Seasonal Variation in the Rhizosphere and Non-Rhizosphere Microbial Community Structures and Functions of Camellia yuhsienensis Hu
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Experimental Design and Sampling
2.3. The Measure of Soil Physiochemical Properties
2.4. Bacterial and Fungal Community Assessment
2.4.1. DNA Extraction and PCR Amplification
2.4.2. Illumina Novaseq6000 Sequencing
2.4.3. Statistical and Bioinformatics Analysis
3. Results
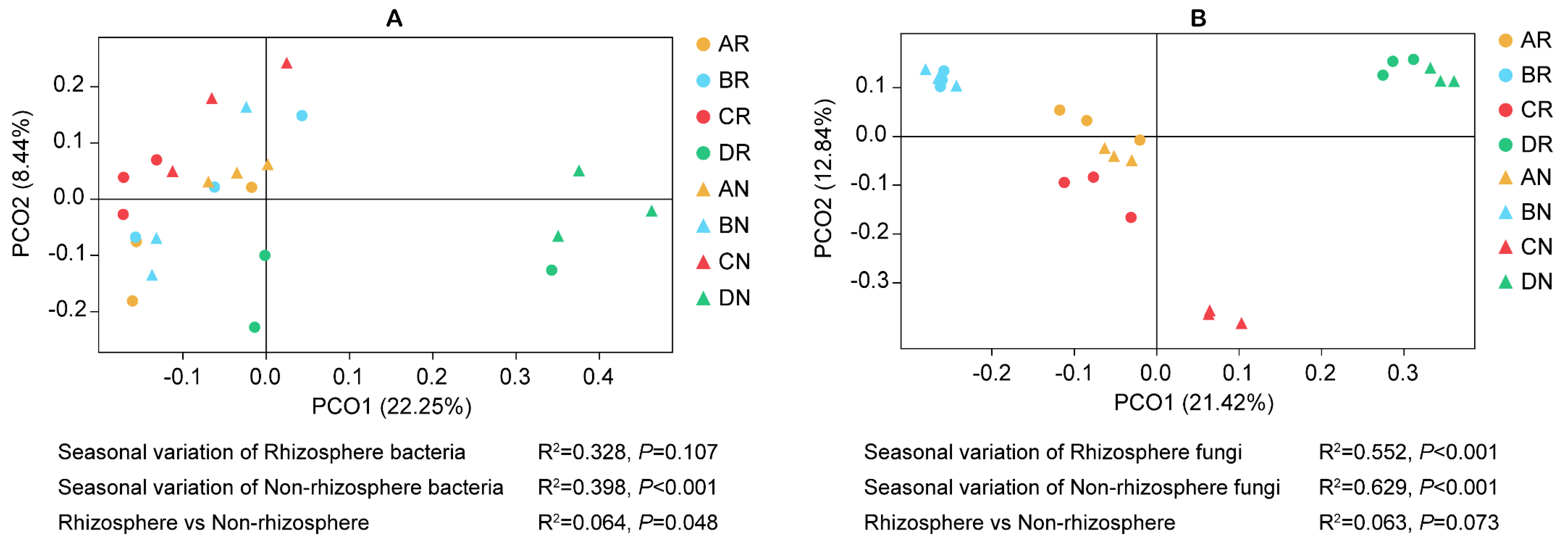
3.1. Soil Microbial Community Structure
3.1.1. Quality Control Report of the High-Throughput Sequencing
3.1.2. Microbial Composition
3.1.3. Microbial Diversity
3.1.4. Microbial Biomarkers in the Rhizosphere
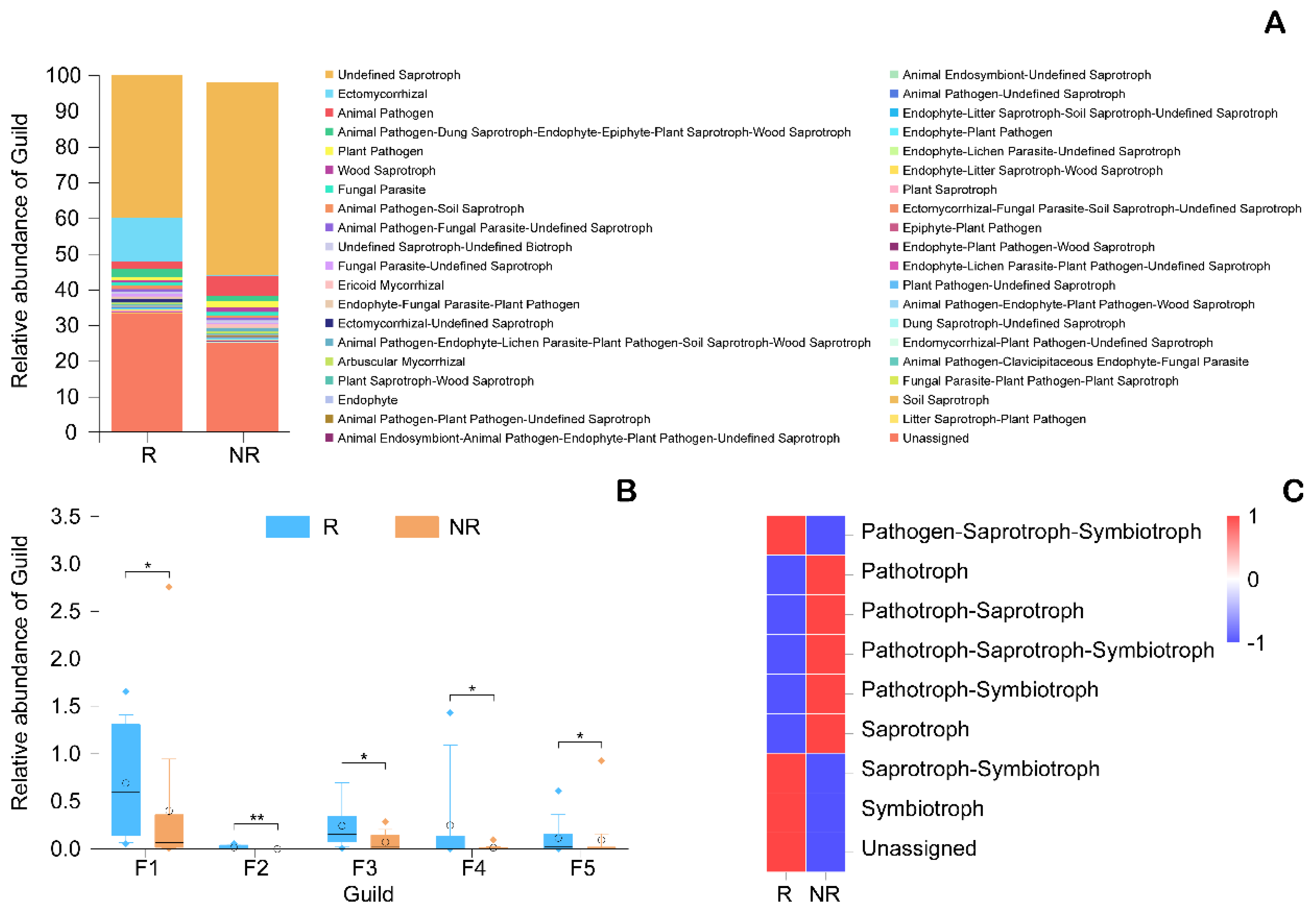
3.2. Functional Analysis of Microorganisms
3.3. Interactions Between Environmental Factors and Microbial Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wood, C.T.; Schlindwein, C.C.D.; Soares, G.L.G.; Araujo, P.B. Feeding rates of Balloniscus sellowii (Crustacea, Isopoda, Oniscidea): The effect of leaf litter decomposition and its relation to the phenolic and flavonoid content. Zookeys 2012, 24, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Progar, R.; Schowalter, T.; Freitag, C.; Morrell, J. Respiration from coarse woody debris as affected by moisture and sapotroph functional diversity in Western Oregon. Oecologia 2000, 124, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, E.; Frank, D. Can Plants Stimulate Soil Microbes and Their Own Nutrient Supply? Evidence from a Grazing Tolerant Grass. Ecology 2001, 82, 2397–2402. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Cleveland, C.C.; Wieder, W.R.; Washenberger, C.L.; Townsend, A.R. Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol. Biochem. 2010, 42, 2153–2160. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; MacLeod, R.A. Observations on the Distinction between Oligotrophic and Eutrophic Marine Bacteria. Appl. Environ. Microb. 1984, 47, 1017–1022. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbioses, 2nd ed.; Academic Press: London, UK, 1997. [Google Scholar]
- Masny, S. Occurrence of Venturia inaequalis races in Poland able to overcome specific apple scab resistance genes. Eur. J. Plant Pathol. 2016, 147, 313–323. [Google Scholar] [CrossRef][Green Version]
- Islam, S.; Akanda, A.M.; Sultana, F.; Hossain, M.M. Chilli rhizosphere fungus Aspergillus spp. PPA1 promotes vegetative growth of cucumber (Cucumis sativus) plants upon root colonisation. Arch. Phytopathol. Plant Protect. 2013, 47, 1231–1238. [Google Scholar] [CrossRef]
- Li, J.; Wu, Z.; Yuan, J. Impact of agro-farming activities on microbial diversity of acidic red soils in a Camellia Oleifera Forest. Rev. Bras. Cienc. Solo 2019, 43, e0190044. [Google Scholar] [CrossRef]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef]
- Marschner, P.; Marino, W.; Lieberei, R. Seasonal effects on microorganisms in the rhizosphere of two tropical plants in a polyculture agroforestry system in Central Amazonia, Brazil. Biol. Fert. Soils 2002, 35, 68–71. [Google Scholar] [CrossRef]
- Lynch, J.M. The Rhizosphere; John Wiley & Sons: New York, NY, USA, 2007. [Google Scholar]
- Jousset, A.; Scheu, S.; Bonkowski, M. Secondary metabolite production facilitates establishment of rhizobacteria by reducing both protozoan predation and the competitive effects of indigenous bacteria. Funct. Ecol. 2008, 22, 714–719. [Google Scholar] [CrossRef]
- Bob, S.; Bakker, A.W.; Bakker, P.A. Interactions of Deleterious and Beneficial Rhizosphere Microorganisms and the Effect of Cropping Practices. Annu. Rev. Phytopathol. 1987, 25, 339–358. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C. Plant responses to plant growth-promoting rhizobacteria. Eur. J. Plant Pathol. 2007, 119, 243–254. [Google Scholar] [CrossRef]
- Kremer, R.J. Deleterious Rhizobacteria. In Plant-Associated Bacteria; Gnanamanickam, S.S., Ed.; Springer: Dordrecht, The Netherlands, 2006; pp. 335–361. [Google Scholar]
- Zhou, S.; Zhang, M.; Zhang, K.; Yang, X.; He, D.; Yin, J.; Wang, C. Effects of reduced nitrogen and suitable soil moisture on wheat (Triticum aestivum L.) rhizosphere soil microbiological, biochemical properties and yield in the Huanghuai Plain, China. J. Integr. Agric. 2020, 19, 234–250. [Google Scholar] [CrossRef]
- Ilyas, N.; Bano, A. Azospirillum strains isolated from roots and rhizosphere soil of wheat (Triticum aestivum L.) grown under different soil moisture conditions. Biol. Fert. Soils 2010, 46, 393–406. [Google Scholar] [CrossRef]
- Taketani, R.G.; Lançoni, M.D.; Kavamura, V.N.; Durrer, A.; Andreote, F.D.; Melo, I.S. Dry Season Constrains Bacterial Phylogenetic Diversity in a Semi-Arid Rhizosphere System. Microb. Ecol. 2016, 73, 153–161. [Google Scholar] [CrossRef]
- Neumann, G.; Römheld, V. The release of root exudates as affected by the plant physiological status. In The Rhizosphere; CRC Press: New York, NY, USA, 2000; pp. 57–110. [Google Scholar]
- López-Gutiérrez, J.C.; Toro, M.; López-Hernández, D. Arbuscular mycorrhiza and enzymatic activities in the rhizosphere of Trachypogon plumosus Ness. in three acid savanna soils. Agric. Ecosyst. Environ. 2004, 103, 405–411. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, X.; Shen, Y. Comparison of soil microbial community catabolic diversity between rhizosphere and bulk soil induced by tillage or residue retention. J. Soil Sci. Plant Nutr. 2013, 13, 187–199. [Google Scholar] [CrossRef][Green Version]
- Mestre, M.C.; Rosa, C.A.; Safar, S.V.B.; Libkind, D.; Fontenla, S.B. Yeast communities associated with the bulk-soil, rhizosphere and ectomycorrhizosphere of a Nothofagus pumilio forest in northwestern Patagonia, Argentina. FEMS Microbiol. Ecol. 2011, 78, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Hernesmaa, A.; Björklöf, K.; Kiikkilä, O.; Fritze, H.; Haahtela, K.; Romantschuk, M. Structure and function of microbial communities in the rhizosphere of Scots pine after tree-felling. Soil Biol. Biochem. 2005, 37, 777–785. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Duta, S.; Yu, S.-M.; Jeong, S.C.; Lee, Y.H. Taxonomic and Functional Changes of Bacterial Communities in the Rhizosphere of Kimchi Cabbage after Seed Bacterization with Proteus vulgaris JBLS202. Plant Pathol. J. 2018, 34, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Shu, Q.; Duan, L.; Chen, C.; Zheng, H. Resistance of Main Cultivars of Oil Tea to Colletotrichum gloeosporioides. J. Anhui Agric. Univ. 2004, 31, 480–483. [Google Scholar] [CrossRef]
- Cook, R.J.; Thomashow, L.S.; Weller, D.M.; Fujimoto, D.; Mazzola, M.; Bangera, G.; Kim, D.S. Molecular Mechanisms of Defense by Rhizobacteria against Root Disease. Proc. Natl. Acad. Sci. USA 1995, 92, 4197–4201. [Google Scholar] [CrossRef]
- Zhang, P.; Cui, Z.; Guo, M.; Xi, R. Characteristics of the soil microbial community in the forestland of Camellia oleifera. PeerJ 2020, 8, e9117. [Google Scholar] [CrossRef]
- Hartmann, A.; Schmid, M.; Tuinen, D.V.; Berg, G. Plant-driven selection of microbes. Plant Soil 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Zhu, J. A Study on the Peroxidase in Camellia Species and Their Resistance to Anthracnoso Causing Fungus. J. Fujian Coll. For. 1990, 10, 368–375. Available online: http://en.cnki.com.cn/Article_en/CJFDTotal-FJLB199004011.htm (accessed on 2 August 2020).
- Liu, F.; Zeng, S. Short-term Effects of Prescribed Burning on Soil under Middle Aged Pinus Massoniana Plantation. Chin. J. Soil Sci. 2017, 48, 1025–1031. [Google Scholar] [CrossRef]
- Wang, X.; Zabowski, D. Nutrient composition of Douglas-fir rhizosphere and bulk soil solutions. Plant Soil 1998, 200, 13–20. [Google Scholar] [CrossRef]
- Bouyoucos, G.J. Evaporating the water with burning alcohol as a rapid means of determining moisture content of soils. Soil Sci. 1937, 44, 377–384. [Google Scholar] [CrossRef]
- Bureau, T.S.F. Determination of pH Value in Forest Soil; Standards Press of China: Beijing, China, 1999; Volume LY/T1239-1999. [Google Scholar]
- Bureau, T.S.F. Determination of Organic Matter in Forest Soil and Calculation Carbon-Nitrogen Ratio; Standards Press of China: Beijing, China, 1999; Volume LY/T1237-1999. [Google Scholar]
- Bureau, T.S.F. Determination of Total Nitrogen in Forest Soil; Standards Press of China: Beijing, China, 1999; Volume LY/T1228-1999. [Google Scholar]
- Bureau, T.S.F. Determination of Hydrolyzable Nitrogen in Forest Soil; Standards Press of China: Beijing, China, 1999; Volume LY/T1229-1999. [Google Scholar]
- Bureau, T.S.F. Determination of Total Phosphorus in Forest Soil; Standards Press of China: Beijing, China, 1999; Volume LY/T1232-1999. [Google Scholar]
- Bureau, T.S.F. Determination of Total Potassium in Forest Soil; Standards Press of China: Beijing, China, 1999; Volume LY/T1234-1999. [Google Scholar]
- Sims, J.T. Comparison of Mehlich 1 and Mehlich 3 extractants for P, K, Ca, Mg, Mn, Cu and Zn in Atlantic Coastal Plain soils. Commun. Soil Sci. Plan. 1989, 20, 1707–1726. [Google Scholar] [CrossRef]
- Li, C.; Zhao, Q.; Gao, T.; Wang, H.; Zhang, Z.; Liang, B.; Wei, Z.; Liu, C.; Ma, F. The mitigation effects of exogenous melatonin on replant disease in apple. J. Pineal Res. 2018, 65, e12523. [Google Scholar] [CrossRef] [PubMed]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 1–7. [Google Scholar] [CrossRef]
- Wickham, H. An Introduction to Ggplot: An Implementation of the Grammar of Graphics in R. Statistics 2006. Available online: http://ftp.auckland.ac.nz/software/CRAN/doc/vignettes/ggplot/introduction.pdf (accessed on 2 August 2020).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Krona: Interactive Metagenomic Visualization in a Web Browser. In Encyclopedia of Metagenomics: Genes, Genomes and Metagenomes: Basics, Methods, Databases and Tools; Nelson, K.E., Ed.; Springer: Boston, MA, USA, 2015; pp. 339–346. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.W. Labdsv: Ordination and Multivariate Analysis for Ecology; R Package Version 2.0-1; R Foundation: Vienna, Austria, 2016. [Google Scholar]
- Aßhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2015, 20, 1–8. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.; Simpson, G.L.; Solymos, P. Vegan: Community Ecology Package; R Package Version 2.3-5; R Foundation: Vienna, Austria, 2016. [Google Scholar]
- Omicshare Tools. Available online: https://www.omicshare.com/tools/ (accessed on 3 September 2020).
- Liang, X.; Ren, H.; Li, S.; Leng, X.; Yao, X. Soil Bacterial Community Structure and Co-occurrence Pattern during Vegetation Restoration in Karst Rocky Desertification Area. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Onwuka, B.M. Effects of soil temperature on Some Soil properties and plant growth. J. Agric. Sci. Technol. 2016, 6, 89–93. [Google Scholar] [CrossRef]
- Wu, C.; Liu, G.; Ma, G.; Liu, Q.; Yu, F.; Huang, C.; Zhao, Z.; Liang, L. Study of the differences in soil properties between the dry season and rainy season in the Mun River Basin. Catena 2019, 182, 104103. [Google Scholar] [CrossRef]
- Aon, M.A.; Colaneri, A.C., II. Temporal and spatial evolution of enzymatic activities and physico-chemical properties in an agricultural soil. Appl. Soil. Ecol. 2001, 18, 255–270. [Google Scholar] [CrossRef]
- Upton, R.N.; Bach, E.M.; Hofmockel, K.S. Spatio-temporal microbial community dynamics within soil aggregates. Soil Biol. Biochem. 2019, 132, 58–68. [Google Scholar] [CrossRef]
- Shi, J.; Yuan, X.; Lin, H.; Yang, Y.; Li, Z. Differences in Soil Properties and Bacterial Communities between the Rhizosphere and Bulk Soil and among Different Production Areas of the Medicinal Plant Fritillaria thunbergii. Int. J. Mol. Sci. 2011, 12, 3770–3785. [Google Scholar] [CrossRef]
- Yuan, J. Study on Adaptive Mechanism of Camellia Oleifera to Low-phosphorus Environment; Beijing Forestry University: Beijing, China, 2013. [Google Scholar]
- Nouri, E.; Breuillin-Sessoms, F.; Feller, U.; Reinhardt, D. Phosphorus and nitrogen regulate arbuscular mycorrhizal symbiosis in Petunia hybrida. PLoS ONE 2014, 9, e90841. [Google Scholar] [CrossRef]
- Breuillin, F.; Schramm, J.; Hajirezaei, M.; Ahkami, A.; Favre, P.; Druege, U.; Hause, B.; Bucher, M.; Kretzschmar, T.; Bossolini, E.; et al. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 2010, 64, 1002–1017. [Google Scholar] [CrossRef]
- Liu, J.; Wu, L.; Chena, D.; Lib, M.; Weib, C. Soil quality assessment of different Camellia oleifera stands in mid-subtropical China. Appl. Soil Ecol. 2017, 113, 29–35. [Google Scholar] [CrossRef]
- Liu, J.; Wu, L.; Chen, D.; Yu, Z.; Wei, C. Development of a soil quality index for Camellia oleifera forestland yield under three different parent materials in Southern China. Soil Till. Res. 2018, 176, 45–50. [Google Scholar] [CrossRef]
- Gu, S.; Hu, Q.; Cheng, Y.; Bai, L.; Liu, Z.; Xiao, W.; Gong, Z.; Wu, Y.; Feng, K.; Deng, Y.; et al. Application of organic fertilizer improves microbial community diversity and alters microbial network structure in tea (Camellia sinensis) plantation soils. Soil Till. Res. 2019, 195, 104356. [Google Scholar] [CrossRef]
- Liu, J.; Yu, S.; You, L.; Lian, L.-N.; Wang, H.; Zeng, J.; Cheng, L.; Qin, J.; Liu, X.; Hu, D. Response of Fine Root Growth of Camellia oleifera to Potassium Application Level. For. Res. 2019, 32, 31–38. [Google Scholar] [CrossRef]
- Yuan, J. Studies on Nutrition Diagnosis and Fertilization in Camellia Oleifera; Central South University of Forestry and Technology: Changsha, China, 2010. [Google Scholar]
- Janssen, P.H. Chthoniobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons Inc., in association with Bergey’s Manual Trust: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Board, T.E. Rhodanobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc., in association with Bergey’s Manual Trust: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Kim, S.-J.; Ahn, J.-H.; Lee, T.-H.; Weon, H.-Y.; Hong, S.-B.; Seok, S.-J.; Whang, K.-S.; Kwon, S.-W. Reyranella soli sp. nov., isolated from forest soil, and emended description of the genus Reyranella Pagnier et al. 2011. Int. J. Syst. Evol. Microbiol. 2013, 63, 3164–3167. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-C.; Whang, K.-S. Reyranella graminifolii sp. nov., isolated from bamboo (Phyllostachys bambusoides) litter. Int. J. Syst. Evol. Microbiol. 2014, 64, 2503–2507. [Google Scholar] [CrossRef]
- Albuquerque, L.; Rainey, F.A.; Costa, M.S.D. Aquicella. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc., in Association with Bergey’s Manual Trust: New York, NY, USA, 2018. [Google Scholar] [CrossRef]
- Dedysh, S.N. Granulicella. In Bergey’s Manual of Systematics of Archaea and Bacteria; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; John Wiley & Sons, Inc., in Association with Bergey’s Manual Trust: New York, NY, USA, 2017. [Google Scholar] [CrossRef]
- Fuerst, J.A.; Lee, K.; Butler, M.K. Gemmata. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc., in Association with Bergey’s Manual Trust: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Huo, Y.; Jong-Pyo, K.; Park, J.K.; Li, J.; Chen, L.; Yang, D.-C. Rhodanobacter ginsengiterrae sp. nov., an antagonistic bacterium against root rot fungal pathogen Fusarium solani, isolated from ginseng rhizospheric soil. Arch. Microbiol. 2018, 200, 1457–1463. [Google Scholar] [CrossRef]
- Green, S.; Prakash, O.; Jasrotia, P.; Overholt, W.; Cardenas, E.; Hubbard, D.; Tiedje, J.M.; Watson, D.B.; Schadt, C.W.; Brooks, S.C.; et al. Denitrifying Bacteria from the Genus Rhodanobacter Dominate Bacterial Communities in the Highly Contaminated Subsurface of a Nuclear Legacy Waste Site. Appl. Environ. Microb. 2012, 78, 1039–1047. [Google Scholar] [CrossRef]
- Prakash, O.; Green, S.; Jasrotia, P.; Overholt, W.; Canion, A.; Watson, D.B.; Brooks, S.C.; Kostka, J.E. Rhodanobacter denitrificans sp. nov., isolated from nitrate-rich zones of a contaminated aquifer. Int. J. Syst. Evol. Micr. 2012, 62, 2457–2462. [Google Scholar] [CrossRef]
- Li, R.; Zheng, J.; Ni, B.; Chen, K.; Jiang, J. Biodegradation of Pentachloronitrobenzene by Labrys portucalensis pcnb-21 Isolated from Polluted Soil. Pedosphere 2011, 21, 31–36. [Google Scholar] [CrossRef]
- Moreira, I.S.; Amorim, C.L.; Carvalho, M.D.F.; Castro, P.M.L. Co-metabolic degradation of chlorobenzene by the fluorobenzene degrading wild strain Labrys portucalensis. Int. Biodeter. Biodegr. 2012, 72, 76–81. [Google Scholar] [CrossRef]
- White, D.C.; Sutton, S.D.; Ringelberg, D.B. The genus Sphingomonas: Physiology and ecology. Curr. Opin. Biotechnol. 1996, 7, 301–306. [Google Scholar] [CrossRef]
- Berg, G.; Ballin, G. Bacterial Antagonists to Verticillium dahliae Kleb. J. Phytopathol. 1994, 141, 99–110. [Google Scholar] [CrossRef]
- Takeuchi, M.; Sawada, H.; Oyaizu, H.; Yokota, A. Phylogenetic Evidence for Sphingomonas and Rhizomonas as Nonphotosynthetic Members of the Alpha-4 Subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 1994, 44, 308–314. [Google Scholar] [CrossRef]
- Schmidt, S.; Wittich, R.-M.; Erdmann, D.; Wilkes, H.; Wittko, F.; Fortnagel, P. Biodegradation of diphenyl ether and its monohalogenated derivatives by Sphingomonas sp. strain SS3. Appl. Environ. Microb. 1992, 58, 2744–2750. [Google Scholar] [CrossRef]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Marra, R.; Woo, S.L.; Lorito, M. Trichoderma–plant–pathogen interactions. Soil Biol. Biochem. 2008, 40, 1–10. [Google Scholar] [CrossRef]
- Mukherjee, M.; Mukherjee, P.K.; Horwitz, B.; Zachow, C.; Berg, G.; Zeilinger, S. Trichoderma–Plant–Pathogen Interactions: Advances in Genetics of Biological Control. Indian J. Microbiol. 2012, 52, 522–529. [Google Scholar] [CrossRef]
- Domsch, K.H.; Gams, W.; Anderson, T.-H. Compendium of Soil Fungi; Academic Press: London, UK, 1980. [Google Scholar]
- Sakuradani, E.; Kobayashi, M.; Ashikari, T.; Shimizu, S. Identification of Δ12-fatty acid desaturase from arachidonic acid-producing Mortierella fungus by heterologous expression in the yeast Saccharomyces cerevisiae and the fungus Aspergillus oryzae. Eur. J. Biochem. 1999, 261, 812–820. [Google Scholar] [CrossRef]
- Higashiyama, K.; Fujikawa, S.; Park, E.Y.; Shimizu, S. Production of arachidonic acid by Mortierella fungi. Biotechnol. Bioproc. Eng. 2002, 7, 252–262. [Google Scholar] [CrossRef]
- Eroshin, V.K.; Dedyukhina, E.G. Effect of lipids from Mortierella hygrophilaon plant resistance to phytopathogens. World J. Microb. Biot. 2002, 18, 165–167. [Google Scholar] [CrossRef]
- Brodeur, J. Host specificity in biological control: Insights from from opportunistic pathogens. Evol. Appl. 2012, 5, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Goettel, M.S.; Koike, M.; Kim, J.J.; Aiuchi, D.; Shinya, R.; Brodeur, J. Potential of Lecanicillium spp. for management of insects, nematodes and plant diseases. J. Invertebr. Pathol. 2008, 98, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Yang, J.; Tao, N.; Liang, L.; Mi, Q. Cloning of the gene Lecanicillium psalliotae chitinase Lpchi1 and identification of its potential role in the biocontrol of root-knot nematode Meloidogyne incognita. Appl. Microbiol. Biot. 2007, 76, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S. Oil tea root-knot nematodes (Meloidogyne Spp.) and indentification of their pathogens. J. Cent. South For. Univ. 1986, 6, 163–168. [Google Scholar] [CrossRef]




| Kingdom | Sample | Sobs | Chao 1 | Shannon |
|---|---|---|---|---|
| Bacteria | AR | 1780 ± 390 a | 2218.8 ± 434.27 a | 8.12 ± 0.1 a |
| BR | 1392 ± 395 a | 1757.83 ± 330.26 abc | 7.31 ± 0.74 ab | |
| CR | 1727 ± 116 a | 2165.43 ± 222.65 ab | 7.72 ± 0.44 ab | |
| DR | 1271 ± 398 a | 1540.55 ± 410.06 c | 7.97 ± 0.24 a | |
| AN | 1422 ± 159 a | 1857.35 ± 83.64 abc | 7.69 ± 0.42 ab | |
| BN | 1641 ± 314 a | 1973.68 ± 340.82 abc | 7.62 ± 0.82 ab | |
| CN | 1265 ± 324 a | 1672.15 ± 199.73 bc | 7.6 ± 1.05 ab | |
| DN | 609 ± 99 b | 882.488 ± 105.93 d | 6.64 ± 0.74 b | |
| Fungus | AR | 497 ± 113 ab | 683.65 ± 67.99 ab | 4.34 ± 1.41 bc |
| BR | 609 ± 109 a | 803.52 ± 94.73 a | 5.81 ± 0.45 ab | |
| CR | 423 ± 84 abc | 597.05 ± 54.6 abc | 3.24 ± 2.31 c | |
| DR | 417 ± 25 abc | 584.27 ± 22.39 abcd | 5.29 ± 0.17 abc | |
| AN | 389 ± 80 abc | 538.32 ± 114.33 bcd | 5.08 ± 0.1 bc | |
| BN | 620 ± 133 a | 759.81 ± 154.94 ab | 6.36 ± 0.05 a | |
| CN | 213 ± 32 c | 315.57 ± 26.62 d | 3.85 ± 1.56 bc | |
| DN | 299 ± 21 bc | 451.79 ± 6.08 cd | 3.16 ± 0.27 c |
| Environmental Factors | Rhizosphere | Non-Rhizosphere | ||||
|---|---|---|---|---|---|---|
| Sobs | Chao 1 | Shannon | Sobs | Chao 1 | Shannon | |
| AP | 0.068 | 0.098 | −0.150 | 0.388 | 0.452 | 0.260 |
| TP | −0.130 | −0.197 | 0.119 | 0.437 | 0.351 | 0.211 |
| AK | 0.069 | 0.105 | 0.512 | 0.532 | 0.521 | 0.281 |
| TK | 0.252 | 0.140 | 0.303 | −0.198 | −0.187 | 0.017 |
| AMN | 0.273 | 0.252 | 0.276 | −0.436 | −0.492 | −0.180 |
| NN | −0.161 | −0.159 | −0.099 | −0.788 ** | −0.822 ** | −0.379 |
| AHN | −0.471 | −0.485 | −0.296 | −0.229 | −0.169 | −0.258 |
| TN | −0.117 | −0.018 | −0.171 | 0.540 | 0.617 * | 0.357 |
| TOC | −0.097 | −0.041 | 0.220 | 0.163 | 0.168 | −0.054 |
| pH | 0.267 | 0.315 | 0.118 | 0.270 | 0.282 | 0.471 |
| SWC | 0.360 | 0.457 | 0.356 | −0.556 | −0.491 | −0.358 |
| Tem | −0.121 | −0.166 | 0.137 | 0.849 ** | 0.866 ** | 0.491 |
| C/N | −0.110 | −0.126 | 0.272 | −0.336 | −0.402 | −0.372 |
| C/P | −0.096 | 0.017 | −0.015 | −0.148 | −0.073 | −0.237 |
| N/P | −0.089 | −0.013 | −0.230 | 0.347 | 0.454 | 0.282 |
| Environmental Factors | Rhizosphere | Not-Rhizosphere | ||||
|---|---|---|---|---|---|---|
| Sobs | Chao 1 | Shannon | Sobs | Chao 1 | Shannon | |
| AP | 0.210 | 0.273 | −0.006 | 0.255 | 0.277 | 0.414 |
| TP | 0.760 ** | 0.758 ** | 0.145 | 0.660 * | 0.586 * | 0.582 * |
| AK | 0.183 | 0.239 | 0.057 | 0.475 | 0.471 | 0.464 |
| TK | −0.551 | −0.703 * | −0.181 | −0.514 | −0.563 | −0.510 |
| AMN | −0.432 | −0.555 | −0.391 | −0.507 | −0.561 | −0.543 |
| NN | −0.521 | −0.620 * | 0.099 | −0.479 | −0.387 | −0.696 * |
| AHN | −0.411 | −0.440 | 0.219 | −0.411 | −0.412 | −0.259 |
| TN | −0.229 | −0.122 | −0.041 | 0.197 | 0.266 | 0.289 |
| TOC | −0.339 | −0.253 | 0.163 | 0.389 | 0.434 | 0.226 |
| pH | 0.311 | 0.348 | 0.000 | −0.071 | −0.091 | 0.003 |
| SWC | −0.419 | −0.432 | −0.242 | −0.683 * | −0.603 * | −0.776 ** |
| Tem | 0.550 | 0.654 * | −0.012 | 0.541 | 0.448 | 0.738 ** |
| C/N | −0.296 | −0.234 | 0.187 | 0.205 | 0.200 | −0.056 |
| C/P | −0.550 | −0.492 | 0.010 | −0.025 | 0.099 | −0.198 |
| N/P | −0.637 * | −0.601 * | −0.123 | −0.108 | −0.027 | 0.023 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Luo, Z.; Zhang, C.; Qu, X.; Chen, M.; Song, T.; Yuan, J. Seasonal Variation in the Rhizosphere and Non-Rhizosphere Microbial Community Structures and Functions of Camellia yuhsienensis Hu. Microorganisms 2020, 8, 1385. https://doi.org/10.3390/microorganisms8091385
Li J, Luo Z, Zhang C, Qu X, Chen M, Song T, Yuan J. Seasonal Variation in the Rhizosphere and Non-Rhizosphere Microbial Community Structures and Functions of Camellia yuhsienensis Hu. Microorganisms. 2020; 8(9):1385. https://doi.org/10.3390/microorganisms8091385
Chicago/Turabian StyleLi, Jun, Ziqiong Luo, Chenhui Zhang, Xinjing Qu, Ming Chen, Ting Song, and Jun Yuan. 2020. "Seasonal Variation in the Rhizosphere and Non-Rhizosphere Microbial Community Structures and Functions of Camellia yuhsienensis Hu" Microorganisms 8, no. 9: 1385. https://doi.org/10.3390/microorganisms8091385
APA StyleLi, J., Luo, Z., Zhang, C., Qu, X., Chen, M., Song, T., & Yuan, J. (2020). Seasonal Variation in the Rhizosphere and Non-Rhizosphere Microbial Community Structures and Functions of Camellia yuhsienensis Hu. Microorganisms, 8(9), 1385. https://doi.org/10.3390/microorganisms8091385





