A Protective Vaccine against Johne’s Disease in Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Vaccine Preparation
2.3. Study Design
2.4. Body Temperature and Weight
2.5. Fecal Culture
2.6. Necropsy, Tissue Collection and Culturing
2.7. Antibody Response
2.8. Intradermal Skin Test
2.9. IFN-γ Release Assay
2.10. Cytokine Profiling
2.11. Statistical Analysis
3. Results
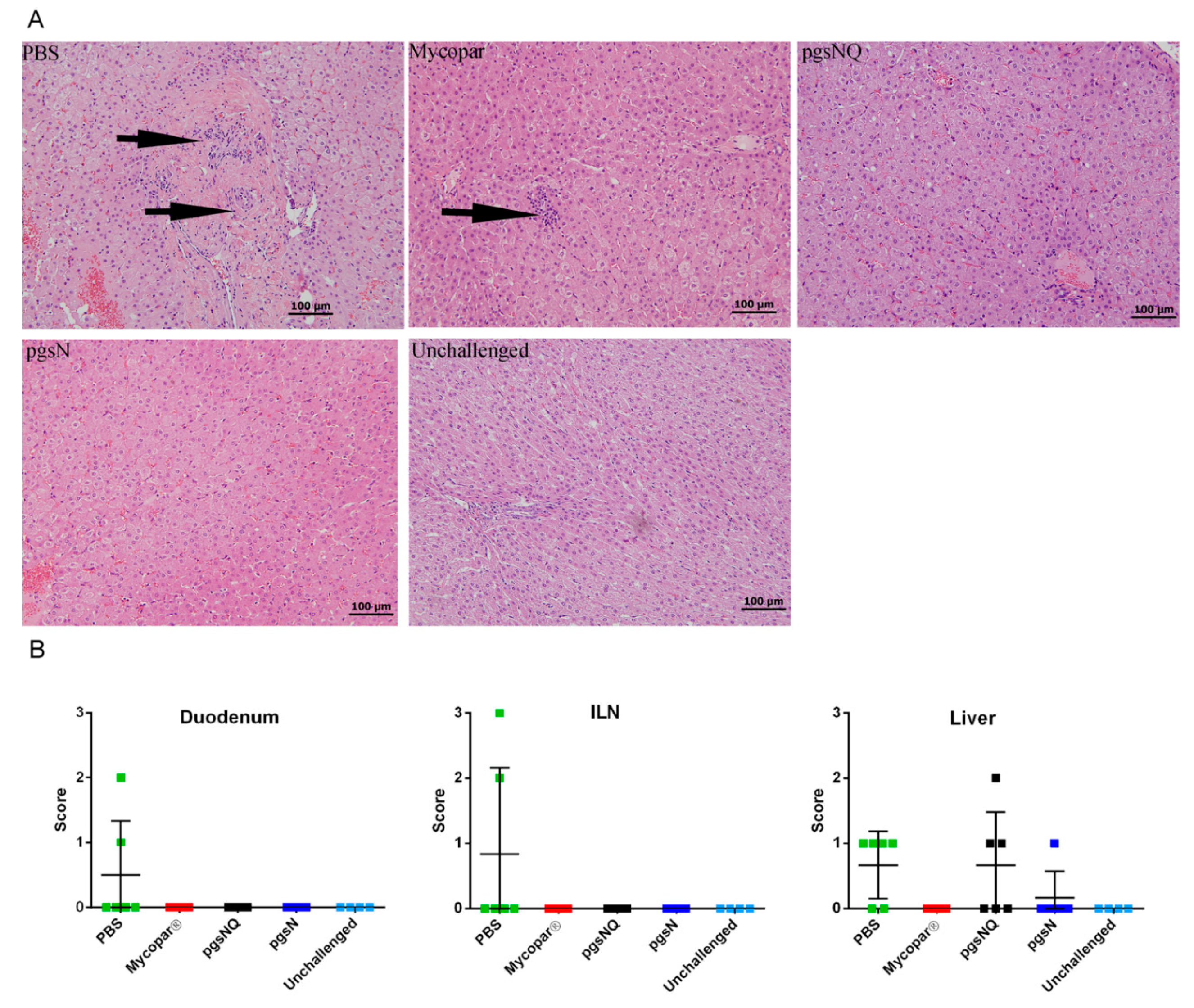
3.1. Safety of Live Attenuated Vaccine Candidates
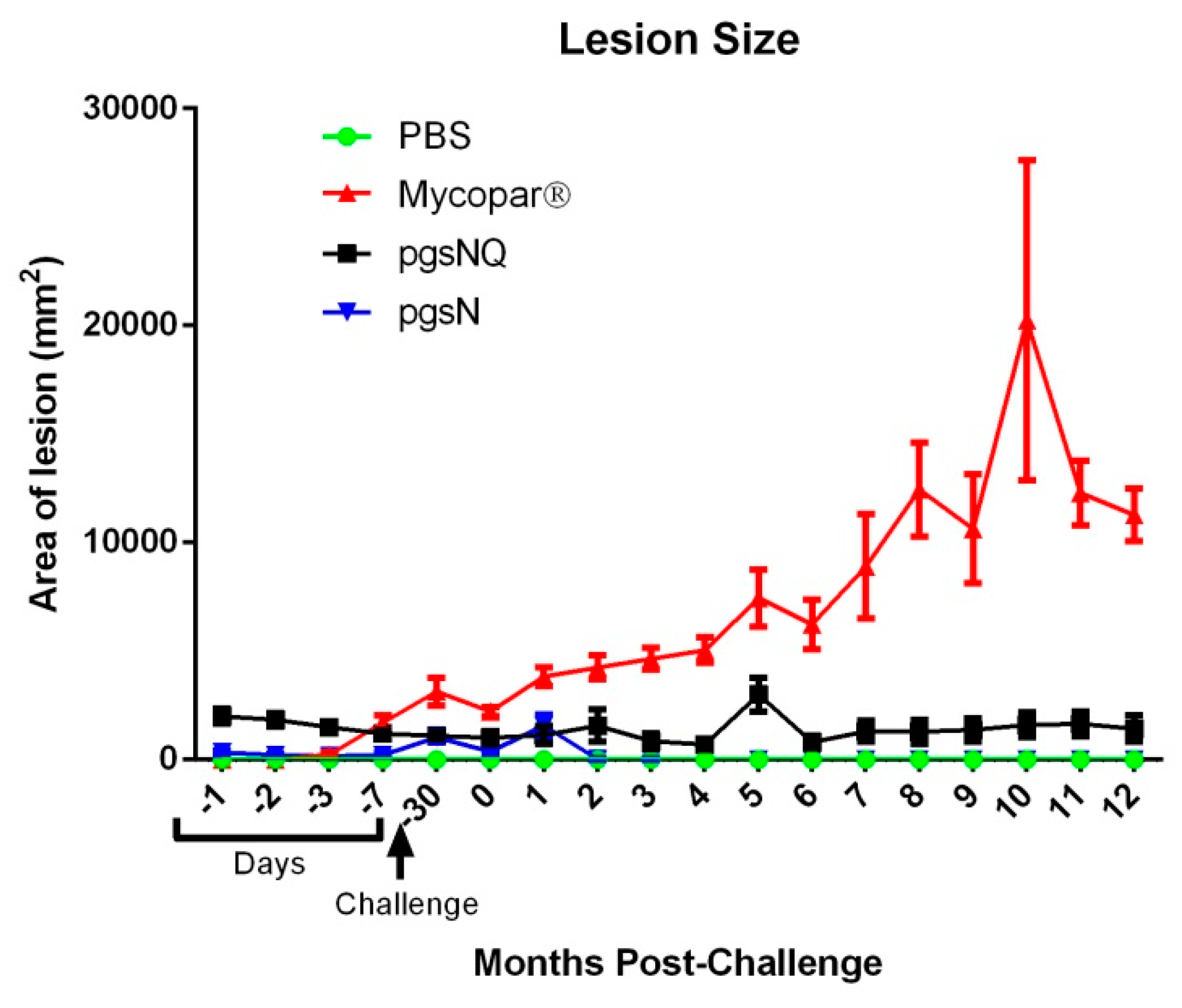
3.2. Live Vaccines Reduced Key Parameters of Johne’s Disease
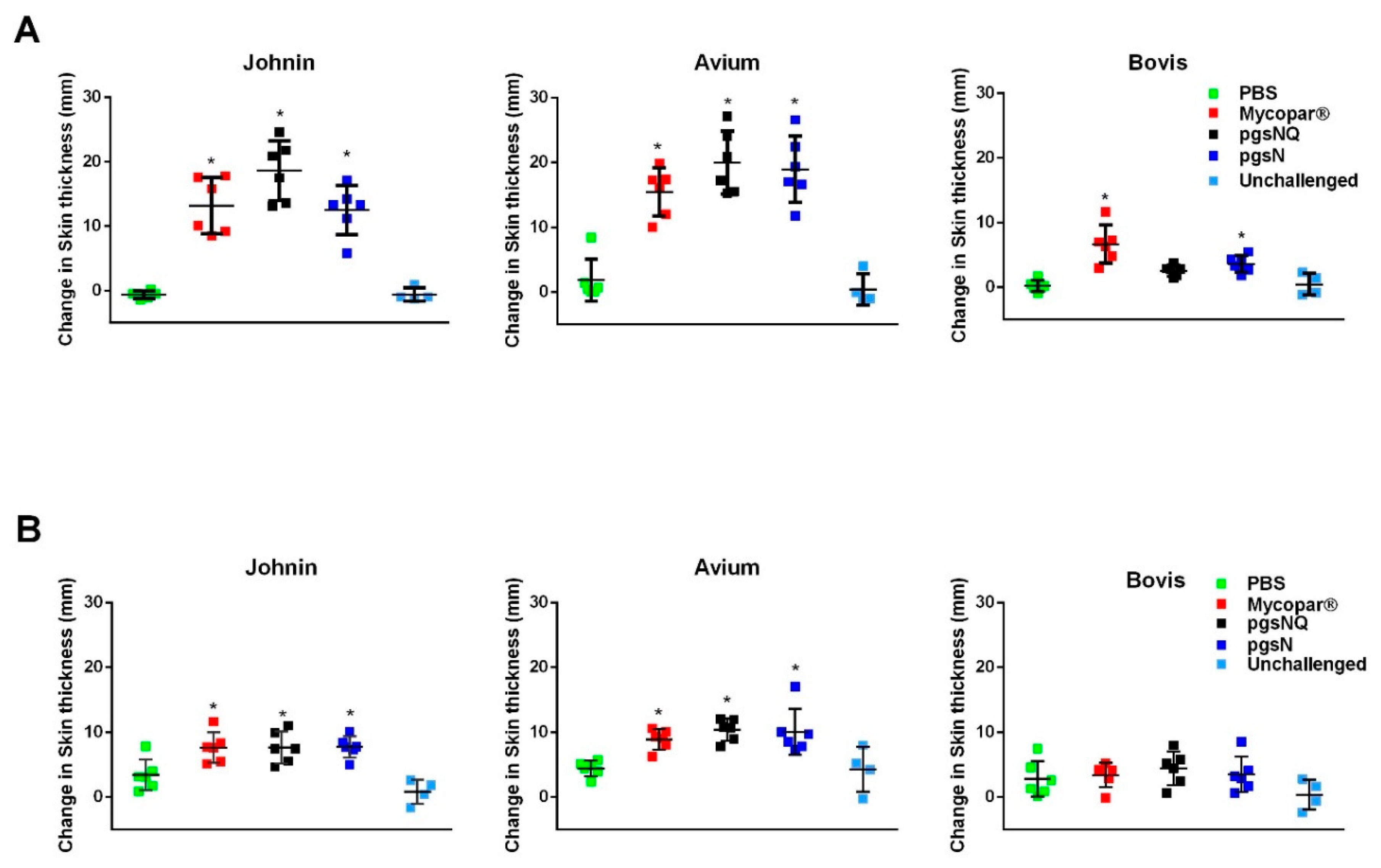
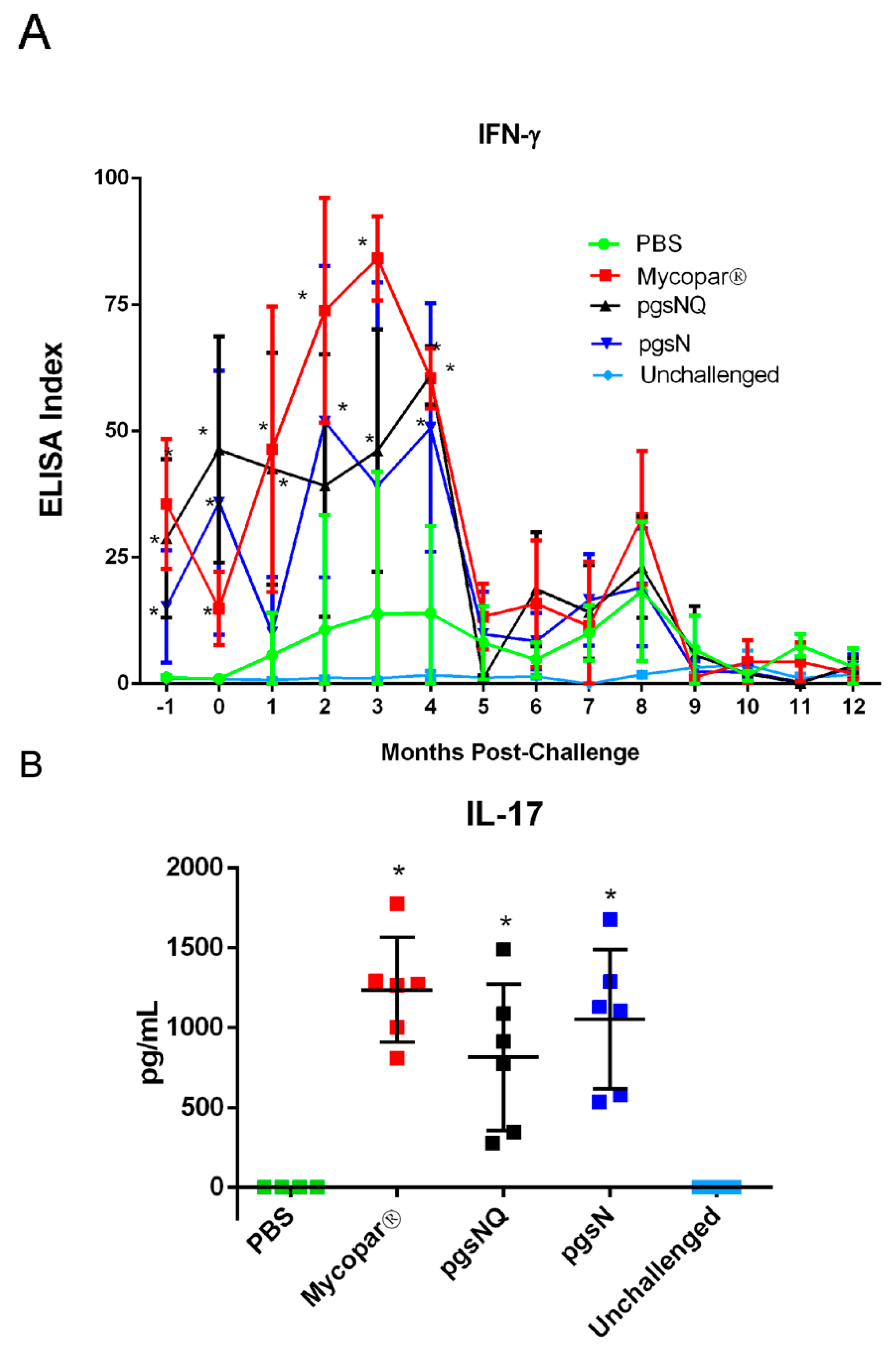
3.3. Robust Cellular Immunity Generated by Live Vaccine Candidates
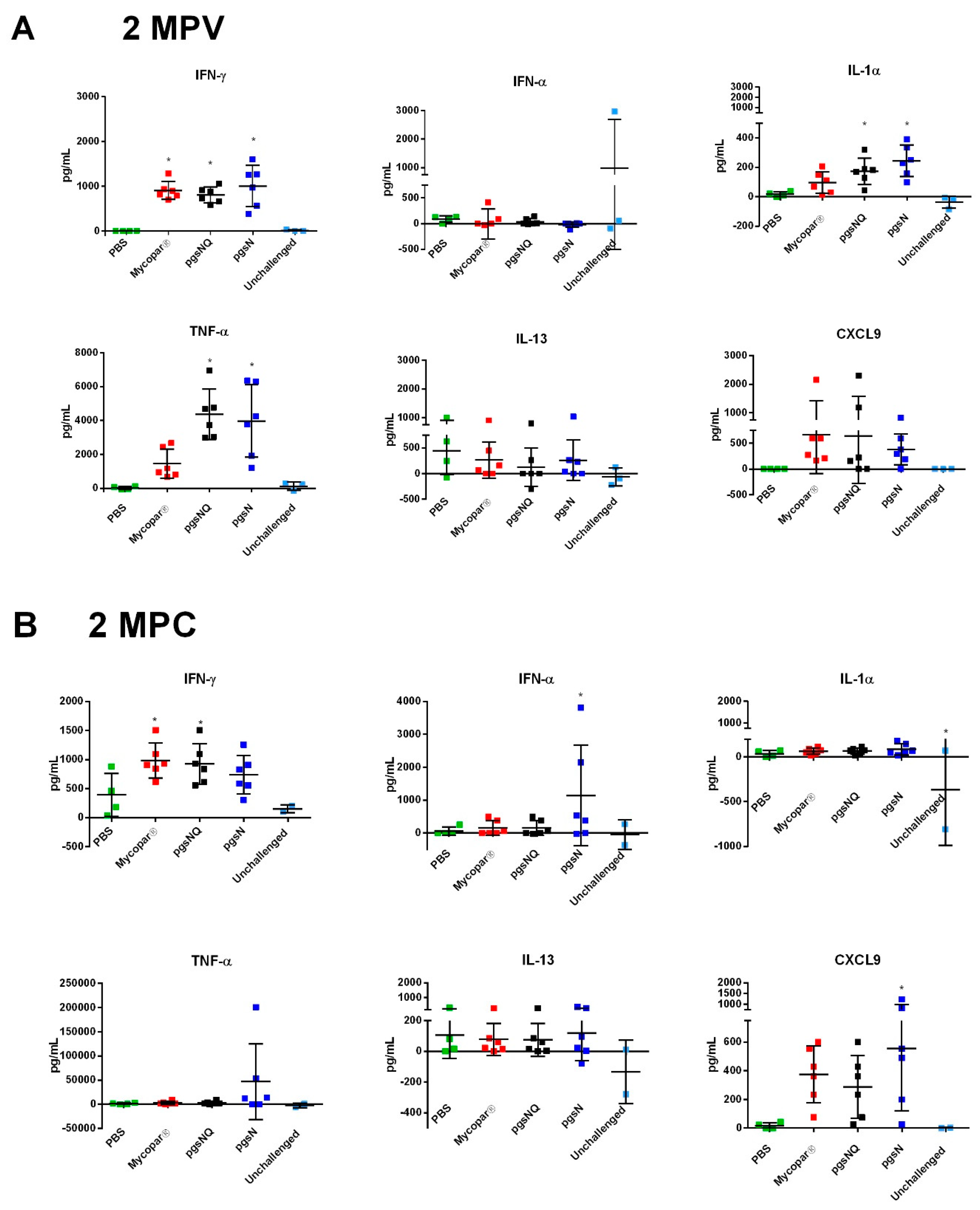
3.4. Cytokine and Chemokine Responses to LAV Candidates
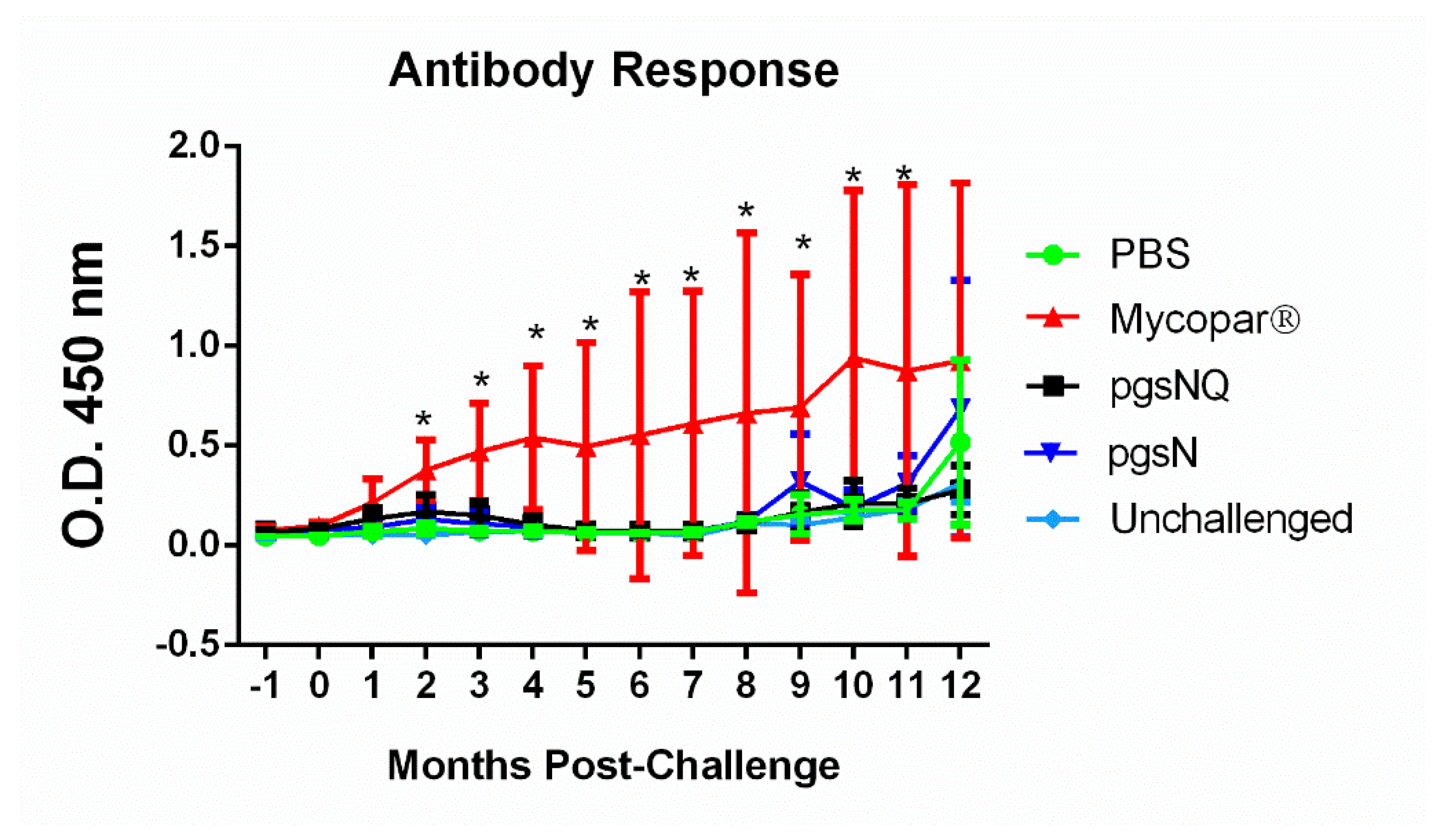
3.5. Humoral Immunity Generated by Live Vaccine Candidates
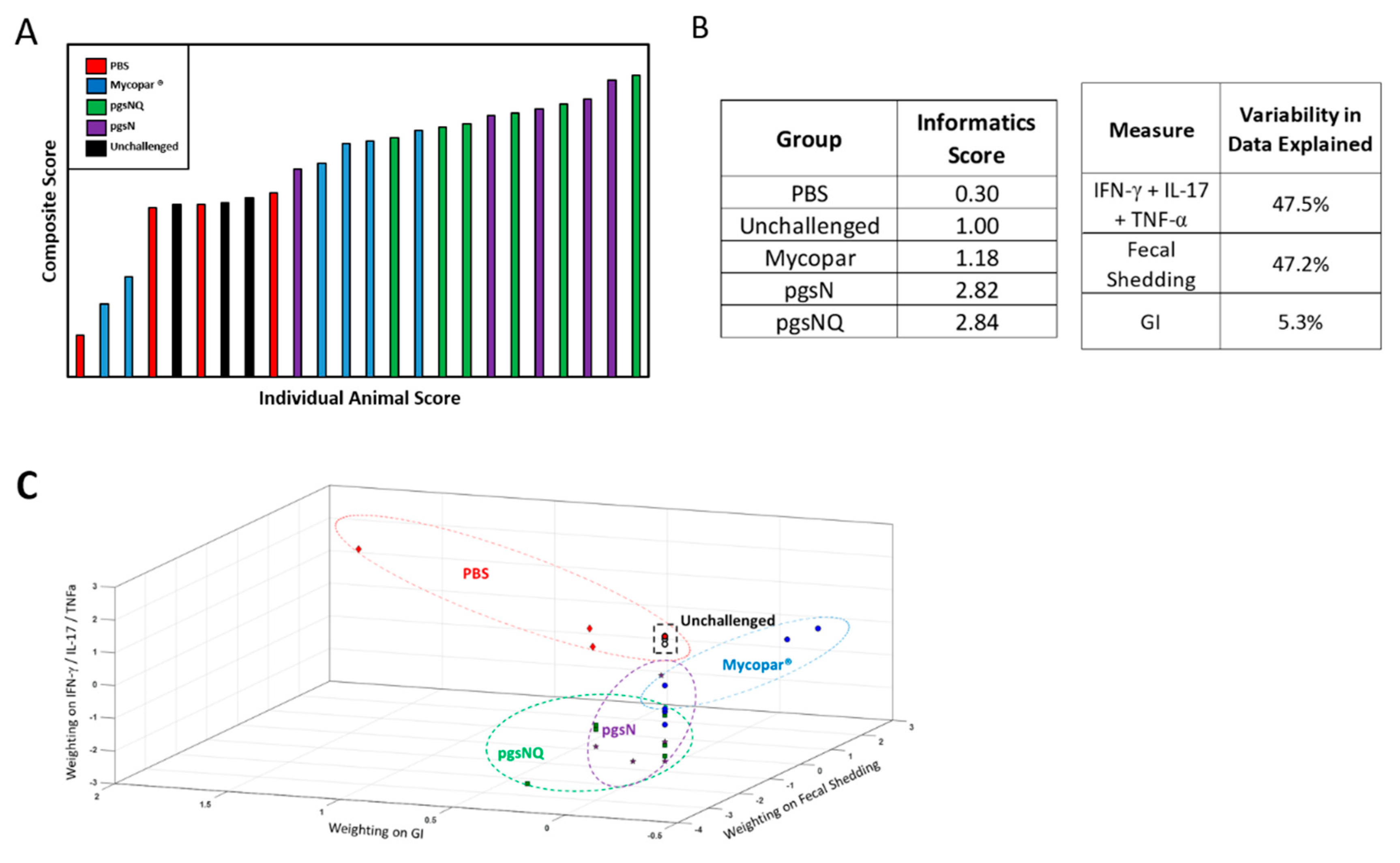
3.6. Composite Analysis of Vaccine Efficacy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Collins, M.T.; Sockett, D.C.; Goodger, W.J.; Conrad, T.A.; Thomas, C.B.; Carr, D.J. Herd prevalence and geographic distribution of, and risk factors for, bovine paratuberculosis in Wisconsin. J. Am. Vet. Med. Assoc. 1994, 204, 636–641. [Google Scholar] [PubMed]
- Timms, V.J.; Daskalopoulos, G.; Mitchell, H.M.; Neilan, B.A. The Association of Mycobacterium avium subsp. paratuberculosis with Inflammatory Bowel Disease. PLoS ONE 2016, 11, e0148731. [Google Scholar] [CrossRef] [PubMed]
- Losinger, W.C. Economic impact of reduced milk production associated with Johne’s disease on dairy operations in the USA. J. Dairy Res. 2005, 72, 425–432. [Google Scholar] [CrossRef]
- Whittington, R.J.; Sergeant, E.S. Progress towards understanding the spread, detection and control of Mycobacterium avium subsp paratuberculosis in animal populations. Aust. Vet. J. 2001, 79, 267–278. [Google Scholar] [CrossRef]
- Lombard, J.E.; Gardner, I.A.; Jafarzadeh, S.R.; Fossler, C.P.; Harris, B.; Capsel, R.T.; Wagner, B.A.; Johnson, W.O. Herd-level prevalence of Mycobacterium avium subsp. paratuberculosis infection in United States dairy herds in 2007. Prev. Vet. Med. 2013, 108, 234–238. [Google Scholar] [CrossRef]
- Kalis, C.H.; Hesselink, J.W.; Barkema, H.W.; Collins, M.T. Use of long-term vaccination with a killed vaccine to prevent fecal shedding of Mycobacterium avium subsp paratuberculosis in dairy herds. Am. J. Vet. Res. 2001, 62, 270–274. [Google Scholar] [CrossRef]
- Shippy, D.C.; Lemke, J.J.; Berry, A.; Nelson, K.; Hines, M.E., II; Talaat, A.M. Superior Protection from Live-Attenuated Vaccines Directed against Johne’s Disease. Clin. Vaccine Immunol. 2017, 24, e00478-16. [Google Scholar] [CrossRef] [PubMed]
- Knust, B.; Patton, E.; Ribeiro-Lima, J.; Bohn, J.J.; Wells, S.J. Evaluation of the effects of a killed whole-cell vaccine against Mycobacterium avium subsp paratuberculosis in 3 herds of dairy cattle with natural exposure to the organism. J. Am. Vet. Med. Assoc. 2013, 242, 663–669. [Google Scholar] [CrossRef]
- Stabel, J.R.; Waters, W.R.; Bannantine, J.P.; Lyashchenko, K. Mediation of Host Immune Responses after Immunization of Neonatal Calves with a Heat-Killed Mycobacterium avium subsp. paratuberculosis Vaccine. Clin. Vaccine Immunol. 2011, 18, 2079–2089. [Google Scholar] [CrossRef]
- Muskens, J.; van Zijderveld, F.; Eger, A.; Bakker, D. Evaluation of the long-term immune response in cattle after vaccination against paratuberculosis in two Dutch dairy herds. Vet. Microbiol. 2002, 86, 269–278. [Google Scholar] [CrossRef]
- Kohler, H.; Gyra, H.; Zimmer, K.; Drager, K.G.; Burkert, B.; Lemser, B.; Hausleithner, D.; Cubler, K.; Klawonn, W.; Hess, R.G. Immune reactions in cattle after immunization with a Mycobacterium paratuberculosis vaccine and implications for the diagnosis of M. paratuberculosis and M. bovis infections. J. Vet. Med. B Infect. Dis. Vet. Public Health 2001, 48, 185–195. [Google Scholar] [CrossRef]
- Mullerad, J.; Hovav, A.H.; Nahary, R.; Fishman, Y.; Bercovier, H. Immunogenicity of a 16.7 kDa Mycobacterium paratuberculosis antigen. Microb. Pathog. 2003, 34, 81–90. [Google Scholar] [CrossRef]
- Santema, W.; Hensen, S.; Rutten, V.; Koets, A. Heat shock protein 70 subunit vaccination against bovine paratuberculosis does not interfere with current immunodiagnostic assays for bovine tuberculosis. Vaccine 2009, 27, 2312–2319. [Google Scholar] [CrossRef]
- Santema, W.; Rutten, V.; Segers, R.; Poot, J.; Hensen, S.; Heesterbeek, H.; Koets, A. Postexposure subunit vaccination against chronic enteric mycobacterial infection in a natural host. Infect. Immun. 2013, 81, 1990–1995. [Google Scholar] [CrossRef] [PubMed]
- Kathaperumal, K.; Kumanan, V.; McDonough, S.; Chen, L.H.; Park, S.U.; Moreira, M.A.; Akey, B.; Huntley, J.; Chang, C.F.; Chang, Y.F. Evaluation of immune responses and protective efficacy in a goat model following immunization with a coctail of recombinant antigens and a polyprotein of Mycobacterium avium subsp. paratuberculosis. Vaccine 2009, 27, 123–135. [Google Scholar] [CrossRef]
- Park, K.T.; Allen, A.J.; Barrington, G.M.; Davis, W.C. Deletion of relA abrogates the capacity of Mycobacterium avium paratuberculosis to establish an infection in calves. Front. Cell. Infect. Microbiol. 2014, 4, 64. [Google Scholar] [CrossRef]
- Park, K.T.; Allen, A.J.; Bannantine, J.P.; Seo, K.S.; Hamilton, M.J.; Abdellrazeq, G.S.; Rihan, H.M.; Grimm, A.; Davis, W.C. Evaluation of two mutants of Mycobacterium avium subsp. paratuberculosis as candidates for a live attenuated vaccine for Johne’s disease. Vaccine 2011, 29, 4709–4719. [Google Scholar] [CrossRef]
- Faisal, S.M.; Chen, J.W.; Yan, F.; Chen, T.T.; Useh, N.M.; Yan, W.; Guo, S.; Wang, S.J.; Glaser, A.L.; McDonough, S.P.; et al. Evaluation of a Mycobacterium avium subsp. paratuberculosis leuD mutant as a vaccine candidate against challenge in a caprine model. Clin. Vaccine Immunol. 2013, 20, 572–581. [Google Scholar] [CrossRef]
- Ghosh, P.; Shippy, D.C.; Talaat, A.M. Superior protection elicited by live-attenuated vaccines in the murine model of paratuberculosis. Vaccine 2015, 33, 7262–7270. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Steinberg, H.; Talaat, A.M. Virulence and Immunity Orchestrated by the Global Gene Regulator sigL in Mycobacterium avium subsp. paratuberculosis. Infect. Immun. 2014, 82, 3066–3075. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Wu, C.-W.; Talaat, A.M. Key Role for the Alternative Sigma Factor, SigH, in the Intracellular Life of Mycobacterium avium subsp. paratuberculosis during Macrophage Stress. Infect. Immun. 2013, 81, 2242–2257. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Schmoller, S.K.; Shin, S.J.; Talaat, A.M. Defining the stressome of Mycobacterium avium subsp. paratuberculosis in vitro and in naturally infected cows. J. Bacteriol. 2007, 189, 7877–7886. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.X.; Xie, Y.; Ye, Y.P. Advances in saponin-based adjuvants. Vaccine 2009, 27, 1787–1796. [Google Scholar] [CrossRef]
- Bannantine, J.P.; Hines, M.E., II; Bermudez, L.E.; Talaat, A.M.; Sreevatsan, S.; Stabel, J.R.; Chang, Y.F.; Coussens, P.M.; Barletta, R.G.; Davis, W.C.; et al. A rational framework for evaluating the next generation of vaccines against Mycobacterium avium subspecies paratuberculosis. Front. Cell. Infect. Microbiol. 2014, 4, 126. [Google Scholar] [CrossRef]
- Ghosh, P.; Hsu, C.; Alyamani, E.J.; Shehata, M.M.; Al-Dubaib, M.A.; Al-Naeem, A.; Hashad, M.; Mahmoud, O.M.; Alharbi, K.B.; Al-Busadah, K.; et al. Genome-wide analysis of the emerging infection with Mycobacterium avium subspecies paratuberculosis in the Arabian camels (Camelus dromedarius). PLoS ONE 2012, 7, e31947. [Google Scholar] [CrossRef]
- Shin, S.J.; Wu, C.W.; Steinberg, H.; Talaat, A.M. Identification of novel virulence determinants in Mycobacterium paratuberculosis by screening a library of insertional mutants. Infect. Immun. 2006, 74, 3825–3833. [Google Scholar] [CrossRef]
- Hsu, C.Y.; Wu, C.W.; Talaat, A.M. Genome-Wide Sequence Variation among Mycobacterium avium Subspecies paratuberculosis Isolates: A Better Understanding of Johne’s Disease Transmission Dynamics. Front. Microbiol. 2011, 2, 236. [Google Scholar] [CrossRef] [PubMed]
- Andriotis, A.N.; Mpourmpakis, G.; Broderick, S.; Rajan, K.; Datta, S.; Sunkara, M.; Menon, M. Informatics guided discovery of surface structure-chemistry relationships in catalytic nanoparticles. J. Chem. Phys. 2014, 140, 094705. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, P.V.; Broderick, S.R.; Rajan, K. Identifying the ‘inorganic gene’ for high-temperature piezoelectric perovskites through statistical learning. Proc. Math. Phys. Eng. Sci. 2011, 467, 2271–2290. [Google Scholar] [CrossRef]
- Broderick, S.R.; Rajan, K. Discovering Electronic Signatures for Phase Stability of Intermetallics via Machine Learning. In Information Science for Materials Discovery and Design. In Information Science for Materials Discovery and Design; Springer: New York, NY, USA, 2016; pp. 223–238. [Google Scholar]
- Ericksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Vikstrom, C. Multi- and Megavariate Data Analysis: Basic Principles and Applications; Umetrics Ab: Umea, Sweden, 2013. [Google Scholar]
- Groenendaal, H.; Zagmutt, F.J.; Patton, E.A.; Wells, S.J. Cost-benefit analysis of vaccination against Mycobacterium avium ssp. paratuberculosis in dairy cattle, given its cross-reactivity with tuberculosis tests. J. Dairy Sci. 2015, 98, 6070–6084. [Google Scholar] [CrossRef]
- Kuenstner, J.T.; Naser, S.; Chamberlin, W.; Borody, T.; Graham, D.Y.; McNees, A.; Hermon-Taylor, J.; Hermon-Taylor, A.; Dow, C.T.; Thayer, W.; et al. The Consensus from the Mycobacterium avium ssp. paratuberculosis (MAP) Conference 2017. Front. Public Health 2017, 5, 208. [Google Scholar] [CrossRef]
- Singh, S.V.; Kuenstner, J.T.; Davis, W.C.; Agarwal, P.; Kumar, N.; Singh, D.; Gupta, S.; Chaubey, K.K.; Kumar, A.; Misri, J.; et al. Concurrent Resolution of Chronic Diarrhea Likely Due to Crohn’s Disease and Infection with Mycobacterium avium paratuberculosis. Front. Med. 2016, 3, 49. [Google Scholar] [CrossRef]
- Singh, S.V.; Sohal, J.S.; Singh, P.K.; Singh, A.V. Genotype profiles of Mycobacterium avium subspecies paratuberculosis isolates recovered from animals, commercial milk, and human beings in North India. Int. J. Infect. Dis. 2009, 13, e221–e227. [Google Scholar] [CrossRef][Green Version]
- Shankar, H.; Singh, S.V.; Singh, P.K.; Singh, A.V.; Sohal, J.S.; Greenstein, R.J. Presence, characterization, and genotype profiles of Mycobacterium avium subspecies paratuberculosis from unpasteurized individual and pooled milk, commercial pasteurized milk, and milk products in India by culture, PCR, and PCR-REA methods. Int. J. Infect. Dis. 2010, 14, e121–e126. [Google Scholar] [CrossRef][Green Version]
- Bastida, F.; Juste, R.A. Paratuberculosis control: A review with a focus on vaccination. J. Immune Based Vaccines 2011, 9, 8. [Google Scholar] [CrossRef]
- Tewari, D.; Hovingh, E.; Linscott, R.; Martel, E.; Lawrence, J.; Wolfgang, D.; Griswold, D. Mycobacterium avium subsp. paratuberculosis antibody response, fecal shedding, and antibody cross-reactivity to Mycobacterium bovis in M. avium subsp. paratuberculosis-infected cattle herds vaccinated against Johne’s disease. Clin. Vaccine Immunol. 2014, 21, 698–703. [Google Scholar] [CrossRef]
- Reddacliff, L.; Eppleston, J.; Windsor, P.; Whittington, R.; Jones, S. Efficacy of a killed vaccine for the control of paratuberculosis in Australian sheep flocks. Vet. Microbiol. 2006, 115, 77–90. [Google Scholar] [CrossRef]
- Uzonna, J.E.; Chilton, P.; Whitlock, R.H.; Habecker, P.L.; Scott, P.; Sweeney, R.W. Efficacy of commercial and field-strain Mycobacterium paratuberculosis vaccinations with recombinant IL-12 in a bovine experimental infection model. Vaccine 2003, 21, 3101–3109. [Google Scholar] [CrossRef]
- Patterson, C.J.; LaVenture, M.; Hurley, S.S.; Davis, J.P. Accidental self-inoculation with Mycobacterium paratuberculosis bacterin (Johne’s bacterin) by veterinarians in Wisconsin. J. Am. Vet. Med. Assoc. 1988, 192, 1197–1199. [Google Scholar] [PubMed]
- Lei, L.; Plattner, B.L.; Hostetter, J.M. Live Mycobacterium avium subsp. paratuberculosis and a killed-bacterium vaccine induce distinct subcutaneous granulomas, with unique cellular and cytokine profiles. Clin. Vaccine Immunol. 2008, 15, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Vordermeier, H.M.; Jones, G.J.; Buddle, B.M.; Hewinson, R.G.; Villarreal-Ramos, B. Bovine Tuberculosis in Cattle: Vaccines, DIVA Tests, and Host Biomarker Discovery. Annu. Rev. Anim. Biosci. 2016, 4, 87–109. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, C.; Finn, A. Chemokine receptors and their role in inflammation and infectious diseases. Blood 2000, 95, 3032–3043. [Google Scholar] [CrossRef] [PubMed]
- Aho, A.D.; McNulty, A.M.; Coussens, P.M. Enhanced expression of interleukin-1alpha and tumor necrosis factor receptor-associated protein 1 in ileal tissues of cattle infected with Mycobacterium avium subsp. paratuberculosis. Infect. Immun. 2003, 71, 6479–6486. [Google Scholar] [CrossRef]
- Guarda, G.; Braun, M.; Staehli, F.; Tardivel, A.; Mattmann, C.; Forster, I.; Farlik, M.; Decker, T.; Du Pasquier, R.A.; Romero, P.; et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity 2011, 34, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Livesey, M.; Schmoller, S.K.; Manning, E.J.; Steinberg, H.; Davis, W.C.; Hamilton, M.J.; Talaat, A.M. Invasion and persistence of Mycobacterium avium subsp. paratuberculosis during early stages of Johne’s disease in calves. Infect. Immun. 2007, 75, 2110–2119. [Google Scholar] [CrossRef]
- Corbett, C.S.; Barkema, H.W.; De Buck, J. Quantifying fecal shedding of Mycobacterium avium ssp. paratuberculosis from calves after experimental infection and exposure. J. Dairy Sci. 2018, 101, 1478–1487. [Google Scholar] [CrossRef]
- Phanse, Y.; Carrillo-Conde, B.R.; Ramer-Tait, A.E.; Broderick, S.; Kong, C.S.; Rajan, K.; Flick, R.; Mandell, R.B.; Narasimhan, B.; Wannemuehler, M.J. A systems approach to designing next generation vaccines: Combining alpha-galactose modified antigens with nanoparticle platforms. Sci. Rep. 2014, 4, 3775. [Google Scholar] [CrossRef]
- Ulery, B.D.; Petersen, L.K.; Phanse, Y.; Kong, C.S.; Broderick, S.R.; Kumar, D.; Ramer-Tait, A.E.; Carrillo-Conde, B.; Rajan, K.; Wannemuehler, M.J.; et al. Rational design of pathogen-mimicking amphiphilic materials as nanoadjuvants. Sci. Rep. 2011, 1, 198. [Google Scholar] [CrossRef]
- Phanse, Y.; Carrillo-Conde, B.R.; Ramer-Tait, A.E.; Roychoudhury, R.; Broderick, S.; Pohl, N.; Rajan, K.; Narasimhan, B.; Wannemuehler, M.J.; Bellaire, B.H. Functionalization promotes pathogen-mimicking characteristics of polyanhydride nanoparticle adjuvants. J. Biomed. Mater. Res. A 2017, 105, 2762–2771. [Google Scholar] [CrossRef]
| Treatment Group | Fecal Shedding (CFU/gm) by Months Post Challenge(Mean [% Positive]) | ||
|---|---|---|---|
| 10 | 11 | 12 | |
| PBS | 10 (50%) | 0 | 3 (17%) |
| Mycopar | 2 (33%) | 0 | 0 |
| pgsN | 0 | 5 (17%) | 0 |
| pgsNQ | 0 | 0 | 0 |
| Unchallenged | 0 | 0 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phanse, Y.; Wu, C.-W.; Venturino, A.J.; Hansen, C.; Nelson, K.; Broderick, S.R.; Steinberg, H.; Talaat, A.M. A Protective Vaccine against Johne’s Disease in Cattle. Microorganisms 2020, 8, 1427. https://doi.org/10.3390/microorganisms8091427
Phanse Y, Wu C-W, Venturino AJ, Hansen C, Nelson K, Broderick SR, Steinberg H, Talaat AM. A Protective Vaccine against Johne’s Disease in Cattle. Microorganisms. 2020; 8(9):1427. https://doi.org/10.3390/microorganisms8091427
Chicago/Turabian StylePhanse, Yashdeep, Chia-Wei Wu, Amanda J. Venturino, Chungyi Hansen, Kathryn Nelson, Scott R Broderick, Howard Steinberg, and Adel M. Talaat. 2020. "A Protective Vaccine against Johne’s Disease in Cattle" Microorganisms 8, no. 9: 1427. https://doi.org/10.3390/microorganisms8091427
APA StylePhanse, Y., Wu, C.-W., Venturino, A. J., Hansen, C., Nelson, K., Broderick, S. R., Steinberg, H., & Talaat, A. M. (2020). A Protective Vaccine against Johne’s Disease in Cattle. Microorganisms, 8(9), 1427. https://doi.org/10.3390/microorganisms8091427





