltra, S.; et al. The oil-contaminated soil diazotroph Azoarcus olearius DQS-4T is genetically and phenotypically similar to the model grass endophyte Azoarcus sp. BH72. Environ. Microbiol. Rep. 2017, 9, 223–238. [Google Scholar] [CrossRef]
- Reinhold, B.; Hurek, T.; Niemann, E.G.; Fendrik, I. Close association of Azospirillum and diazotrophic rods with different root zones of kallar grass. Appl. Environ. Microbiol. 1986, 52, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Hurek, T.; Reinhold-Hurek, B.; Van Montagu, M.; Kellenberger, E. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J. Bacteriol. 1994, 176, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Stein, T.; Hayen-Schneg, N.; Fendrik, I. Contribution of BNF by Azoarcus sp. BH72 in Sorghum vulgare. Soil Biol. Biochem. 1997, 29, 969–971. [Google Scholar] [CrossRef]
- Egener, T.; Hurek, T.; Reinhold-Hurek, B. Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol. Plant Microbe Interact. 1999, 12, 813–819. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Azoarcus spp. and their interactions with grass roots. Plant Soil 1997, 194, 57–64. [Google Scholar] [CrossRef]
- Hurek, T.; Handley, L.L.; Reinhold-Hurek, B.; Piché, Y. Azoarcus grass endophytes contribute fixed nitrogen to the plant in an unculturable state. Mol. Plant Microbe Interact. 2002, 15, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Llamosas, H.; Prandoni, N.; Fernández-Pascual, M.; Fajardo, S.; Morcillo, C.; Díaz, E.; Carmona, M. Azoarcus sp. CIB, an anaerobic biodegrader of aromatic compounds shows an endophytic lifestyle. PLoS ONE 2014, 9, e110771. [Google Scholar]
- Rabus, R.; Wöhlbrand, L.; Thies, D.; Meyer, M.; Reinhold-Hurek, B.; Kämpfer, P. Aromatoleum gen. nov. a novel genus accommodating the phylogenetic lineage including Azoarcus evansii and related species, and proposal of Aromatoleum aromaticum sp. nov. Aromatoleum petrolei sp. nov. Aromatoleum bremense sp. nov. Aromatoleum toluolicum sp. nov. and Aromatoleum diolicum sp. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 982–997. [Google Scholar]
- López-Barragán, M.J.; Carmona, M.; Zamarro, M.T.; Thiele, M.; Boll, M.; Fuchs, G.; Garcia, J.L.; Díaz, E. The bzd gene cluster, coding for anaerobic benzoate catabolism in Azoarcus sp. CIB. J. Bacteriol. 2004, 186, 5462–5774. [Google Scholar]
- Valderrama, J.A.; Durante-Rodríguez, G.; Blázquez, B.; García, J.L.; Carmona, M.; Díaz, E. Bacterial degradation of benzoate: Cross-regulation between aerobic and anaerobic pathways. J. Biol. Chem. 2012, 287, 10494–10508. [Google Scholar] [CrossRef] [PubMed]
- Juárez, J.F.; Zamarro, M.T.; Eberlein, C.; Boll, M.; Carmona, M.; Díaz, E. Characterization of the mbd cluster encoding the anaerobic 3-methylbenzoyl-CoA central pathway. Environ. Microbiol. 2013, 15, 148–166. [Google Scholar] [CrossRef]
- Blázquez, B.; Carmona, M.; Díaz, E. Transcriptional regulation of the peripheral pathway for the anaerobic catabolism of toluene and m-xylene in Azoarcus sp. CIB. Front. Microbiol. 2018, 9, 506. [Google Scholar] [CrossRef]
- Durante-Rodríguez, G.; Fernández-Llamosas, H.; Alonso-Fernandes, E.; Fernández-Muñiz, M.N.; Muñoz-Olivas, R.; Díaz, E.; Carmona, M. ArxA from Azoarcus sp. CIB, an anaerobic arsenite oxidase from an obligate heterotrophic and mesophilic bacterium. Front. Microbiol. 2019, 10, 1699. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Llamosas, H.; Castro, L.; Blázquez, M.L.; Díaz, E.; Carmona, M. Biosynthesis of selenium nanoparticles by Azoarcus sp. CIB. Microb. Cell Fact. 2017, 15, 109. [Google Scholar]
- Xue, S.G.; Shi, L.; Wu, C.; Wu, H.; Qin, Y.; Pan, W.; Hartley, W.; Cui, M. Cadmium, lead, and arsenic contamination in paddy soils of a mining area and their exposure effects on human HEPG2 and keratinocyte cell-lines. Environ. Res. 2017, 156, 23–30. [Google Scholar] [CrossRef]
- Cameron, R.E. Guide to Site and Soil Description for Hazardous Waste Site Characterization; EPA: Washington, DC, USA, 1992; Volume 1. [Google Scholar]
- Wan, L.; Zhang, H. Cadmium toxicity: Effects on cytoskeleton, vesicular trafficking and cell wall construction. Plant Signal. Behav. 2012, 7, 345–348. [Google Scholar] [CrossRef]
- Shao, J.F.; Fujii-Kashino, M.; Yamaji, N.; Fukuoka, S.; Shen, R.F.; Ma, J.F. Isolation and characterization of a rice line with high Cd accumulation for potential use in phytoremediation. Plant. Soil 2017, 410, 357–368. [Google Scholar] [CrossRef]
- Kögel-Knabner, I.; Amelung, W.; Cao, Z.; Fiedler, S.; Frenzel, P.; Jahn, R.; Kalbitz, K.; Kölbl, A.; Schloter, M. Biogeochemistry of paddy soils. Geoderma 2010, 157, 1–14. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, D.; Liu, Y.; Zhang, Y.; Wei1, X.; Xu, B.; Bocharnikova, E.; Matichenkov, V. Cadmium phytoextraction from contaminated paddy soil as influenced by EDTA and Si fertilizer. Environ. Sci. Pollut. Res. 2019, 26, 23638–23644. [Google Scholar] [CrossRef]
- De Lorenzo, V.; Timmis, K.N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994, 235, 386–405. [Google Scholar] [PubMed]
- Manoil, C.; Beckwith, J. TnphoA: A transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 1985, 82, 8129–8133. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Coenye, T.; Sturz, A.V.; Vandamme, P.; Barka, E.A.; Salles, J.F.; van Elsas, J.D.; Fauré, D.; Reiter, B.; Glick, B.R.; et al. Burkholderia phytofirmans sp. nov. a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. Microbiol. 2005, 55, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Silva-Rocha, R.; Martínez-García, E.; Calles, B.; Chavarría, M.; Arce-Rodríguez, A.; de Las Heras, A.; Páez-Espino, A.D.; Durante-Rodríguez, G.; Kim, J.; Nikel, P.I.; et al. The standard european vector architecture (SEVA): A coherent platform for the analysis and development of complex prokaryotic phenotypes. Nucleic Acids Res. 2013, 41, D666–D675. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Ruiz, E.; Hernáez, M.J.; Martínez-Pérez, O.; Santero, E. Identification and functional characterization of Sphingomonas macrogolitabida strain TFA genes involved in the first two steps of the tetralin catabolic pathway. J. Bacteriol. 2003, 185, 2026–2030. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Dworkin, M.; Foster, J.W. Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 1958, 75, 592–603. [Google Scholar] [CrossRef]
- Sambrook, J.; Rusell, D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2011. [Google Scholar]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant 2003, 118, 10–15. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- McCord, J.M.; Fridovich, I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar]
- Smyth, E.M.; McCarthy, J.; Nevin, R.; Khan, M.R.; Dow, J.M.; O’Gara, F.; Dooham, F.M. In vitro analyses are not reliable predictors of the plant growth promotion capability of bacteria: A Pseudomonas fluorescens strain that promotes the growth and yield of wheat. J. Appl. Microbiol. 2011, 111, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Belimov, A.; Hontzeas, N.; Safronova, V.I.; Demchinskaya, S.V.; Piluzza, G.; Bullitta, S.; Glick, B.R. Cadmium tolerant plant growth-promoting bacteria associated with roots of Indian mustard (Brassica juncea L. Czern.). Soil Biol. Biochem. 2005, 37, 241–250. [Google Scholar] [CrossRef]
- Nikolic, B.; Schwab, H.; Sessitsch, A. Metagenomic analysis of the 1-aminocyclopropane-1-carboxylate deaminase gene (acdS) operon of an uncultured bacterial endophyte colonizing Solanum tuberosum L. Arch. Microbiol. 2011, 193, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Mitter, B.; Petric, A.; Shin, M.W.; Chain, P.S.G.; Hauberg-Lotte, L.; Reinhold-Hurek, B.; Nowak, J.; Sessitsch, A. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front. Plant Sci. 2013, 4, 120. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Tavares, M.J.; Glick, B.R.; Rossi, M.J. Improvement of Cupriavidus taiwanensis nodulation and plant growth promoting abilities by the expression of an exogenous ACC deaminase gene. Curr. Microbiol. 2018, 75, 961–965. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Tavares, M.J.; Glick, B.R.; Rossi., M.J. The expression of an exogenous ACC deaminase by the endophyte Serratia grimesii BXF1 promotes the early nodulation and growth of common bean. Lett. Appl. Microbiol. 2018, 66, 252–259. [Google Scholar]
- Nascimento, F.X.; Brígido, C.; Glick, B.R.; Oliveira, S.; Alho, L. Mesorhizobium ciceri LMS-1 expressing an exogenous 1-aminocyclopropane-1-carboxylate (ACC) deaminase increases its nodulation abilities and chickpea plant resistance to soil constraints. Lett. Appl. Microbiol. 2012, 55, 15–21. [Google Scholar] [CrossRef]
- Arshad, M.; Saleem, M.; Hussain, S. Perspectives of bacterial ACC deaminase in phytoremediation. Trends Biotechnol. 2007, 25, 356–362. [Google Scholar] [CrossRef]
- Treesubsuntorn, C.; Dhurakit, P.; Khaksar, G.; Thiravetyan, P. Effect of microorganisms on reducing cadmium uptake and toxicity in rice (Oryza sativa L.). Environ. Sci. Pollut. Res. 2017, 1, 1–12. [Google Scholar] [CrossRef]
- Danish, S.; Zafar-ul-Hye, M.; Mohsin, F.; Hussain, M. ACC-deaminase producing plant growth promoting rhizobacteria and biochar mitigate adverse effects of drought stress on maize growth. PLoS ONE 2020, 15, e0230615. [Google Scholar] [CrossRef]
- Jiang, C.Y.; Sheng, X.F.; Qian, M.; Wang, Q.Y. Isolation and characterization of a heavy metal resistant Burkholderia sp. from heavy metal-contaminated paddy field soil and its potential in promoting plant growth and heavy metal accumulation in metal polluted soil. Chemosphere 2008, 72, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Farwell, A.J.; Vesely, S.; Nero, V.; Rodriguez, H.; McCormack, K.; Shah, S.; Dixon, D.G.; Glick, B.R. Tolerance of transgenic canola plants (Brassica napus) amended with plant growth-promoting bacteria to flooding stress at a metal contaminated field site. Environ. Pollut. 2007, 147, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, R.; Yang, Z.; Zhan, Y.; Ma, Y.; Ping, S.; Zhang, L.; Lin, M.; Yan, Y. 1-aminocyclopropane-1-carboxylate deaminase from Pseudomonas stutzeri A1501 facilitates the growth of rice in the presence of salt or heavy metals. J. Microbiol. Biotechnol. 2015, 25, 1119–1128. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.; Zhang, X.; Dong, L.; Zhang, J.; Wei, Y.; Feng, Y.; Lu, L. Improved plant growth and Zn accumulation in grains of rice (Oryza sativa L.) by inoculation of endophytic microbes isolated from a Zn hyperaccumulator, Sedum alfredii H. J. Agric. Food Chem. 2014, 62, 1783–1791. [Google Scholar] [CrossRef] [PubMed]
- Arteca, R.N.; Arteca, J.M. Heavy-metal-induced ethylene production in Arabidopsis thaliana. J. Plant Physiol. 2007, 164, 1480–1488. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B. The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 2014, 5, 685. [Google Scholar] [CrossRef]
- Satpathy, D.; Reddy, M.V.; Dhal, S.P. Risk assessment of heavy metals contamination in paddy soil, plants, and grains (Oryza sativa L.) at the east coast of India. Biomed. Res. Int. 2014, 2014, 545473. [Google Scholar] [CrossRef]
- He, L.Y.; Chen, Z.J.; Ren, G.D.; Zhang, Y.F.; Qian, M.; Sheng, X.F. Increased cadmium and lead uptake of a cadmium hyperaccumulator tomato by cadmium-resistant bacteria. Ecotox. Environ. Saf. 2009, 72, 1343–1348. [Google Scholar] [CrossRef]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Fallmann, K.; Puschenreiter, M. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef]
- Lux, A.; Martinka, M.; Vaculík, M.; White, P.J. Root responses to cadmium in the rhizosphere: A review. J. Exp. Bot. 2011, 62, 21–37. [Google Scholar] [CrossRef]
- Clemens, S.; Ma, J.F. Toxic heavy metal and metalloid accumulation in crop plants and foods. Ann. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Sun, G.X.; Lei, M.; Teng, M.; Liu, Y.X.; Chen, N.C.; Wang, L.H.; Carey, A.M.; Deacon, C.; Raab, A.; et al. High percentage inorganic arsenic content of mining impacted and nonimpacted Chinese rice. Environ. Sci. Technol. 2008, 42, 5008–5013. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.H.; Chen, C.; Zhu, Q.H.; Huang, D.Y. Effects of soil acidification and liming on the phytoavailability of cadmium in paddy soils of central subtropical China. Environ. Pollut. 2016, 219, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Borujeni, S.R.; Khavazi, K.; Asgharzadeh, A.; Borujeni, I.R. Use of bacterial ACC deaminase to increase oil (especially polyaromatic hydrocarbons) phytoremediation efficiency for maize (Zea mays) seedlings. Int. J. Phytoremediation 2018, 16, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Weyens, N.; Croes, S.; Dupae, J.; Newman, L.; van der Lelie, D.; Carleer, R.; Vangronsveld, J. Endophytic bacteria improve phytoremediation of Ni and TCE co-contamination. Environ. Pollut. 2010, 158, 2422–2427. [Google Scholar] [CrossRef]
- Weyens, N.; Beckers, B.; Schellingen, K.; Ceulemans, R.; van der Lelie, D.; Newman, L.; Taghavi, S.; Carleer, R.; Vangronsveld, J. The potential of the Ni-resistant TCE-degrading Pseudomonas putida W619-TCE to reduce phytotoxicity and improve phytoremediation efficiency of poplar cuttings on a Ni-TCE co-contamination. Int. J. Phytoremediation 2015, 17, 40–48. [Google Scholar] [CrossRef]


| Strain or Plasmid | Relevant Genotype and Characteristic(s) | Reference or Source |
|---|---|---|
| E. coli strains | ||
| DH10B | F’, mcrA, Δ(mrr hsdRMS-mcrBC), Φ80lacZΔM15, ΔlacX74, deoR, recA1, araD139, Δ(ara-leu)7697, galU, galK, rpsL (Smr), endA1, nupG | Life Technologies |
| S17-1λpir | Tpr Smr recA thi hsdRM+ RP4::2-Tc::Mu::Km λpir phage lysogen | [44] |
| CC118 | Δ(ara-leu), araD, ΔlacX7, galE, galK, phoA20, rpoB thi-1, rpsE, (Spr), (Rfr), argE, (Am), recA1 | [45] |
| Azoarcus strains | ||
| CIB | Wild type strain | [32] |
| Paraburkholderia strains | ||
| P. phytofirmans PsJN | Wild type strain | [46] |
| Plasmids | ||
| pSEVA237 | Kmr, ori pBBR1, harbors the gfp gene under the control of the PlexA promoter | [47] |
| pSEVA237acdS | Kmr, pSEVA237 derivative that includes a AscI fragment of 1172 bp containing the gene acdS under the control of Ptac promoter | This work |
| pIZ1016 | Gmr, ori pBBR1MCS-5 derivative vector for cloning and expression harboring the Ptac promoter and the lacI gene | [48] |
| pIZacdS | Gmr, pIZ1016 derivative containing the gene acdS from P. phytofirmans under the control of the Ptac promoter | This work |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Llamosas, H.; Ibero, J.; Thijs, S.; Imperato, V.; Vangronsveld, J.; Díaz, E.; Carmona, M. Enhancing the Rice Seedlings Growth Promotion Abilities of Azoarcus sp. CIB by Heterologous Expression of ACC Deaminase to Improve Performance of Plants Exposed to Cadmium Stress. Microorganisms 2020, 8, 1453. https://doi.org/10.3390/microorganisms8091453
Fernández-Llamosas H, Ibero J, Thijs S, Imperato V, Vangronsveld J, Díaz E, Carmona M. Enhancing the Rice Seedlings Growth Promotion Abilities of Azoarcus sp. CIB by Heterologous Expression of ACC Deaminase to Improve Performance of Plants Exposed to Cadmium Stress. Microorganisms. 2020; 8(9):1453. https://doi.org/10.3390/microorganisms8091453
Chicago/Turabian StyleFernández-Llamosas, Helga, Juan Ibero, Sofie Thijs, Valeria Imperato, Jaco Vangronsveld, Eduardo Díaz, and Manuel Carmona. 2020. "Enhancing the Rice Seedlings Growth Promotion Abilities of Azoarcus sp. CIB by Heterologous Expression of ACC Deaminase to Improve Performance of Plants Exposed to Cadmium Stress" Microorganisms 8, no. 9: 1453. https://doi.org/10.3390/microorganisms8091453
APA StyleFernández-Llamosas, H., Ibero, J., Thijs, S., Imperato, V., Vangronsveld, J., Díaz, E., & Carmona, M. (2020). Enhancing the Rice Seedlings Growth Promotion Abilities of Azoarcus sp. CIB by Heterologous Expression of ACC Deaminase to Improve Performance of Plants Exposed to Cadmium Stress. Microorganisms, 8(9), 1453. https://doi.org/10.3390/microorganisms8091453







