Non-Tuberculous Mycobacteria: Molecular and Physiological Bases of Virulence and Adaptation to Ecological Niches
Abstract
1. Introduction: Mycobacteria and the Host
2. Mycobacteria Diversity and Evolution
2.1. General Characteristics of the Mycobacterium Genus
2.2. Phylogeny and Evolution of the Mycobacterium Genus
2.3. Evolutionary Drivers of the Mycobacterium Genus
3. Mycobacteria Ecology: The Underlying Resilience Biology
3.1. Genetic Variability
3.2. Transcriptional Regulation
3.3. Mycobacterial Outer Membrane
3.4. Slow Growth
3.5. Biofilm Formation and Quorum-Sensing
3.6. Resistance and Degradation of Recalcitrant Compounds
3.7. Antimicrobial Resistance
3.8. Protozoa-Mycobacteria Symbiosis
4. Mycobacteria Ecological Niches
4.1. Water
4.2. Soil
4.3. Plants
4.4. Dust and Air
4.5. Extreme Environments
4.6. Healthcare Settings
5. Mycobacteria Infection
5.1. Human Infection
Clinically-Relevant Species
5.2. Animal Infection
6. Detection, Identification, and Differentiation Tools for Mycobacteria
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Parte, A.C. LPSN—List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Shinnick, T.M.; Good, R.C. Mycobacterial taxonomy. Eur. J. Clin. Microbiol. Infect. Dis. 1994, 13, 884–901. [Google Scholar] [CrossRef] [PubMed]
- Balloux, F.; van Dorp, L. Q&A: What are pathogens, and what have they done to and for us? BMC Biol. 2017, 15, 91. [Google Scholar] [CrossRef]
- Springer, B.; Stockman, L.; Teschner, K.; Roberts, G.D.; Böttger, E.C. Two-laboratory collaborative study on identification of mycobacteria: Molecular versus phenotypic methods. J. Clin. Microbiol. 1996, 34, 296–303. [Google Scholar] [CrossRef]
- Christianson, L.C.; Dewlett, H.J. Pulmonary disease in adults associated with unclassified mycobacteria. Am. J. Med. 1960, 29, 980–991. [Google Scholar] [CrossRef]
- Lillis, J.V.; Ansdell, V.E.; Ruben, K.; Simpson, E.L.; Tumbaga, G.; Ansdell, D.; Bremmer, S.; Kurtz, S.E.; White, C.R.; Blauvelt, A.; et al. Sequelae of World War II: An Outbreak of Chronic Cutaneous Nontuberculous Mycobacterial Infection among Satowanese Islanders. Clin. Infect. Dis. 2009, 48, 1541–1546. [Google Scholar] [CrossRef]
- Johnson, M.M.; Odell, J.A. Nontuberculous mycobacterial pulmonary infections. J. Thorac. Dis. 2014, 6, 210–220. [Google Scholar] [CrossRef]
- Jeon, D. Infection Source and Epidemiology of Nontuberculous Mycobacterial Lung Disease. Tuberc. Respir. Dis. 2019, 82, 94–101. [Google Scholar] [CrossRef]
- Claudio, P.; Claudio, S. Extrapulmonary Infections Associated with Nontuberculous Mycobacteria in Immunocompetent Persons. Emerg. Infect. Dis. J. 2009, 15, 1351. [Google Scholar] [CrossRef]
- Sousa, S.; Borges, V.; Joao, I.; Gomes, J.P.; Jordao, L. Nontuberculous Mycobacteria Persistence in a Cell Model Mimicking Alveolar Macrophages. Microorganisms 2019, 7, 113. [Google Scholar] [CrossRef]
- Hänsch, H.; Smith, D.A.; Mielke, M.; Hahn, H.; Bancroft, G.; Ehlers, S. Mechanisms of granuloma formation in murine Mycobacterium avium infection: The contribution of CD4+ T cells. Int. Immunol. 1996, 8, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Magee, J.G.; Ward, A.C. Mycobacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Rainey, F., Kämpfer, P., Trujillo, M., Chun, J., DeVos, P., Hedlund, B., Dedysh, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–84. [Google Scholar] [CrossRef]
- Gupta, R.S.; Lo, B.; Son, J. Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Böddinghaus, B.; Rogall, T.; Flohr, T.; Blöcker, H.; Böttger, E.C. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 1990, 28, 1751. [Google Scholar] [CrossRef]
- Roth, A.; Fischer, M.; Hamid, M.E.; Michalke, S.; Ludwig, W.; Mauch, H. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 1998, 36, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Telenti, A.; Marchesi, F.; Balz, M.; Bally, F.; Böttger, E.C.; Bodmer, T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 1993, 31, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Fedrizzi, T.; Meehan, C.J.; Grottola, A.; Giacobazzi, E.; Fregni Serpini, G.; Tagliazucchi, S.; Fabio, A.; Bettua, C.; Bertorelli, R.; De Sanctis, V.; et al. Genomic characterization of Nontuberculous Mycobacteria. Sci. Rep. 2017, 7, 45258. [Google Scholar] [CrossRef]
- Wee, W.Y.; Dutta, A.; Choo, S.W. Comparative genome analyses of mycobacteria give better insights into their evolution. PLoS ONE 2017, 12, e0172831. [Google Scholar] [CrossRef]
- Movahedzadeh, F.; Bitter, W. Ins and outs of mycobacterial plasmids. Methods Mol. Biol. (Clifton N. J.) 2009, 465, 217–228. [Google Scholar] [CrossRef]
- Jucker, M.T.; Falkinham, J.O. Epidemiology of Infection by Nontuberculous Mycobacteria: IX. Evidence for Two DNA Homology Groups among Small Plasmids in Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum. Am. Rev. Respir. Dis. 1990, 142, 858–862. [Google Scholar] [CrossRef]
- Jensen, A.G.; Bennedsen, J.; Rosdahl, V.T. Plasmid Profiles of Mycobacterium avium/intracellulare Isolated from Patients with AIDS or Cervical Lymphadenitis and from Environmental Samples. Scand. J. Infect. Dis. 1989, 21, 645–649. [Google Scholar] [CrossRef]
- Picardeau, M.; Le Dantec, C.; Vincent, V. Analysis of the internal replication region of a mycobacterial linear plasmid. Microbiology 2000, 146, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Kirby, C.; Waring, A.; Griffin, T.J., IV; Falkinham, J.O., III; Grindley, N.D.F.; Derbyshire, K.M. Cryptic plasmids of Mycobacterium avium: Tn552 to the rescue. Mol. Microbiol. 2002, 43, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.; Drancourt, M.; Tuller, T.; Pontarotti, P. Genome-wide analysis of horizontally acquired genes in the genus Mycobacterium. Sci. Rep. 2018, 8, 14817. [Google Scholar] [CrossRef] [PubMed]
- Reva, O.; Korotetskiy, I.; Ilin, A. Role of the horizontal gene exchange in evolution of pathogenic Mycobacteria. BMC Evol. Biol. 2015, 15, S2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stinear, T.P.; Seemann, T.; Harrison, P.F.; Jenkin, G.A.; Davies, J.K.; Johnson, P.D.R.; Abdellah, Z.; Arrowsmith, C.; Chillingworth, T.; Churcher, C.; et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008, 18, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Ripoll, F.; Pasek, S.; Schenowitz, C.; Dossat, C.; Barbe, V.; Rottman, M.; Macheras, E.; Heym, B.; Herrmann, J.-L.; Daffé, M.; et al. Non Mycobacterial Virulence Genes in the Genome of the Emerging Pathogen Mycobacterium abscessus. PLoS ONE 2009, 4, e5660. [Google Scholar] [CrossRef]
- Erardi, F.X.; Failla, M.L.; Falkinham, J.O., 3rd. Plasmid-encoded copper resistance and precipitation by Mycobacterium scrofulaceum. Appl. Environ. Microbiol. 1987, 53, 1951–1954. [Google Scholar] [CrossRef]
- Meissner, P.S.; Falkinham, J.O., 3rd. Plasmid-encoded mercuric reductase in Mycobacterium scrofulaceum. J. Bacteriol. 1984, 157, 669–672. [Google Scholar] [CrossRef]
- Stinear, T.P.; Mve-Obiang, A.; Small, P.L.C.; Frigui, W.; Pryor, M.J.; Brosch, R.; Jenkin, G.A.; Johnson, P.D.R.; Davies, J.K.; Lee, R.E.; et al. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. USA 2004, 101, 1345. [Google Scholar] [CrossRef]
- Falkinham, J.O., 3rd; Iseman, M.D.; de Haas, P.; van Soolingen, D. Mycobacterium avium in a shower linked to pulmonary disease. J. Water Health 2008, 6, 209–213. [Google Scholar] [CrossRef]
- Reis, A.C.; Albuquerque, T.; Botelho, A.; Cunha, M.V. Polyclonal infection as a new scenario in Mycobacterium caprae epidemiology. Vet. Microbiol. 2020, 240, 108533. [Google Scholar] [CrossRef] [PubMed]
- Derbyshire, K.M.; Gray, T.A. Distributive Conjugal Transfer: New Insights into Horizontal Gene Transfer and Genetic Exchange in Mycobacteria. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Labidi, A.; David, H.L.; Roulland-Dussoix, D. Resitriction endonuclease mapping and cloning of Mycobacterium fortuitum var. fortuitum plasmid pAL5000. Ann. De L’institut Pasteur/Microbiol. 1985, 136, 209–215. [Google Scholar] [CrossRef]
- Gavigan, J.A.; Aínsa, J.A.; Pérez, E.; Otal, I.; Martín, C. Isolation by genetic labeling of a new mycobacterial plasmid, pJAZ38, from Mycobacterium fortuitum. J. Bacteriol. 1997, 179, 4115. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qin, M.; Taniguchi, H.; Mizuguchi, Y. Analysis of the replication region of a mycobacterial plasmid, pMSC262. J. Bacteriol. 1994, 176, 419. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beggs, M.L.; Crawford, J.T.; Eisenach, K.D. Isolation and sequencing of the replication region of Mycobacterium avium plasmid pLR7. J. Bacteriol. 1995, 177, 4836–4840. [Google Scholar] [CrossRef] [PubMed]
- Leão, S.C.; Matsumoto, C.K.; Carneiro, A.; Ramos, R.T.; Nogueira, C.L.; Lima, J.D.; Lima, K.V.; Lopes, M.L.; Schneider, H.; Azevedo, V.A.; et al. The detection and sequencing of a broad-host-range conjugative IncP-1β plasmid in an epidemic strain of Mycobacterium abscessus subsp. bolletii. PLoS ONE 2013, 8, e60746. [Google Scholar] [CrossRef]
- Ummels, R.; Abdallah, A.M.; Kuiper, V.; Aâjoud, A.; Sparrius, M.; Naeem, R.; Spaink, H.P.; van Soolingen, D.; Pain, A.; Bitter, W. Identification of a Novel Conjugative Plasmid in Mycobacteria That Requires Both Type IV and Type VII Secretion. mBio 2014, 5, e01744-14. [Google Scholar] [CrossRef]
- Flentie, K.; Garner, A.L.; Stallings, C.L. The Mycobacterium tuberculosis transcription machinery: Ready to respond to host attacks. J. Bacteriol. 2016. [Google Scholar] [CrossRef]
- Sachdeva, P.; Misra, R.; Tyagi, A.K.; Singh, Y. The sigma factors of Mycobacterium tuberculosis: Regulation of the regulators. Febs J. 2010, 277, 605–626. [Google Scholar] [CrossRef]
- Waagmeester, A.; Thompson, J.; Reyrat, J.-M. Identifying sigma factors in Mycobacterium smegmatis by comparative genomic analysis. Trends Microbiol. 2005, 13, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Hurst-Hess, K.; Biswas, R.; Yang, Y.; Rudra, P.; Lasek-Nesselquist, E.; Ghosh, P. Mycobacterial SigA and SigB Cotranscribe Essential Housekeeping Genes during Exponential Growth. mBio 2019, 10, e00273-19. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, S.; Hümpel, A.; McLellan, A.D.; Cook, G.M. The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology 2008, 154, 2786–2795. [Google Scholar] [CrossRef] [PubMed]
- Sechi, L.A.; Felis, G.E.; Ahmed, N.; Paccagnini, D.; Usai, D.; Ortu, S.; Molicotti, P.; Zanetti, S. Genome and transcriptome scale portrait of sigma factors in Mycobacterium avium subsp. paratuberculosis. Infect. Genet. Evol. 2007, 7, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Wu, C.-w.; Talaat, A.M. Key Role for the Alternative Sigma Factor, SigH, in the Intracellular Life of Mycobacterium avium subsp. paratuberculosis during Macrophage Stress. Infect. Immun. 2013, 81, 2242–2257. [Google Scholar] [CrossRef]
- Manganelli, R. Sigma Factors: Key Molecules in Mycobacterium tuberculosis Physiology and Virulence. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Pettersson, B.M.F.; Das, S.; Behra, P.R.K.; Jordan, H.R.; Ramesh, M.; Mallick, A.; Root, K.M.; Cheramie, M.N.; de la Cruz Melara, I.; Small, P.L.C.; et al. Comparative Sigma Factor-mRNA Levels in Mycobacterium marinum under Stress Conditions and during Host Infection. PLoS ONE 2015, 10, e0139823. [Google Scholar] [CrossRef]
- Behra, P.R.K.; Das, S.; Pettersson, B.M.F.; Shirreff, L.; DuCote, T.; Jacobsson, K.-G.; Ennis, D.G.; Kirsebom, L.A. Extended insight into the Mycobacterium chelonae-abscessus complex through whole genome sequencing of Mycobacterium salmoniphilum outbreak and Mycobacterium salmoniphilum-like strains. Sci. Rep. 2019, 9, 4603. [Google Scholar] [CrossRef]
- Lewis, A.; Falkinham, J., III. Microaerobic growth and anaerobic survival of Mycobacterium avium, Mycobacterium intracellulare and Mycobacterium scrofulaceum. Int. J. Mycobacteriol. 2015, 4, 25–30. [Google Scholar] [CrossRef]
- Chiaradia, L.; Lefebvre, C.; Parra, J.; Marcoux, J.; Burlet-Schiltz, O.; Etienne, G.; Tropis, M.; Daffé, M. Dissecting the mycobacterial cell envelope and defining the composition of the native mycomembrane. Sci. Rep. 2017, 7, 12807. [Google Scholar] [CrossRef]
- Zuber, B.; Chami, M.; Houssin, C.; Dubochet, J.; Griffiths, G.; Daffé, M. Direct Visualization of the Outer Membrane of Mycobacteria and Corynebacteria in Their Native State. J. Bacteriol. 2008, 190, 5672. [Google Scholar] [CrossRef] [PubMed]
- Cabruja, M.; Mondino, S.; Tsai, Y.T.; Lara, J.; Gramajo, H.; Gago, G. A conditional mutant of the fatty acid synthase unveils unexpected cross talks in mycobacterial lipid metabolism. Open Biol. 2017, 7, 160277. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, S.; Crick, D.C.; McNeil, M.R.; Brennan, P.J. Unique Structural Features of the Peptidoglycan of Mycobacterium leprae. J. Bacteriol. 2008, 190, 655–661. [Google Scholar] [CrossRef]
- Raymond, J.B.; Mahapatra, S.; Crick, D.C.; Pavelka, M.S. Identification of the namH Gene, Encoding the Hydroxylase Responsible for the N-Glycolylation of the Mycobacterial Peptidoglycan. J. Biol. Chem. 2005, 280, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.; Lloyd, G.; Joe, M.; Lowary, T.L.; Reynolds, E.; Walters-Morgan, H.; Bhatt, A.; Lovering, A.; Besra, G.S.; Alderwick, L.J. Lcp1 Is a Phosphotransferase Responsible for Ligating Arabinogalactan to Peptidoglycan in Mycobacterium tuberculosis. mBio 2016, 7, e00972-16. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.M.; Golchin, S.A.; Veyrier, F.J.; Domenech, P.; Boneca, I.G.; Azad, A.K.; Rajaram, M.V.S.; Schlesinger, L.S.; Divangahi, M.; Reed, M.B.; et al. N-Glycolylated Peptidoglycan Contributes to the Immunogenicity but Not Pathogenicity of Mycobacterium tuberculosis. J. Infect. Dis. 2013, 209, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Wietzerbin, J.; Das, B.C.; Petit, J.F.; Lederer, E.; Leyh-Bouille, M.; Ghuysen, J.M. Occurrence of D-alanyl-(D)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of Mycobacteria. Biochemistry 1974, 13, 3471–3476. [Google Scholar] [CrossRef]
- Kana, B.D.; Gordhan, B.G.; Downing, K.J.; Sung, N.; Vostroktunova, G.; Machowski, E.E.; Tsenova, L.; Young, M.; Kaprelyants, A.; Kaplan, G.; et al. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol. Microbiol. 2008, 67, 672–684. [Google Scholar] [CrossRef]
- Xie, J.; Hodgkinson, J.W.; Katzenback, B.A.; Kovacevic, N.; Belosevic, M. Characterization of three Nod-like receptors and their role in antimicrobial responses of goldfish (Carassius auratus L.) macrophages to Aeromonas salmonicida and Mycobacterium marinum. Dev. Comp. Immunol. 2013, 39, 180–187. [Google Scholar] [CrossRef]
- Faller, M.; Niederweis, M.; Schulz, G.E. The Structure of a Mycobacterial Outer-Membrane Channel. Science 2004, 303, 1189–1192. [Google Scholar] [CrossRef]
- Alahari, A.; Saint, N.; Campagna, S.; Molle, V.; Molle, G.; Kremer, L. The N-Terminal Domain of OmpATb Is Required for Membrane Translocation and Pore-Forming Activity in Mycobacteria. J. Bacteriol. 2007, 189, 6351. [Google Scholar] [CrossRef] [PubMed]
- Danilchanka, O.; Pires, D.; Anes, E.; Niederweis, M. The Mycobacterium tuberculosis outer membrane channel protein CpnT confers susceptibility to toxic molecules. Antimicrob. Agents Chemother. 2015, 59, 2328–2336. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Barsch, A.; Niehaus, K.; Pühler, A.; Tauch, A.; Kalinowski, J. The glycosylated cell surface protein Rpf2, containing a resuscitation-promoting factor motif, is involved in intercellular communication of Corynebacterium glutamicum. Arch. Microbiol. 2004, 182, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Speer, A.; Sun, J.; Danilchanka, O.; Meikle, V.; Rowland, J.L.; Walter, K.; Buck, B.R.; Pavlenok, M.; Hölscher, C.; Ehrt, S.; et al. Surface hydrolysis of sphingomyelin by the outer membrane protein Rv0888 supports replication of Mycobacterium tuberculosis in macrophages. Mol. Microbiol. 2015, 97, 881–897. [Google Scholar] [CrossRef]
- Boddingius, J.; Dijkman, H. Subcellular localization of Mycobacterium leprae-specific phenolic glycolipid (PGL-I) antigen in human leprosy lesions and in M. leprae isolated from armadillo liver. Microbiology 1990, 136, 2001–2012. [Google Scholar] [CrossRef]
- Schwebach, J.R.; Glatman-Freedman, A.; Gunther-Cummins, L.; Dai, Z.; Robbins, J.B.; Schneerson, R.; Casadevall, A. Glucan Is a Component of the Mycobacterium tuberculosis Surface That Is Expressed In Vitro and In Vivo. Infect. Immun. 2002, 70, 2566. [Google Scholar] [CrossRef][Green Version]
- Lemassu, A.; Ortalo-Magné, A.; Bardou, F.; Silve, G.; Lanéelle, M.-A.; Daffé, M. Extracellular and surface-exposed polysaccharides of non-tuberculous mycobacteria. Microbiology 1996, 142, 1513–1520. [Google Scholar] [CrossRef]
- Steed, K.A.; Falkinham, J.O., 3rd. Effect of growth in biofilms on chlorine susceptibility of Mycobacterium avium and Mycobacterium intracellulare. Appl. Environ. Microbiol. 2006, 72, 4007–4011. [Google Scholar] [CrossRef]
- Drapal, M.; Wheeler, P.R.; Fraser, P.D. Metabolite analysis of Mycobacterium species under aerobic and hypoxic conditions reveals common metabolic traits. Microbiology 2016, 162, 1456–1467. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Keevil, C.W.; Lappin-Scott, H.M. Mycobacterium fortuitum and Mycobacterium chelonae biofilm formation under high and low nutrient conditions. J. Appl. Microbiol. 1998, 85, 60S–69S. [Google Scholar] [CrossRef]
- Lipner, E.M.; Knox, D.; French, J.; Rudman, J.; Strong, M.; Crooks, J.L. A Geospatial Epidemiologic Analysis of Nontuberculous Mycobacterial Infection: An Ecological Study in Colorado. Ann. Am. Thorac. Soc. 2017, 14, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Falkinham, J.O. Nontuberculous Mycobacteria from Household Plumbing of Patients with Nontuberculous Mycobacteria Disease. Emerg. Infect. Dis. J. 2011, 17, 419. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; de Carvalho, C.C.C.R.; Stevenson, A.; Grant, I.R.; Hallsworth, J.E. Extraordinary solute-stress tolerance contributes to the environmental tenacity of mycobacteria. Environ. Microbiol. Rep. 2015, 7, 746–764. [Google Scholar] [CrossRef] [PubMed]
- Archuleta, R.J.; Yvonne Hoppes, P.; Primm, T.P. Mycobacterium avium enters a state of metabolic dormancy in response to starvation. Tuberculosis 2005, 85, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Primm, T.P.; Lucero, C.A.; Falkinham, J.O. Health Impacts of Environmental Mycobacteria. Clin. Microbiol. Rev. 2004, 17, 98–106. [Google Scholar] [CrossRef]
- Sharbati, S.; Schramm, K.; Rempel, S.; Wang, H.; Andrich, R.; Tykiel, V.; Kunisch, R.; Lewin, A. Characterisation of porin genes from Mycobacterium fortuitumand their impact on growth. BMC Microbiol. 2009, 9, 31. [Google Scholar] [CrossRef]
- Rock, J.M.; Lang, U.F.; Chase, M.R.; Ford, C.B.; Gerrick, E.R.; Gawande, R.; Coscolla, M.; Gagneux, S.; Fortune, S.M.; Lamers, M.H. DNA replication fidelity in Mycobacterium tuberculosis is mediated by an ancestral prokaryotic proofreader. Nat. Genet. 2015, 47, 677–681. [Google Scholar] [CrossRef]
- Sharma, I.M.; Petchiappan, A.; Chatterji, D. Quorum sensing and biofilm formation in mycobacteria: Role of c-di-GMP and methods to study this second messenger. Iubmb Life 2014, 66, 823–834. [Google Scholar] [CrossRef]
- Kearns, D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8, 634–644. [Google Scholar] [CrossRef]
- Recht, J.; Martínez, A.; Torello, S.; Kolter, R. Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 2000, 182, 4348–4351. [Google Scholar] [CrossRef]
- Falkinham, J.O., 3rd; Norton, C.D.; LeChevallier, M.W. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other Mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 2001, 67, 1225–1231. [Google Scholar] [CrossRef] [PubMed]
- Nessar, R.; Reyrat, J.-M.; Davidson, L.B.; Byrd, T.F. Deletion of the mmpL4b gene in the Mycobacterium abscessus glycopeptidolipid biosynthetic pathway results in loss of surface colonization capability, but enhanced ability to replicate in human macrophages and stimulate their innate immune response. Microbiology 2011, 157, 1187–1195. [Google Scholar] [CrossRef]
- Tsai, S.-H.; Shen, G.-H.; Lin, C.-H.; Liau, J.-R.; Lai, H.-C.; Hu, S.-T. Mab_3168c, a Putative Acetyltransferase, Enhances Adherence, Intracellular Survival and Antimicrobial Resistance of Mycobacterium abscessus. PLoS ONE 2013, 8, e67563. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, Y.; Danelishvili, L.; Wu, M.; Hidaka, E.; Katsuyama, T.; Stang, B.; Petrofsky, M.; Bildfell, R.; Bermudez, L.E. The ability to form biofilm influences Mycobacterium avium invasion and translocation of bronchial epithelial cells. Cell. Microbiol. 2006, 8, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Johansen, T.B.; Agdestein, A.; Olsen, I.; Nilsen, S.F.; Holstad, G.; Djønne, B. Biofilm formation by Mycobacterium avium isolates originating from humans, swine and birds. BMC Microbiol. 2009, 9, 159. [Google Scholar] [CrossRef]
- Carter, G.; Wu, M.; Drummond, D.C.; Bermudez, L.E. Characterization of biofilm formation by clinical isolates of Mycobacterium avium. J. Med. Microbiol. 2003, 52, 747–752. [Google Scholar] [CrossRef]
- Geier, H.; Mostowy, S.; Cangelosi, G.A.; Behr, M.A.; Ford, T.E. Autoinducer-2 Triggers the Oxidative Stress Response in Mycobacterium avium, Leading to Biofilm Formation. Appl. Environ. Microbiol. 2008, 74, 1798–1804. [Google Scholar] [CrossRef]
- Rose, S.J.; Bermudez, L.E. Identification of Bicarbonate as a Trigger and Genes Involved with Extracellular DNA Export in Mycobacterial Biofilms. mBio 2016, 7, e01597-16. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Danelishvili, L.; Wu, M.; MacNab, M.; Bermudez, L.E. Mycobacterium avium Genes Associated with the Ability to Form a Biofilm. Appl. Environ. Microbiol. 2006, 72, 819–825. [Google Scholar] [CrossRef]
- Rose, S.J.; Bermudez, L.E. Mycobacterium avium Biofilm Attenuates Mononuclear Phagocyte Function by Triggering Hyperstimulation and Apoptosis during Early Infection. Infect. Immun. 2014, 82, 405–412. [Google Scholar] [CrossRef]
- Ren, H.; Dover, L.G.; Islam, S.T.; Alexander, D.C.; Chen, J.M.; Besra, G.S.; Liu, J. Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum. Mol. Microbiol. 2007, 63, 1345–1359. [Google Scholar] [CrossRef] [PubMed]
- Mohandas, P.; Budell, W.C.; Mueller, E.; Au, A.; Bythrow, G.V.; Quadri, L.E.N. Pleiotropic consequences of gene knockouts in the phthiocerol dimycocerosate and phenolic glycolipid biosynthetic gene cluster of the opportunistic human pathogen Mycobacterium marinum. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]
- Marsollier, L.; Brodin, P.; Jackson, M.; Korduláková, J.; Tafelmeyer, P.; Carbonnelle, E.; Aubry, J.; Milon, G.; Legras, P.; André, J.-P.S.; et al. Impact of Mycobacterium ulcerans Biofilm on Transmissibility to Ecological Niches and Buruli Ulcer Pathogenesis. PLoS Pathog. 2007, 3, e62. [Google Scholar] [CrossRef] [PubMed]
- Pidot, S.J.; Porter, J.L.; Tobias, N.J.; Anderson, J.; Catmull, D.; Seemann, T.; Kidd, S.; Davies, J.K.; Reynolds, E.; Dashper, S.; et al. Regulation of the 18 kDa heat shock protein in Mycobacterium ulcerans: An alpha-crystallin orthologue that promotes biofilm formation. Mol. Microbiol. 2010, 78, 1216–1231. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Kumar, A. The extracellular matrix of mycobacterial biofilms: Could we shorten the treatment of mycobacterial infections? Microb. Cell (Grazaustria) 2019, 6, 105–122. [Google Scholar] [CrossRef]
- Mathew, R.; Mukherjee, R.; Balachandar, R.; Chatterji, D. Deletion of the rpoZ gene, encoding the ω subunit of RNA polymerase, results in pleiotropic surface-related phenotypes in Mycobacterium smegmatis. Microbiology 2006, 152, 1741–1750. [Google Scholar] [CrossRef][Green Version]
- Pacheco, S.A.; Hsu, F.-F.; Powers, K.M.; Purdy, G.E. MmpL11 Protein Transports Mycolic Acid-containing Lipids to the Mycobacterial Cell Wall and Contributes to Biofilm Formation in Mycobacterium smegmatis. J. Biol. Chem. 2013, 288, 24213–24222. [Google Scholar] [CrossRef]
- Chen, J.M.; German, G.J.; Alexander, D.C.; Ren, H.; Tan, T.; Liu, J. Roles of Lsr2 in Colony Morphology and Biofilm Formation of Mycobacterium smegmatis. J. Bacteriol. 2006, 188, 633–641. [Google Scholar] [CrossRef]
- Yang, Y.; Thomas, J.; Li, Y.; Vilchèze, C.; Derbyshire, K.M.; Jacobs, W.R., Jr.; Ojha, A.K. Defining a temporal order of genetic requirements for development of mycobacterial biofilms. Mol. Microbiol. 2017, 105, 794–809. [Google Scholar] [CrossRef]
- Richards, J.P.; Ojha, A.K. Mycobacterial Biofilms. Microbiol. Spectr. 2014, 2, MGM2–MGM4. [Google Scholar]
- Wolff, K.A.; de la Peña, A.H.; Nguyen, H.T.; Pham, T.H.; Amzel, L.M.; Gabelli, S.B.; Nguyen, L. A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development. PLoS Pathog. 2015, 11, e1004839. [Google Scholar] [CrossRef] [PubMed]
- Vargas, D.; Hageman, S.; Gulati, M.; Nobile, C.J.; Rawat, M. S-nitrosomycothiol reductase and mycothiol are required for survival under aldehyde stress and biofilm formation in Mycobacterium smegmatis. Iubmb Life 2016, 68, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Ojha, A.; Hatfull, G.F. The role of iron in Mycobacterium smegmatis biofilm formation: The exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol. Microbiol. 2007, 66, 468–483. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control. 2019, 8, 76. [Google Scholar] [CrossRef]
- Nguyen, K.T.; Piastro, K.; Gray, T.A.; Derbyshire, K.M. Mycobacterial Biofilms Facilitate Horizontal DNA Transfer between Strains of Mycobacterium smegmatis. J. Bacteriol. 2010, 192, 5134–5142. [Google Scholar] [CrossRef]
- Santos, C.L.; Correia-Neves, M.; Moradas-Ferreira, P.; Mendes, M.V. A Walk into the LuxR Regulators of Actinobacteria: Phylogenomic Distribution and Functional Diversity. PLoS ONE 2012, 7, e46758. [Google Scholar] [CrossRef]
- Polkade, A.V.; Mantri, S.S.; Patwekar, U.J.; Jangid, K. Quorum Sensing: An Under-Explored Phenomenon in the Phylum Actinobacteria. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef][Green Version]
- Banaiee, N.; Jacobs, W.R.; Ernst, J.D. Regulation of Mycobacterium tuberculosis whiB3 in the Mouse Lung and Macrophages. Infect. Immun. 2006, 74, 6449–6457. [Google Scholar] [CrossRef]
- Gupta, K.R.; Baloni, P.; Indi, S.S.; Chatterji, D. Regulation of Growth, Cell Shape, Cell Division, and Gene Expression by Second Messengers (p)ppGpp and Cyclic Di-GMP in Mycobacterium smegmatis. J. Bacteriol. 2016, 198, 1414–1422. [Google Scholar] [CrossRef]
- Norton, C.D.; LeChevallier, M.W.; Falkinham, J.O. Survival of Mycobacterium avium in a model distribution system. Water Res. 2004, 38, 1457–1466. [Google Scholar] [CrossRef]
- Carson, L.A.; Petersen, N.J.; Favero, M.S.; Aguero, S.M. Growth characteristics of atypical mycobacteria in water and their comparative resistance to disinfectants. Appl. Environ. Microbiol. 1978, 36, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ogawa, M.; Fukuda, K.; Miyamoto, H.; Taniguchi, H. Isolation and identification of mycobacteria from soils at an illegal dumping site and landfills in Japan. Microbiol. Immunol. 2006, 50, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Zhang, T.; Li, B.; Wang, Z.; Ju, F.; Liang, Y.-t. Mycobacterial species and their contribution to cholesterol degradation in wastewater treatment plants. Sci. Rep. 2019, 9, 836. [Google Scholar] [CrossRef] [PubMed]
- Wick, L.Y.; Pasche, N.; Bernasconi, S.M.; Pelz, O.; Harms, H. Characterization of Multiple-Substrate Utilization by Anthracene-Degrading Mycobacterium frederiksbergense LB501T. Appl. Environ. Microbiol. 2003, 69, 6133. [Google Scholar] [CrossRef]
- Wick, L.Y.; Pelz, O.; Bernasconi, S.M.; Andersen, N.; Harms, H. Influence of the growth substrate on ester-linked phospho- and glycolipid fatty acids of PAH-degrading Mycobacterium sp. LB501T. Environ. Microbiol. 2003, 5, 672–680. [Google Scholar] [CrossRef]
- Wick, L.; de Munain, A.; Springael, D.; Harms, H. Responses of Mycobacterium sp. LB501T to the low bioavailability of solid anthracene. Appl. Microbiol. Biotechnol. 2002, 58, 378–385. [Google Scholar] [CrossRef]
- Heitkamp, M.A.; Cerniglia, C.E. Mineralization of polycyclic aromatic hydrocarbons by a bacterium isolated from sediment below an oil field. Appl. Environ. Microbiol. 1988, 54, 1612–1614. [Google Scholar] [CrossRef]
- Khan, A.A.; Wang, R.F.; Cao, W.W.; Doerge, D.R.; Wennerstrom, D.; Cerniglia, C.E. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl Environ. Microbiol. 2001, 67, 3577–3585. [Google Scholar] [CrossRef]
- Moody, J.D.; Freeman, J.P.; Doerge, D.R.; Cerniglia, C.E. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 2001, 67, 1476–1483. [Google Scholar] [CrossRef]
- Moody, J.D.; Freeman, J.P.; Fu, P.P.; Cerniglia, C.E. Degradation of Benzo[a]pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 2004, 70, 340. [Google Scholar] [CrossRef]
- Kim, S.J.; Kweon, O.; Freeman, J.P.; Jones, R.C.; Adjei, M.D.; Jhoo, J.W.; Edmondson, R.D.; Cerniglia, C.E. Molecular cloning and expression of genes encoding a novel dioxygenase involved in low- and high-molecular-weight polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Appl Environ. Microbiol. 2006, 72, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Kweon, O.; Jones, R.C.; Freeman, J.P.; Edmondson, R.D.; Cerniglia, C.E. Complete and Integrated Pyrene Degradation Pathway in Mycobacterium vanbaalenii PYR-1 Based on Systems Biology. J. Bacteriol. 2007, 189, 464. [Google Scholar] [CrossRef]
- Kweon, O.; Kim, S.-J.; Jones, R.C.; Freeman, J.P.; Adjei, M.D.; Edmondson, R.D.; Cerniglia, C.E. A Polyomic Approach to Elucidate the Fluoranthene-Degradative Pathway in Mycobacterium vanbaalenii PYR-1. J. Bacteriol. 2007, 189, 4635. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kweon, O.; Jones, R.C.; Edmondson, R.D.; Cerniglia, C.E. Genomic analysis of polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Biodegradation 2008, 19, 859–881. [Google Scholar] [CrossRef]
- Kweon, O.; Kim, S.-J.; Holland, R.D.; Chen, H.; Kim, D.-W.; Gao, Y.; Yu, L.-R.; Baek, S.; Baek, D.-H.; Ahn, H.; et al. Polycyclic Aromatic Hydrocarbon Metabolic Network in Mycobacterium vanbaalenii PYR-1. J. Bacteriol. 2011, 193, 4326. [Google Scholar] [CrossRef] [PubMed]
- Brezna, B.; Kweon, O.; Stingley, R.L.; Freeman, J.P.; Khan, A.A.; Polek, B.; Jones, R.C.; Cerniglia, C.E. Molecular characterization of cytochrome P450 genes in the polycyclic aromatic hydrocarbon degrading Mycobacterium vanbaalenii PYR-1. Appl. Microbiol. Biotechnol. 2006, 71, 522–532. [Google Scholar] [CrossRef]
- Capyk, J.K.; Kalscheuer, R.; Stewart, G.R.; Liu, J.; Kwon, H.; Zhao, R.; Okamoto, S.; Jacobs, W.R.; Eltis, L.D.; Mohn, W.W. Mycobacterial Cytochrome P450 125 (Cyp125) Catalyzes the Terminal Hydroxylation of C27 Steroids. J. Biol. Chem. 2009, 284, 35534–35542. [Google Scholar] [CrossRef]
- Funhoff, E.G.; Bauer, U.; García-Rubio, I.; Witholt, B.; van Beilen, J.B. CYP153A6, a Soluble P450 Oxygenase Catalyzing Terminal-Alkane Hydroxylation. J. Bacteriol. 2006, 188, 5220–5227. [Google Scholar] [CrossRef]
- Poupin, P.; Truffaut, N.; Combourieu, B.; Besse, P.; Sancelme, M.; Veschambre, H.; Delort, A.M. Degradation of morpholine by an environmental Mycobacterium strain involves a cytochrome P-450. Appl. Environ. Microbiol. 1998, 64, 159–165. [Google Scholar] [CrossRef]
- Erardi, F.X.; Failla, M.L.; Falkinham, J.O., 3rd. Accumulation and transport of cadmium by tolerant and susceptible strains of Mycobacterium scrofulaceum. Antimicrob. Agents Chemother. 1989, 33, 350–355. [Google Scholar] [CrossRef]
- Prammananan, T.; Sander, P.; Brown, B.A.; Frischkorn, K.; Onyi, G.O.; Zhang, Y.; Böttger, E.C.; Wallace, R.J., Jr. A Single 16S Ribosomal RNA Substitution Is Responsible for Resistance to Amikacin and Other 2-Deoxystreptamine Aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 1998, 177, 1573–1581. [Google Scholar] [CrossRef] [PubMed]
- Rominski, A.; Schulthess, B.; Müller, D.M.; Keller, P.M.; Sander, P. Effect of β-lactamase production and β-lactam instability on MIC testing results for Mycobacterium abscessus. J. Antimicrob. Chemother. 2017, 72, 3070–3078. [Google Scholar] [CrossRef] [PubMed]
- Rominski, A.; Selchow, P.; Becker, K.; Brülle, J.K.; Dal Molin, M.; Sander, P. Elucidation of Mycobacterium abscessus aminoglycoside and capreomycin resistance by targeted deletion of three putative resistance genes. J. Antimicrob. Chemother. 2017, 72, 2191–2200. [Google Scholar] [CrossRef] [PubMed]
- Miranda-CasoLuengo, A.A.; Staunton, P.M.; Dinan, A.M.; Lohan, A.J.; Loftus, B.J. Functional characterization of the Mycobacterium abscessus genome coupled with condition specific transcriptomics reveals conserved molecular strategies for host adaptation and persistence. BMC Genom. 2016, 17, 553. [Google Scholar] [CrossRef]
- Walker, J.; Moore, G.; Collins, S.; Parks, S.; Garvey, M.I.; Lamagni, T.; Smith, G.; Dawkin, L.; Goldenberg, S.; Chand, M. Microbiological problems and biofilms associated with Mycobacterium chimaera in heater–cooler units used for cardiopulmonary bypass. J. Hosp. Infect. 2017, 96, 209–220. [Google Scholar] [CrossRef]
- Mougari, F.; Bouziane, F.; Crockett, F.; Nessar, R.; Chau, F.; Veziris, N.; Sapriel, G.; Raskine, L.; Cambau, E. Selection of Resistance to Clarithromycin in Mycobacterium abscessus Subspecies. Antimicrob. Agents Chemother. 2017, 61, e00943-16. [Google Scholar] [CrossRef]
- Halstrom, S.; Price, P.; Thomson, R. Environmental mycobacteria as a cause of human infection. Int. J. Mycobacteriol. 2015, 4, 81–91. [Google Scholar] [CrossRef]
- Wu, M.-L.; Aziz, D.B.; Dartois, V.; Dick, T. NTM drug discovery: Status, gaps and the way forward. Drug Discov. Today 2018, 23, 1502–1519. [Google Scholar] [CrossRef]
- Rindi, L. Efflux Pump Inhibitors Against Nontuberculous Mycobacteria. Int. J. Mol. Sci. 2020, 21, 4191. [Google Scholar] [CrossRef]
- Schmalstieg, A.M.; Srivastava, S.; Belkaya, S.; Deshpande, D.; Meek, C.; Leff, R.; van Oers, N.S.C.; Gumbo, T. The Antibiotic Resistance Arrow of Time: Efflux Pump Induction Is a General First Step in the Evolution of Mycobacterial Drug Resistance. Antimicrob. Agents Chemother. 2012, 56, 4806–4815. [Google Scholar] [CrossRef]
- Vianna, J.S.; Machado, D.; Ramis, I.B.; Silva, F.P.; Bierhals, D.V.; Abril, M.A.; von Groll, A.; Ramos, D.F.; Lourenço, M.C.S.; Viveiros, M.; et al. The Contribution of Efflux Pumps in Mycobacterium abscessus Complex Resistance to Clarithromycin. Antibiotics 2019, 8, 153. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, A.V.; Richard, M.; Roquet-Banères, F.; Viljoen, A.; Kremer, L. The TetR Family Transcription Factor MAB_2299c Regulates the Expression of Two Distinct MmpS-MmpL Efflux Pumps Involved in Cross-Resistance to Clofazimine and Bedaquiline in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2019, 63, e01000-19. [Google Scholar] [CrossRef] [PubMed]
- Madsen, C.T.; Jakobsen, L.; Douthwaite, S. Mycobacterium smegmatis Erm(38) Is a Reluctant Dimethyltransferase. Antimicrob. Agents Chemother. 2005, 49, 3803–3809. [Google Scholar] [CrossRef] [PubMed]
- Nash, K.A.; Zhang, Y.; Brown-Elliott, B.A.; Wallace, R.J., Jr. Molecular basis of intrinsic macrolide resistance in clinical isolates of Mycobacterium fortuitum. J. Antimicrob. Chemother. 2005, 55, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Nash, K.A.; Andini, N.; Zhang, Y.; Brown-Elliott, B.A.; Wallace, R.J. Intrinsic Macrolide Resistance in Rapidly Growing Mycobacteria. Antimicrob. Agents Chemother. 2006, 50, 3476–3478. [Google Scholar] [CrossRef]
- Nash, K.A.; Brown-Elliott, B.A.; Wallace, R.J. A Novel Gene, erm(41), Confers Inducible Macrolide Resistance to Clinical Isolates of Mycobacterium abscessus but Is Absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 2009, 53, 1367–1376. [Google Scholar] [CrossRef]
- Roberts, M.C. Resistance to macrolide, lincosamide, streptogramin, ketolide, and oxazolidinone antibiotics. Mol. Biotechnol. 2004, 28, 47. [Google Scholar] [CrossRef]
- Brown-Elliott, B.A.; Vasireddy, S.; Vasireddy, R.; Iakhiaeva, E.; Howard, S.T.; Nash, K.; Parodi, N.; Strong, A.; Gee, M.; Smith, T.; et al. Utility of Sequencing the erm(41) Gene in Isolates of Mycobacterium abscessus subsp. abscessus with Low and Intermediate Clarithromycin MICs. J. Clin. Microbiol. 2015, 53, 1211–1215. [Google Scholar] [CrossRef]
- Lee, S.H.; Park, Y.K.; Kim, C.K.; Kim, H.J. Isolation and characterizations of clarithromycin-resistant Mycobacterium avium clinical isolates. J. Clin. Lab. Anal. 2011, 25, 33–36. [Google Scholar] [CrossRef]
- Meier, A.; Kirschner, P.; Springer, B.; Steingrube, V.A.; Brown, B.A.; Wallace, R.J.; Böttger, E.C. Identification of mutations in 23S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob. Agents Chemother. 1994, 38, 381–384. [Google Scholar] [CrossRef]
- Wallace, R.J.; Meier, A.; Brown, B.A.; Zhang, Y.; Sander, P.; Onyi, G.O.; Böttger, E.C. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob. Agents Chemother. 1996, 40, 1676–1681. [Google Scholar] [CrossRef] [PubMed]
- Lety, M.A.; Nair, S.; Berche, P.; Escuyer, V. A single point mutation in the embB gene is responsible for resistance to ethambutol in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 1997, 41, 2629–2633. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.; McDonnell, G. Relationship between mycobacteria and amoebae: Ecological and epidemiological concerns. Lett. Appl. Microbiol. 2007, 45, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M. Looking in amoebae as a source of mycobacteria. Microb. Pathog. 2014, 77, 119–124. [Google Scholar] [CrossRef]
- Salah, I.B.; Ghigo, E.; Drancourt, M. Free-living amoebae, a training field for macrophage resistance of mycobacteria. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2009, 15, 894–905. [Google Scholar] [CrossRef]
- Lamrabet, O.; Merhej, V.; Pontarotti, P.; Raoult, D.; Drancourt, M. The genealogic tree of mycobacteria reveals a long-standing sympatric life into free-living protozoa. PLoS ONE 2012, 7, e34754. [Google Scholar] [CrossRef]
- Andersson, A.; Ahlinder, J.; Mathisen, P.; Hägglund, M.; Bäckman, S.; Nilsson, E.; Sjödin, A.; Thelaus, J. Predators and nutrient availability favor protozoa-resisting bacteria in aquatic systems. Sci. Rep. 2018, 8, 8415. [Google Scholar] [CrossRef]
- Greub, G.; Raoult, D. Microorganisms Resistant to Free-Living Amoebae. Clin. Microbiol. Rev. 2004, 17, 413–433. [Google Scholar] [CrossRef]
- Danelishvili, L.; Wu, M.; Stang, B.; Harriff, M.; Cirillo, S.; Cirillo, J.; Bildfell, R.; Arbogast, B.; Bermudez, L.E. Identification of Mycobacterium avium pathogenicity island important for macrophage and amoeba infection. Proc. Natl. Acad. Sci. USA 2007, 104, 11038–11043. [Google Scholar] [CrossRef]
- Tenant, R.; Bermudez, L.E. Mycobacterium avium Genes Upregulated Upon Infection of Acanthamoeba castellanii Demonstrate a Common Response to the Intracellular Environment. Curr. Microbiol. 2006, 52, 128–133. [Google Scholar] [CrossRef]
- Vij, R.; Danchik, C.; Crawford, C.; Dragotakes, Q.; Casadevall, A. Variation in Cell Surface Hydrophobicity Strains Influences Interactions with Amoebas. mSphere 2020, 5, e00310-20. [Google Scholar] [CrossRef] [PubMed]
- Alibaud, L.; Pawelczyk, J.; Gannoun-Zaki, L.; Singh, V.K.; Rombouts, Y.; Drancourt, M.; Dziadek, J.; Guerardel, Y.; Kremer, L. Increased phagocytosis of Mycobacterium marinum mutants defective in lipooligosaccharide production: A structure-activity relationship study. J. Biol. Chem. 2013. [Google Scholar] [CrossRef]
- Koliwer-Brandl, H.; Knobloch, P.; Barisch, C.; Welin, A.; Hanna, N.; Soldati, T.; Hilbi, H. Distinct Mycobacterium marinum phosphatases determine pathogen vacuole phosphoinositide pattern, phagosome maturation, and escape to the cytosol. Cell. Microbiol. 2019, 21, e13008. [Google Scholar] [CrossRef] [PubMed]
- Laencina, L.; Dubois, V.; Le Moigne, V.; Viljoen, A.; Majlessi, L.; Pritchard, J.; Bernut, A.; Piel, L.; Roux, A.-L.; Gaillard, J.-L.; et al. Identification of genes required for Mycobacterium abscessus growth in vivo with a prominent role of the ESX-4 locus. Proc. Natl. Acad. Sci. USA 2018, 115, E1002–E1011. [Google Scholar] [CrossRef] [PubMed]
- Dubois, V.; Pawlik, A.; Bories, A.; Le Moigne, V.; Sismeiro, O.; Legendre, R.; Varet, H.; Rodríguez-Ordóñez, M.d.P.; Gaillard, J.-L.; Coppée, J.-Y.; et al. Mycobacterium abscessus virulence traits unraveled by transcriptomic profiling in amoeba and macrophages. PLoS Pathog. 2019, 15, e1008069. [Google Scholar] [CrossRef] [PubMed]
- Bakala N’ Goma, J.C.; Le Moigne, V.; Soismier, N.; Laencina, L.; Le Chevalier, F.; Roux, A.-L.; Poncin, I.; Serveau–Avesque, C.; Rottman, M.; Gaillard, J.-L.; et al. Mycobacterium abscessus Phospholipase C expression is induced during co-culture within amoeba and enhances M. abscessus virulence in mice. Infect. Immun. 2014. [Google Scholar] [CrossRef]
- Strahl, E.D.; Gillaspy, G.E.; Falkinham, J.O., 3rd. Fluorescent acid-fast microscopy for measuring phagocytosis of Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum by Tetrahymena pyriformis and their intracellular growth. Appl Environ. Microbiol. 2001, 67, 4432–4439. [Google Scholar] [CrossRef][Green Version]
- Biet, F.; Boschiroli, M.L. Non-tuberculous mycobacterial infections of veterinary relevance. Res. Vet. Sci. 2014, 97, S69–S77. [Google Scholar] [CrossRef]
- Torvinen, E.; Meklin, T.; Torkko, P.; Suomalainen, S.; Reiman, M.; Katila, M.-L.; Paulin, L.; Nevalainen, A. Mycobacteria and fungi in moisture-damaged building materials. Appl. Environ. Microbiol. 2006, 72, 6822–6824. [Google Scholar] [CrossRef]
- Eaton, T.; Falkinham, J.O., 3rd; von Reyn, C.F. Recovery of Mycobacterium avium from cigarettes. J. Clin. Microbiol. 1995, 33, 2757–2758. [Google Scholar] [CrossRef]
- Shelton, B.G.; Flanders, W.D.; Morris, G.K. Mycobacterium sp. as a possible cause of hypersensitivity pneumonitis in machine workers. Emerg. Infect. Dis. 1999, 5, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, I.A.; Molina, E.; Tello, M.; Elguezabal, N.; Juste, R.A.; Garrido, J.M. Detection of Mycobacteria by Culture and DNA-Based Methods in Animal-Derived Food Products Purchased at Spanish Supermarkets. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Sun, W.; Zhang, F.; He, L.; Loganathan, K. Actinomycetes from the South China Sea sponges: Isolation, diversity and potential for aromatic polyketides discovery. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Kuang, W.; Li, J.; Zhang, S.; Long, L. Diversity and distribution of Actinobacteria associated with reef coral Porites lutea. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Cook, J.L. Nontuberculous mycobacteria: Opportunistic environmental pathogens for predisposed hosts. Br. Med. Bull. 2010, 96, 45–59. [Google Scholar] [CrossRef]
- Walsh, C.M.; Gebert, M.J.; Delgado-Baquerizo, M.; Maestre, F.T.; Fierer, N. A Global Survey of Mycobacterial Diversity in Soil. Appl. Environ. Microbiol. 2019, 85, e01180-19. [Google Scholar] [CrossRef]
- Santos, R.; Oliveira, F.; Fernandes, J.; Gonçalves, S.; Macieira, F.; Cadete, M. Detection and identification of mycobacteria in the Lisbon water distribution system. Water Sci. Technol. 2005, 52, 177–180. [Google Scholar] [CrossRef]
- Torvinen, E.; Suomalainen, S.; Lehtola, M.J.; Miettinen, I.T.; Zacheus, O.; Paulin, L.; Katila, M.-L.; Martikainen, P.J. Mycobacteria in Water and Loose Deposits of Drinking Water Distribution Systems in Finland. Appl. Environ. Microbiol. 2004, 70, 1973–1981. [Google Scholar] [CrossRef]
- Hoefsloot, W.; van Ingen, J.; Andrejak, C.; Angeby, K.; Bauriaud, R.; Bemer, P.; Beylis, N.; Boeree, M.J.; Cacho, J.; Chihota, V.; et al. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: An NTM-NET collaborative study. Eur. Respir. J. 2013, 42, 1604–1613. [Google Scholar] [CrossRef]
- Jacobs, J.M.; Stine, C.B.; Baya, A.M.; Kent, M.L. A review of mycobacteriosis in marine fish. J. Fish. Dis. 2009, 32, 119–130. [Google Scholar] [CrossRef]
- Thomson, R.; Tolson, C.; Sidjabat, H.; Huygens, F.; Hargreaves, M. Mycobacterium abscessus isolated from municipal water—A potential source of human infection. BMC Infect. Dis. 2013, 13, 241. [Google Scholar] [CrossRef] [PubMed]
- Lumb, R.; Stapledon, R.; Scroop, A.; Bond, P.; Cunliffe, D.; Goodwin, A.; Doyle, R.; Bastian, I. Investigation of spa pools associated with lung disorders caused by Mycobacterium avium complex in immunocompetent adults. Appl. Environ. Microbiol. 2004, 70, 4906–4910. [Google Scholar] [CrossRef] [PubMed]
- Gira, A.K.; Reisenauer, A.H.; Hammock, L.; Nadiminti, U.; Macy, J.T.; Reeves, A.; Burnett, C.; Yakrus, M.A.; Toney, S.; Jensen, B.J.; et al. Furunculosis due to Mycobacterium mageritense associated with footbaths at a nail salon. J. Clin. Microbiol. 2004, 42, 1813–1817. [Google Scholar] [CrossRef] [PubMed]
- Thomson, R.; Tolson, C.; Carter, R.; Coulter, C.; Huygens, F.; Hargreaves, M. Isolation of nontuberculous mycobacteria (NTM) from household water and shower aerosols in patients with pulmonary disease caused by NTM. J. Clin. Microbiol. 2013, 51, 3006–3011. [Google Scholar] [CrossRef]
- Cooksey, R.C.; Jhung, M.A.; Yakrus, M.A.; Butler, W.R.; Adékambi, T.; Morlock, G.P.; Williams, M.; Shams, A.M.; Jensen, B.J.; Morey, R.E.; et al. Multiphasic Approach Reveals Genetic Diversity of Environmental and Patient Isolates of Mycobacterium mucogenicum and Mycobacterium phocaicum Associated with an Outbreak of Bacteremias at a Texas Hospital. Appl. Environ. Microbiol. 2008, 74, 2480–2487. [Google Scholar] [CrossRef]
- Chang, C.-T.; Wang, L.-Y.; Liao, C.-Y.; Huang, S.-P. Identification of nontuberculous mycobacteria existing in tap water by PCR-restriction fragment length polymorphism. Appl. Environ. Microbiol. 2002, 68, 3159–3161. [Google Scholar] [CrossRef][Green Version]
- Iivanainen, E.K.; Martikainen, P.J.; Väänänen, P.K.; Katila, M.L. Environmental factors affecting the occurrence of mycobacteria in brook waters. Appl. Environ. Microbiol. 1993, 59, 398–404. [Google Scholar] [CrossRef]
- Adrados, B.; Julián, E.; Codony, F.; Torrents, E.; Luquin, M.; Morató, J. Prevalence and Concentration of Non-tuberculous Mycobacteria in Cooling Towers by Means of Quantitative PCR: A Prospective Study. Curr. Microbiol. 2011, 62, 313–319. [Google Scholar] [CrossRef]
- Amha, Y.M.; Anwar, M.Z.; Kumaraswamy, R.; Henschel, A.; Ahmad, F. Mycobacteria in Municipal Wastewater Treatment and Reuse: Microbial Diversity for Screening the Occurrence of Clinically and Environmentally Relevant Species in Arid Regions. Environ. Sci. Technol. 2017, 51, 3048–3056. [Google Scholar] [CrossRef]
- Pryor, M.; Springthorpe, S.; Riffard, S.; Brooks, T.; Huo, Y.; Davis, G.; Sattar, S.A. Investigation of opportunistic pathogens in municipal drinking water under different supply and treatment regimes. Water Sci. Technol. 2004, 50, 83–90. [Google Scholar] [CrossRef]
- Parker, B.C.; Ford, M.A.; Gruft, H.; Falkinham, J.O., 3rd. Epidemiology of infection by nontuberculous mycobacteria. IV. Preferential aerosolization of Mycobacterium intracellulare from natural waters. Am. Rev. Respir. Dis. 1983, 128, 652–656. [Google Scholar] [CrossRef]
- Falkinham, I. JO Surrounded by mycobacteria: Nontuberculous mycobacteria in the human environment. J. Appl. Microbiol. 2009, 107, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Dailloux, M.; Laurain, C.; Weber, M.; Hartemann, P. Water and nontuberculous mycobacteria. Water Res. 1999, 33, 2219–2228. [Google Scholar] [CrossRef]
- Vaerewijck, M.J.M.; Huys, G.; Palomino, J.C.; Swings, J.; Portaels, F. Mycobacteria in drinking water distribution systems: Ecology and significance for human health. FEMS Microbiol. Rev. 2005, 29, 911–934. [Google Scholar] [CrossRef] [PubMed]
- Bohrerova, Z.; Linden, K.G. Ultraviolet and Chlorine Disinfection of Mycobacterium in Wastewater: Effect of Aggregation. Water Environ. Res. 2006, 78, 565–571. [Google Scholar] [CrossRef]
- Radomski, N.; Betelli, L.; Moilleron, R.; Haenn, S.; Moulin, L.; Cambau, E.; Rocher, V.; Gonçalves, A.; Lucas, F.S. Mycobacterium Behavior in Wastewater Treatment Plant, A Bacterial Model Distinct From Escherichia coli and Enterococci. Environ. Sci. Technol. 2011, 45, 5380–5386. [Google Scholar] [CrossRef] [PubMed]
- Le Dantec, C.; Duguet, J.-P.; Montiel, A.; Dumoutier, N.; Dubrou, S.; Vincent, V. Occurrence of Mycobacteria in Water Treatment Lines and in Water Distribution Systems. Appl. Environ. Microbiol. 2002, 68, 5318–5325. [Google Scholar] [CrossRef]
- Jacobs, J.; Rhodes, M.; Sturgis, B.; Wood, B. Influence of Environmental Gradients on the Abundance and Distribution of Mycobacterium spp. in a Coastal Lagoon Estuary. Appl. Environ. Microbiol. 2009, 75, 7378–7384. [Google Scholar] [CrossRef]
- De Groote, M.A.; Pace, N.R.; Fulton, K.; Falkinham, J.O., 3rd. Relationships between Mycobacterium isolates from patients with pulmonary mycobacterial infection and potting soils. Appl. Environ. Microbiol. 2006, 72, 7602–7606. [Google Scholar] [CrossRef]
- Iivanainen, E.K.; Martikainen, P.J.; Räisänen, M.L.; Katila, M.-L. Mycobacteria in boreal coniferous forest soils. FEMS Microbiol. Ecol. 1997, 23, 325–332. [Google Scholar] [CrossRef]
- Thorel, M.-F.; Falkinham, J.O., III; Moreau, R.G. Environmental mycobacteria from alpine and subalpine habitats. FEMS Microbiol. Ecol. 2004, 49, 343–347. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kankya, C.; Muwonge, A.; Djønne, B.; Munyeme, M.; Opuda-Asibo, J.; Skjerve, E.; Oloya, J.; Edvardsen, V.; Johansen, T.B. Isolation of non-tuberculous mycobacteria from pastoral ecosystems of Uganda: Public Health significance. BMC Public Health 2011, 11, 320. [Google Scholar] [CrossRef] [PubMed]
- Davarpanah, M.; Azadi, D.; Shojaei, H. Prevalence and molecular characterization of non-tuberculous mycobacteria in hospital soil and dust of a developing country, Iran. Microbiology 2019, 165, 1306–1314. [Google Scholar] [CrossRef] [PubMed]
- Vacca, D.J.; Bleam, W.F.; Hickey, W.J. Isolation of soil bacteria adapted to degrade humic acid-sorbed phenanthrene. Appl. Environ. Microbiol. 2005, 71, 3797–3805. [Google Scholar] [CrossRef] [PubMed]
- Kopecky, J.; Kyselkova, M.; Omelka, M.; Cermak, L.; Novotna, J.; Grundmann, G.; Moënne-Loccoz, Y.; Sagova-Mareckova, M. Environmental mycobacteria closely related to the pathogenic species evidenced in an acidic forest wetland. Soil Biol. Biochem. 2011, 43, 697–700. [Google Scholar] [CrossRef]
- Norby, B.; Fosgate, G.T.; Manning, E.J.; Collins, M.T.; Roussel, A.J. Environmental mycobacteria in soil and water on beef ranches: Association between presence of cultivable mycobacteria and soil and water physicochemical characteristics. Vet. Microbiol 2007, 124, 153–159. [Google Scholar] [CrossRef]
- Camper, A.K. Involvement of humic substances in regrowth. Int. J. Food Microbiol. 2004, 92, 355–364. [Google Scholar] [CrossRef]
- Bolster, C.H.; Cook, K.L.; Haznedaroglu, B.Z.; Walker, S.L. The transport of Mycobacterium avium subsp. paratuberculosis through saturated aquifer materials. Lett. Appl. Microbiol. 2009, 48, 307–312. [Google Scholar] [CrossRef]
- Bouam, A.; Armstrong, N.; Levasseur, A.; Drancourt, M. Mycobacterium terramassiliense, Mycobacterium rhizamassiliense and Mycobacterium numidiamassiliense sp. nov., three new Mycobacterium simiae complex species cultured from plant roots. Sci. Rep. 2018, 8, 9309. [Google Scholar] [CrossRef]
- Tran, P.M.; Dahl, J.L. Mycobacterium sarraceniae sp. nov. and Mycobacterium helvum sp. nov., isolated from the pitcher plant Sarracenia purpurea. Int. J. Syst. Evol. Microbiol. 2016, 66, 4480–4485. [Google Scholar] [CrossRef]
- Kaevska, M.; Lvoncik, S.; Slana, I.; Kulich, P.; Kralik, P. Microscopy, Culture, and Quantitative Real-Time PCR Examination Confirm Internalization of Mycobacteria in Plants. Appl. Environ. Microbiol. 2014, 80, 3888–3894. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hruska, K.; Kaevska, M. Mycobacteria in water, soil, plants and air: A review. Vet. Med. 2012, 57, 623–679. [Google Scholar] [CrossRef]
- Yoder, S.; Argueta, C.; Holtzman, A.; Aronson, T.; Berlin, O.G.; Tomasek, P.; Glover, N.; Froman, S.; Stelma, G., Jr. PCR comparison of Mycobacterium avium isolates obtained from patients and foods. Appl. Environ. Microbiol. 1999, 65, 2650–2653. [Google Scholar] [CrossRef] [PubMed]
- Zwielehner, J.; Handschur, M.; Michaelsen, A.; Irez, S.; Demel, M.; Denner, E.B.; Haslberger, A.G. DGGE and real-time PCR analysis of lactic acid bacteria in bacterial communities of the phyllosphere of lettuce. Mol. Nutr. Food Res. 2008, 52, 614–623. [Google Scholar] [CrossRef]
- Taber, R.A.; Thielen, M.A.; Falkinham, J.O.; Smith, R.H. Mycobacterium scrofulaceum: A bacterial contaminant in plant tissue culture. Plant. Sci. 1991, 78, 231–236. [Google Scholar] [CrossRef]
- Marsollier, L.; Stinear, T.; Aubry, J.; Saint André, J.P.; Robert, R.; Legras, P.; Manceau, A.-L.; Audrain, C.; Bourdon, S.; Kouakou, H.; et al. Aquatic Plants Stimulate the Growth of and Biofilm Formation by Mycobacterium ulcerans in Axenic Culture and Harbor These Bacteria in the Environment. Appl. Environ. Microbiol. 2004, 70, 1097–1103. [Google Scholar] [CrossRef]
- Lahiri, A.; Kneisel, J.; Kloster, I.; Kamal, E.; Lewin, A. Abundance of Mycobacterium avium ssp. hominissuis in soil and dust in Germany–implications for the infection route. Lett. Appl. Microbiol. 2014, 59, 65–70. [Google Scholar] [CrossRef]
- Choi, S.; Choi, M. Isolation of Nontuberculous Mycobacteria (NTM) from Air Conditioner Dust. Korean J. Clin. Lab. Sci. 2017, 49, 435–438. [Google Scholar] [CrossRef]
- Torvinen, E.; Torkko, P.; Rintala, A.N.H. Real-time PCR detection of environmental mycobacteria in house dust. J. Microbiol. Methods 2010, 82, 78–84. [Google Scholar] [CrossRef]
- Leski, T.A.; Malanoski, A.P.; Gregory, M.J.; Lin, B.; Stenger, D.A. Application of a Broad-Range Resequencing Array for Detection of Pathogens in Desert Dust Samples from Kuwait and Iraq. Appl. Environ. Microbiol. 2011, 77, 4285–4292. [Google Scholar] [CrossRef]
- Mangione, E.J.; Huitt, G.; Lenaway, D.; Beebe, J.; Bailey, A.; Figoski, M.; Rau, M.P.; Albrecht, K.D.; Yakrus, M.A. Nontuberculous mycobacterial disease following hot tub exposure. Emerg. Infect. Dis. 2001, 7, 1039–1042. [Google Scholar] [CrossRef] [PubMed]
- Sarbu, S.; Aerts, J.; Flot, J.-F.; Spanning, R.; Baciu, C.; Ionescu, A.; Kis, B.-M.; Incze, R.; Sikó-Barabási, S.; Para, Z.; et al. Sulfur Cave (Romania), an extreme environment with microbial mats in a CO2-H2S/O2 gas chemocline dominated by mycobacteria. Int. J. Speleol. 2018, 47, 173–187. [Google Scholar] [CrossRef]
- Kusumi, A.; Li, X.S.; Katayama, Y. Mycobacteria Isolated from Angkor Monument Sandstones Grow Chemolithoautotrophically by Oxidizing Elemental Sulfur. Front. Microbiol. 2011, 2, 104. [Google Scholar] [CrossRef] [PubMed]
- Park, S.S.; Decicco, B.T. Autotrophic Growth with Hydrogen of Mycobacterium gordonae and Another Scotochromogenic Mycobacterium. Int. J. Syst. Evol. Microbiol. 1974, 24, 338–345. [Google Scholar] [CrossRef][Green Version]
- Park, S.W.; Hwang, E.H.; Park, H.; Kim, J.A.; Heo, J.; Lee, K.H.; Song, T.; Kim, E.; Ro, Y.T.; Kim, S.W.; et al. Growth of Mycobacteria on Carbon Monoxide and Methanol. J. Bacteriol. 2003, 185, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Schelle, M.W.; Bertozzi, C.R. Sulfate Metabolism in Mycobacteria. ChemBioChem 2006, 7, 1516–1524. [Google Scholar] [CrossRef]
- Bland, C.S.; Ireland, J.M.; Lozano, E.; Alvarez, M.E.; Primm, T.P. Mycobacterial Ecology of the Rio Grande. Appl. Environ. Microbiol. 2005, 71, 5719–5727. [Google Scholar] [CrossRef]
- Walker, J.J.; Spear, J.R.; Pace, N.R. Geobiology of a microbial endolithic community in the Yellowstone geothermal environment. Nature 2005, 434, 1011–1014. [Google Scholar] [CrossRef]
- Santos, R.; Fernandes, J.; Fernandes, N.; Oliveira, F.; Cadete, M. Mycobacterium parascrofulaceum in Acidic Hot Springs in Yellowstone National Park. Appl. Environ. Microbiol. 2007, 73, 5071–5073. [Google Scholar] [CrossRef][Green Version]
- Valverde, A.; Tuffin, M.; Cowan, D.A. Biogeography of bacterial communities in hot springs: A focus on the actinobacteria. Extremophiles 2012, 16, 669–679. [Google Scholar] [CrossRef]
- Pavlik, I.; Gersl, M.; Bartos, M.; Ulmann, V.; Kaucka, P.; Caha, J.; Unc, A.; Hubelova, D.; Konecny, O.; Modra, H. Nontuberculous mycobacteria in the environment of Hranice Abyss, the world’s deepest flooded cave (Hranice karst, Czech Republic). Environ. Sci. Pollut. Res. 2018, 25, 23712–23724. [Google Scholar] [CrossRef] [PubMed]
- Ikner, L.A.; Toomey, R.S.; Nolan, G.; Neilson, J.W.; Pryor, B.M.; Maier, R.M. Culturable Microbial Diversity and the Impact of Tourism in Kartchner Caverns, Arizona. Microb. Ecol. 2007, 53, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Barton, H.; Taylor, N.; Kreate, M.; Springer, A.; Oehrle, S.; Bertog, J. The Impact of Host Rock Geochemistry on Bacterial Community Structure in Oligotrophic Cave Environments. Int. J. Speleol. 2007, 36. [Google Scholar] [CrossRef]
- De Mandal, S.; Panda, A.K.; Lalnunmawii, E.; Bisht, S.S.; Kumar, N.S. Illumina-based analysis of bacterial community in Khuangcherapuk cave of Mizoram, Northeast India. Genom. Data 2015, 5, 13–14. [Google Scholar] [CrossRef]
- Alarico, S.; Nunes-Costa, D.; Silva, A.; Costa, M.; Macedo-Ribeiro, S.; Empadinhas, N. A genuine mycobacterial thermophile: Mycobacterium hassiacum growth, survival and GpgS stability at near-pasteurization temperatures. Microbiology 2020, 166, 474–483. [Google Scholar] [CrossRef]
- Deinhardt-Emmer, S.; Höring, S.; Mura, C.; Hillemann, D.; Hermann, B.; Sachse, S.; Bohnert, J.; Löffler, B. First Time Isolation of Mycobacterium hassiacum From a Respiratory Sample. Clin. Med. Insights Circ. Respir. Pulm. Med. 2018, 12. [Google Scholar] [CrossRef]
- Gebert, M.J.; Delgado-Baquerizo, M.; Oliverio, A.M.; Webster, T.M.; Nichols, L.M.; Honda, J.R.; Chan, E.D.; Adjemian, J.; Dunn, R.R.; Fierer, N. Ecological Analyses of Mycobacteria in Showerhead Biofilms and Their Relevance to Human Health. mBio 2018, 9, e01614-18. [Google Scholar] [CrossRef]
- Campos-Gutiérrez, S.; Ramos-Real, M.J.; Abreu, R.; Jiménez, M.S.; Lecuona, M. Pseudo-outbreak of Mycobacterium fortuitum in a hospital bronchoscopy unit. Am. J. Infect. Control. 2019. [Google Scholar] [CrossRef]
- Griffith, D.E.; Aksamit, T.; Brown-Elliott, B.A.; Catanzaro, A.; Daley, C.; Gordin, F.; Holland, S.M.; Horsburgh, R.; Huitt, G.; Iademarco, M.F.; et al. An Official ATS/IDSA Statement: Diagnosis, Treatment, and Prevention of Nontuberculous Mycobacterial Diseases. Am. J. Respir. Crit. Care Med. 2007, 175, 367–416. [Google Scholar] [CrossRef]
- Weinstein, R.; Stamm, W. Pseudoepidemics in hospital. Lancet 1977, 310, 862–864. [Google Scholar] [CrossRef]
- Barbeau, J.; Buhler, T. Biofilms augment the number of free-living amoebae in dental unit waterlines. Res. Microbiol. 2001, 152, 753–760. [Google Scholar] [CrossRef]
- Falkinham, J.O. Current Epidemiologic Trends of the Nontuberculous Mycobacteria (NTM). Curr. Environ. Health Rep. 2016, 3, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Nishiuchi, Y.; Maekura, R.; Kitada, S.; Tamaru, A.; Taguri, T.; Kira, Y.; Hiraga, T.; Hirotani, A.; Yoshimura, K.; Miki, M.; et al. The recovery of Mycobacterium avium-intracellulare complex (MAC) from the residential bathrooms of patients with pulmonary MAC. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2007, 45, 347–351. [Google Scholar] [CrossRef]
- Fujita, K.; Ito, Y.; Hirai, T.; Maekawa, K.; Imai, S.; Tatsumi, S.; Niimi, A.; Iinuma, Y.; Ichiyama, S.; Mishima, M. Genetic relatedness of Mycobacterium avium-intracellulare complex isolates from patients with pulmonary MAC disease and their residential soils. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2013, 19, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Collier, D.N. Cutaneous infections from coastal and marine bacteria. Derm. Ther. 2002, 15, 1–9. [Google Scholar] [CrossRef]
- Jang, J.; Becq, J.; Gicquel, B.; Deschavanne, P.; Neyrolles, O. Horizontally acquired genomic islands in the tubercle bacilli. Trends Microbiol. 2008, 16, 303–308. [Google Scholar] [CrossRef]
- Ratnatunga, C.N.; Lutzky, V.P.; Kupz, A.; Doolan, D.L.; Reid, D.W.; Field, M.; Bell, S.C.; Thomson, R.M.; Miles, J.J. The Rise of Non-Tuberculosis Mycobacterial Lung Disease. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Lindeboom, J.A.; Prins, J.M.; Bruijnesteijn van Coppenraet, E.S.; Lindeboom, R.; Kuijper, E.J. Cervicofacial Lymphadenitis in Children Caused by Mycobacterium haemophilum. Clin. Infect. Dis. 2005, 41, 1569–1575. [Google Scholar] [CrossRef]
- Zenone, T.; Boibieux, A.; Tigaud, S.; Fredenucci, J.-F.; Vincent, V.; Chidiac, C.; Peyramond, D. Non-tuberculous Mycobacterial Tenosynovitis: A Review. Scand. J. Infect. Dis. 1999, 31, 221–228. [Google Scholar] [CrossRef]
- Franco-Paredes, C.; Marcos, L.A.; Henao-Martínez, A.F.; Rodríguez-Morales, A.J.; Villamil-Gómez, W.E.; Gotuzzo, E.; Bonifaz, A. Cutaneous Mycobacterial Infections. Clin. Microbiol. Rev. 2018, 32, e00069-18. [Google Scholar] [CrossRef]
- Forbes, B.A.; Hall, G.S.; Miller, M.B.; Novak, S.M.; Rowlinson, M.-C.; Salfinger, M.; Somoskövi, A.; Warshauer, D.M.; Wilson, M.L. Practice Guidelines for Clinical Microbiology Laboratories: Mycobacteria. Clin. Microbiol. Rev. 2018, 31, e00038-17. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Buruli Ulcer (Mycobacterium Ulcerans Infection) Fact Sheet; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Shin, J.-I.; Shin, S.J.; Shin, M.-K. Differential Genotyping of Mycobacterium avium Complex and Its Implications in Clinical and Environmental Epidemiology. Microorganisms 2020, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Koh, W.-J. Nontuberculous Mycobacteria—Overview. Microbiol Spectr. 2017, 5, TNMI7-0024-2016. [Google Scholar] [CrossRef]
- Velayati, A.; Rahideh, S.; Nezhad, Z.; Farnia, P.; Mirsaeidi, M. Nontuberculous mycobacteria in Middle East: Current situation and future challenges. Int. J. Mycobacteriol. 2015, 4, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.; Höller, C.; Jacobshagen, A.; Hamouda, O.; Abu Sin, M.; Monnet, D.L.; Plachouras, D.; Eckmanns, T. Contamination during production of heater-cooler units by Mycobacterium chimaera potential cause for invasive cardiovascular infections: Results of an outbreak investigation in Germany, April 2015 to February 2016. Euro Surveill. 2016, 21, 30215. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.C.; Chiang, L.; Elwood, K. Mycobacterium kansasii. Microbiol Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Hoefsloot, W.; Boeree, M.J.; van Ingen, J.; Bendien, S.; Magis, C.; de Lange, W.; Dekhuijzen, P.N.; van Soolingen, D. The rising incidence and clinical relevance of Mycobacterium malmoense: A review of the literature. Int. J. Tuberc. Lung Dis. Off. J. Int. Union Against Tuberc. Lung Dis. 2008, 12, 987–993. [Google Scholar]
- Sassi, M.; Drancourt, M. Genome analysis reveals three genomospecies in Mycobacterium abscessus. BMC Genom. 2014, 15, 359. [Google Scholar] [CrossRef]
- Chopra, S.; Matsuyama, K.; Hutson, C.; Madrid, P. Identification of antimicrobial activity among FDA-approved drugs for combating Mycobacterium abscessus and Mycobacterium chelonae. J. Antimicrob. Chemother. 2011, 66, 1533–1536. [Google Scholar] [CrossRef]
- Rüegg, E.; Cheretakis, A.; Modarressi, A.; Harbarth, S.; Pittet-Cuénod, B. Multisite Infection with Mycobacterium abscessus after Replacement of Breast Implants and Gluteal Lipofilling. Case Rep. Infect. Dis. 2015, 2015, 361340. [Google Scholar] [CrossRef]
- Bosio, S.; Leekha, S.; Gamb, S.I.; Wright, A.J.; Terrell, C.L.; Miller, D.V. Mycobacterium fortuitum prosthetic valve endocarditis: A case for the pathogenetic role of biofilms. Cardiovasc. Pathol. 2012, 21, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.M.; Grogono, D.M.; Greaves, D.; Foweraker, J.; Roddick, I.; Inns, T.; Reacher, M.; Haworth, C.S.; Curran, M.D.; Harris, S.R.; et al. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: A retrospective cohort study. Lancet 2013, 381, 1551–1560. [Google Scholar] [CrossRef]
- Hashish, E.; Merwad, A.; Elgaml, S.; Amer, A.; Kamal, H.; Elsadek, A.; Marei, A.; Sitohy, M. Mycobacterium marinum infection in fish and man: Epidemiology, pathophysiology and management; a review. Vet. Q. 2018, 38, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Borges, A.C.; Pereira, N.; Franco, M.; Vale, L.; Pereira, M.; Cunha, M.V.; Amaro, A.; Albuquerque, T.; Rebelo, M. Implementation of a Zebrafish Health Program in a Research Facility: A 4-Year Retrospective Study. Zebrafish 2016, 13, S-115-S-126. [Google Scholar] [CrossRef]
- Bernstein, C.N.; Blanchard, J.F.; Rawsthorne, P.; Collins, M.T. Population-based case control study of seroprevalence of Mycobacterium paratuberculosis in patients with Crohn’s disease and ulcerative colitis. J. Clin. Microbiol. 2004, 42, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Naser, S.A.; Ghobrial, G.; Romero, C.; Valentine, J.F. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn’s disease. Lancet (Lond. Engl.) 2004, 364, 1039–1044. [Google Scholar] [CrossRef]
- Cho, J.-K.; Choi, Y.M.; Lee, S.S.; Park, H.K.; Cha, R.R.; Kim, W.S.; Kim, J.J.; Lee, J.M.; Kim, H.J.; Ha, C.Y.; et al. Clinical features and outcomes of abdominal tuberculosis in southeastern Korea: 12 years of experience. BMC Infect. Dis. 2018, 18, 699. [Google Scholar] [CrossRef] [PubMed]
- Chongwe, G.; Michelo, C.; Kelly, P. Diagnostic yield of nontuberculous mycobacteria in patients booked for endoscopy at the University Teaching Hospital, Lusaka. BMC Res. Notes 2017, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Herz, U.; Gerhold, K.; Grüber, C.; Braun, A.; Wahn, U.; Renz, H.; Paul, K. BCG infection suppresses allergic sensitization and development of increased airway reactivity in an animal model. J. Allergy Clin. Immunol. 1998, 102, 867–874. [Google Scholar] [CrossRef]
- Hopfenspirger, M.T.; Agrawal, D.K. Airway Hyperresponsiveness, Late Allergic Response, and Eosinophilia Are Reversed with Mycobacterial Antigens in Ovalbumin-Presensitized Mice. J. Immunol. 2002, 168, 2516. [Google Scholar] [CrossRef]
- Huh, H.J.; Song, D.J.; Ki, C.S.; Lee, N.Y. Is Cross-reactivity with Nontuberculous Mycobacteria a Systematic Problem in the Xpert MTB/RIF Assay? Tuberc. Respir. Dis. 2019, 82, 88–89. [Google Scholar] [CrossRef]
- Latorre, I.; De Souza-Galvão, M.; Ruiz-Manzano, J.; Lacoma, A.; Prat, C.; Altet, N.; Ausina, V.; Domínguez, J. Evaluating the non-tuberculous mycobacteria effect in the tuberculosis infection diagnosis. Eur. Respir. J. 2010, 35, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Poyntz, H.C.; Stylianou, E.; Griffiths, K.L.; Marsay, L.; Checkley, A.M.; McShane, H. Non-tuberculous mycobacteria have diverse effects on BCG efficacy against Mycobacterium tuberculosis. Tuberculosis 2014, 94, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.T.; Byrd, T.F. The rapidly growing mycobacteria: Saprophytes and parasites. Microbes Infect. 2000, 2, 1845–1853. [Google Scholar] [CrossRef]
- Mansfield, K.G.; Lackner, A.A. Simian Immunodeficiency Virus–Inoculated Macaques Acquire Mycobacterium avium from Potable Water during AIDS. J. Infect. Dis. 1997, 175, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Pai, H.H.; Chen, W.C.; Peng, C.F. Isolation of non-tuberculous mycobacteria from hospital cockroaches (Periplaneta americana). J. Hosp. Infect. 2003, 53, 224–228. [Google Scholar] [CrossRef]
- Friend, S.C.E.; Russell, E.G. Mycobacterium intracellulare infection in a water monitor. J. Wildl. Dis. 1979, 15, 229–233. [Google Scholar] [CrossRef]
- Florou, M.; Leontides, L.; Kostoulas, P.; Billinis, C.; Sofia, M.; Kyriazakis, I.; Lykotrafitis, F. Isolation of Mycobacterium avium subspecies paratuberculosis from non-ruminant wildlife living in the sheds and on the pastures of Greek sheep and goats. Epidemiol. Infect. 2008, 136, 644–652. [Google Scholar] [CrossRef]
- Cunha, M.V.; Rosalino, L.M.; Leão, C.; Bandeira, V.; Fonseca, C.; Botelho, A.; Reis, A.C. Ecological drivers of Mycobacterium avium subsp. paratuberculosis detection in mongoose (Herpestes ichneumon) using IS900 as proxy. Sci. Rep. 2020, 10, 860. [Google Scholar] [CrossRef]
- Richardson, H.; Rhodes, G.; Henrys, P.; Sedda, L.; Weightman, A.J.; Pickup, R.W. Presence of Mycobacterium avium Subspecies paratuberculosis Monitored Over Varying Temporal and Spatial Scales in River Catchments: Persistent Routes for Human Exposure. Microorganisms 2019, 7, 136. [Google Scholar] [CrossRef]
- Brammer, D.W.; O’Rourke, C.M.; Heath, L.A.; Chrlsp, C.E.; Peter, G.K.; Hofing, G.L. Mycobacterium kansasii infection in squirrel monkeys (Saimiri sciureus sciureus). J. Med. Primatol. 1995, 24, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Hall, P.B.; Bender, L.C.; Garner, M.M. Mycobacteriosis in a black-tailed deer (Odocoileus hemionus columbianus) caused by Mycobacterium kansasii. J. Zoo Wildl. Med. Off. Publ. Am. Assoc. Zoo Vet. 2005, 36, 115–116. [Google Scholar] [CrossRef] [PubMed]
- Schafbuch, R.; Tinkler, S.; Lim, C.K.; Wolking, R.; Ramos-Vara, J. Disseminated mycobacteriosis caused by Mycobacterium kansasii in a pot-bellied pig. J. Vet. Diagn Investig. 2018, 30, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Murai, A.; Maruyama, S.; Nagata, M.; Yuki, M. Mastitis caused by Mycobacterium kansasii infection in a dog. Vet. Clin. Pathol. 2013, 42, 377–381. [Google Scholar] [CrossRef]
- Acosta, B.; Real, F.; Ferrer, O.; Deniz, S.; Poveda, B. Isolation of Mycobacterium kansasii from a tuberculin-positive goat. Vet. Rec. 1998, 142, 195–196. [Google Scholar] [CrossRef]
- Shipley, S.T.; Johnson, D.K.; Roodgar, M.; Smith, D.G.; Montgomery, C.A.; Lloyd, S.M.; Higgins, J.A.; Kriel, E.H.; Klein, H.J.; Porter, W.P.; et al. Mycobacterium kansasii Isolated from Tuberculinpositive Rhesus Macaques (Macaca mulatta) in the Absence of Disease. Comp. Med. 2017, 67, 368–375. [Google Scholar]
- Johnson, C.T.; Winkler, C.E.; Boughton, E.; Penfold, J.W. Mycobacterium kansasii infection in a llama. Vet. Rec. 1993, 133, 243–244. [Google Scholar] [CrossRef]
- Konuk, M.; Korcan, E.; Dülgerbaki, S.; Altindiş, M. Isolation and identification of Mycobacteria from raw milk samples in Afyonkarahisar district of Turkey. Int. J. Food Microbiol. 2007, 115, 343–347. [Google Scholar] [CrossRef]
- Manrique, W.G.; Pereira Figueiredo, M.A.; Charlie-Silva, I.; Antonio de Andrade Belo, M.; Dib, C.C. Spleen melanomacrophage centers response of Nile tilapia during Aeromanas hydrophila and Mycobacterium marinum infections. Fish. Shellfish Immunol. 2019, 95, 514–518. [Google Scholar] [CrossRef]
- dos Santos, N.M.; do Vale, A.; Sousa, M.J.; Silva, M.T. Mycobacterial infection in farmed turbot Scophthalmus maximus. Dis. Aquat. Org. 2002, 52, 87–91. [Google Scholar] [CrossRef]
- Brocklebank, J.; Raverty, S.; Robinson, J. Mycobacteriosis in Atlantic salmon farmed in British Columbia. Can. Vet. J. La Rev. Vet. Can. 2003, 44, 486–489. [Google Scholar]
- Gauthier, D.T.; Rhodes, M.W.; Vogelbein, W.K.; Kator, H.; Ottinger, C.A. Experimental mycobacteriosis in striped bass Morone saxatilis. Dis. Aquat. Org. 2003, 54, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Swaim, L.E.; Connolly, L.E.; Volkman, H.E.; Humbert, O.; Born, D.E.; Ramakrishnan, L. Mycobacterium marinum Infection of Adult Zebrafish Causes Caseating Granulomatous Tuberculosis and Is Moderated by Adaptive Immunity. Infect. Immun. 2006, 74, 6108. [Google Scholar] [CrossRef] [PubMed]
- Diamant, A.; Banet, A.; Ucko, M.; Colorni, A.; Knibb, W.; Kvitt, H. Mycobacteriosis in wild rabbitfish Siganus rivulatus associated with cage farming in the Gulf of Eilat, Red Sea. Dis. Aquat. Org. 2000, 39, 211–219. [Google Scholar] [CrossRef]
- Zhang, D.F.; Ji, C.; Zhang, X.J.; Li, T.T.; Li, A.H.; Gong, X.N. Mixed mycobacterial infections in farmed sturgeons. Aquac. Res. 2015, 46, 1914–1923. [Google Scholar] [CrossRef]
- Ferreira, R.; Fonseca Lde, S.; Afonso, A.M.; da Silva, M.G.; Saad, M.H.; Lilenbaum, W. A report of mycobacteriosis caused by Mycobacterium marinum in bullfrogs (Rana catesbeiana). Vet. J. 2006, 171, 177–180. [Google Scholar] [CrossRef]
- Haridy, M.; Tachikawa, Y.; Yoshida, S.; Tsuyuguchi, K.; Tomita, M.; Maeda, S.; Wada, T.; Ibi, K.; Sakai, H.; Yanai, T. Mycobacterium marinum Infection in Japanese Forest Green Tree Frogs (Rhacophorus arboreus). J. Comp. Pathol. 2014, 151, 277–289. [Google Scholar] [CrossRef]
- Tappe, J.P.; Weitzman, I.; Liu, S.; Dolensek, E.P.; Karp, D. Systemic Mycobacterium marinum infection in a European hedgehog. J. Am. Vet. Med Assoc. 1983, 183, 1280–1281. [Google Scholar]
- Sato, T.; Shibuya, H.; Ohba, S.; Nojiri, T.; Shirai, W. Mycobacteriosis in two captive Florida manatees (Trichechus manatus latirostris). J. Zoo Wildl. Med. Off. Publ. Am. Assoc. Zoo Vet. 2003, 34, 184–188. [Google Scholar] [CrossRef]
- St-Jean, G.; Gagnon, C.A.; Soualhine, H.; Tremblay, M.; Beaulieu, A.-A.; Sylvestre, D. Mycobacterium xenopi systemic infection in a domestic fiery-shouldered conure bird (Pyrrhura egregia). JMM Case Rep. 2018, 5, e005158. [Google Scholar] [CrossRef]
- Jarnagin, J.L.; Richards, W.D.; Muhm, R.L.; Ellis, E.M. The Isolation of Mycobacterium xenopi from Granulomatous Lesions in Swine. Am. Rev. Respir. Dis. 1971, 104, 763–765. [Google Scholar] [CrossRef] [PubMed]
- Davendralingam, N.; Davagnanam, I.; Stidworthy, M.F.; Baldrey, V.; Peters, L.M.; Stapleton, N. Transmission of Mycobacterium xenopi to a pet albino ferret (Mustela putorius furo) from a domestic aquarium. Vet. Rec. 2017, 181, 169. [Google Scholar] [CrossRef]
- Meeks, C.; Levy, J.K.; Crawford, P.C.; Farina, L.L.; Origgi, F.; Alleman, R.; Seddon, O.M.; Salcedo, A.; Hirsch, B.J.; Hirsch, S.G. Chronic Disseminated Mycobacterium xenopi Infection in a Cat with Idiopathic CD4+ T Lymphocytopenia. J. Vet. Intern. Med. 2008, 22, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Gortazar, C.; Torres, M.J.; Acevedo, P.; Aznar, J.; Negro, J.J.; de la Fuente, J.; Vicente, J. Fine-tuning the space, time, and host distribution of mycobacteria in wildlife. BMC Microbiol. 2011, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Ebani, V.V.; Fratini, F.; Bertelloni, F.; Cerri, D.; Tortoli, E. Isolation and identification of mycobacteria from captive reptiles. Res. Vet. Sci. 2012, 93, 1136–1138. [Google Scholar] [CrossRef] [PubMed]
- Gcebe, N.; Michel, A.L.; Hlokwe, T.M. Non-tuberculous Mycobacterium species causing mycobacteriosis in farmed aquatic animals of South Africa. BMC Microbiol. 2018, 18, 32. [Google Scholar] [CrossRef]
- Beran, V.; Matlova, L.; Dvorska, L.; Svastova, P.; Pavlik, I. Distribution of mycobacteria in clinically healthy ornamental fish and their aquarium environment. J. Fish. Dis 2006, 29, 383–393. [Google Scholar] [CrossRef]
- Hamid, M.E. Current Perspectives on Mycobacterium farcinogenes and Mycobacterium senegalense, the Causal Agents of Bovine Farcy. Vet. Med. Int. 2014, 2014, 247906. [Google Scholar] [CrossRef]
- Alfonso, R.; Romero, R.E.; Diaz, A.; Calderon, M.N.; Urdaneta, G.; Arce, J.; Patarroyo, M.E.; Patarroyo, M.A. Isolation and identification of mycobacteria in New World primates maintained in captivity. Vet. Microbiol. 2004, 98, 285–295. [Google Scholar] [CrossRef]
- Cvetnić, Z.; Spicic, S.; Benić, M.; Katalinic-Jankovic, V.; Pate, M.; Krt, B.; Ocepek, M. Mycobacterial infection of pigs in Croatia. Acta Vet. Hung. 2007, 55, 1–9. [Google Scholar] [CrossRef]
- Ueda, K.; Yamamoto, S.; Ohtsuka, Y.; Machii, K.; Yamazaki, S.; Saito, H. Naturally occurring Mycobacterium scrofulaceum infection in a laboratory mouse colony. Jikken Dobutsu. Exp. Anim. 1992, 41, 339–347. [Google Scholar]
- Fischer, O.; Mátlová, L.; Dvorská, L.; Švástová, P.; Bartl, J.; Melichárek, I.; Weston, R.T.; Pavlík, I. Diptera as vectors of mycobacterial infections in cattle and pigs. Med Vet. Entomol. 2001, 15, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, I.A.; Molina, E.; Elguezabal, N.; Pérez, V.; Garrido, J.M.; Juste, R.A. Detection of mycobacteria, Mycobacterium avium subspecies, and Mycobacterium tuberculosis complex by a novel tetraplex real-time PCR assay. J. Clin. Microbiol 2015, 53, 930–940. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; de Klerk-Lorist, L.M.; Henton, M.M.; Lane, E.; Parsons, S.; Gey van Pittius, N.C.; Kotze, A.; van Helden, P.D.; Tanner, M. Mixed infections of Corynebacterium pseudotuberculosis and non-tuberculous mycobacteria in South African antelopes presenting with tuberculosis-like lesions. Vet. Microbiol. 2011, 147, 340–345. [Google Scholar] [CrossRef]
- Sattar, A.; Zakaria, Z.; Abu, J.; Aziz, S.A.; Gabriel, R.-P. Evaluation of six decontamination procedures for isolation of Mycobacterium avium complex from avian feces. PLoS ONE 2018, 13, e0202034. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, D.; Perry, A.; Appleby, M.R.; Lee, D.; Davison, J.; Johnston, A.; Jones, A.L.; Nelson, A.; Bourke, S.J.; Thomas, M.F.; et al. An evaluation of methods for the isolation of nontuberculous mycobacteria from patients with cystic fibrosis, bronchiectasis and patients assessed for lung transplantation. BMC Pulm. Med. 2019, 19, 19. [Google Scholar] [CrossRef] [PubMed]
- Pinel, I.S.M.; Moed, D.H.; Vrouwenvelder, J.S.; van Loosdrecht, M.C.M. Bacterial community dynamics and disinfection impact in cooling water systems. Water Res. 2020, 172, 115505. [Google Scholar] [CrossRef]
- Hu, Y.; Yu, X.; Zhao, D.; Li, R.; Liu, Y.; Ge, M.; Hu, H. Isolation of nontuberculous mycobacteria from soil using Middlebrook 7H10 agar with increased malachite green concentration. AMB Express 2017, 7, 69. [Google Scholar] [CrossRef]
- Müller, S.; Nebe-von-Caron, G. Functional single-cell analyses: Flow cytometry and cell sorting of microbial populations and communities. FEMS Microbiol. Rev. 2010, 34, 554–587. [Google Scholar] [CrossRef]
- Bhalla, G.S.; Sarao, M.S.; Kalra, D.; Bandyopadhyay, K.; John, A.R. Methods of phenotypic identification of non-tuberculous mycobacteria. Pr. Lab. Med. 2018, 12, e00107. [Google Scholar] [CrossRef]
- Khosravi, A.D.; Hashemzadeh, M.; Hashemi Shahraki, A.; Teimoori, A. Differential Identification of Mycobacterial Species Using High-Resolution Melting Analysis. Front. Microbiol 2017, 8, 2045. [Google Scholar] [CrossRef] [PubMed]
- Ceyssens, P.J.; Soetaert, K.; Timke, M.; Van den Bossche, A.; Sparbier, K.; De Cremer, K.; Kostrzewa, M.; Hendrickx, M.; Mathys, V. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Combined Species Identification and Drug Sensitivity Testing in Mycobacteria. J. Clin. Microbiol. 2017, 55, 624–634. [Google Scholar] [CrossRef] [PubMed]
- López-Hernández, Y.; Patiño-Rodríguez, O.; García-Orta, S.T.; Pinos-Rodríguez, J.M. Mass spectrometry applied to the identification of Mycobacterium tuberculosis and biomarker discovery. J. Appl. Microbiol. 2016, 121, 1485–1497. [Google Scholar] [CrossRef] [PubMed]
- Alcolea-Medina, A.; Fernandez, M.T.C.; Montiel, N.; García, M.P.L.; Sevilla, C.D.; North, N.; Lirola, M.J.M.; Wilks, M. An improved simple method for the identification of Mycobacteria by MALDI-TOF MS (Matrix-Assisted Laser Desorption- Ionization mass spectrometry). Sci. Rep. 2019, 9, 20216. [Google Scholar] [CrossRef] [PubMed]
- Minnikin, D.E.; Minnikin, S.M.; Parlett, J.H.; Goodfellow, M.; Magnusson, M. Mycolic acid patterns of some species of Mycobacterium. Arch. Microbiol. 1984, 139, 225–231. [Google Scholar] [CrossRef]
- Brennan, P.J.; Heifets, M.; Ullom, B.P. Thin-layer chromatography of lipid antigens as a means of identifying nontuberculous mycobacteria. J. Clin. Microbiol. 1982, 15, 447–455. [Google Scholar] [CrossRef]
- Kellogg, J.A.; Bankert, D.A.; Withers, G.S.; Sweimler, W.; Kiehn, T.E.; Pfyffer, G.E. Application of the Sherlock Mycobacteria Identification System using high-performance liquid chromatography in a clinical laboratory. J. Clin. Microbiol. 2001, 39, 964–970. [Google Scholar] [CrossRef][Green Version]
- Baliga, S.; Murphy, C.; Sharon, L.; Shenoy, S.; Biranthabail, D.; Weltman, H.; Miller, S.; Ramasamy, R.; Shah, J. Rapid method for detecting and differentiating Mycobacterium tuberculosis complex and non-tuberculous mycobacteria in sputum by fluorescence in situ hybridization with DNA probes. Int. J. Infect. Dis. Ijid Off. Publ. Int. Soc. Infect. Dis. 2018, 75, 1–7. [Google Scholar] [CrossRef]
- Shah, J.; Weltman, H.; Narciso, P.; Murphy, C.; Poruri, A.; Baliga, S.; Sharon, L.; York, M.; Cunningham, G.; Miller, S.; et al. Dual color fluorescence in situ hybridization (FISH) assays for detecting Mycobacterium tuberculosis and Mycobacterium avium complexes and related pathogens in cultures. PLoS ONE 2017, 12, e0174989. [Google Scholar] [CrossRef]
- Kim, N.; Lee, S.H.; Yi, J.; Chang, C.L. Evaluation of dual-color fluorescence in situ hybridization with peptide nucleic acid probes for the detection of Mycobacterium tuberculosis and non-tuberculous mycobacteria in clinical specimens. Ann. Lab. Med. 2015, 35, 500–505. [Google Scholar] [CrossRef]
- Lehtola, M.J.; Torvinen, E.; Miettinen, I.T.; Keevil, C.W. Fluorescence In Situ Hybridization Using Peptide Nucleic Acid Probes for Rapid Detection of Mycobacterium avium subsp. avium and Mycobacterium avium subsp. paratuberculosis in Potable-Water Biofilms. Appl. Environ. Microbiol. 2006, 72, 848–853. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lu, L.L.; Smith, M.T.; Yu, K.K.Q.; Luedemann, C.; Suscovich, T.J.; Grace, P.S.; Cain, A.; Yu, W.H.; McKitrick, T.R.; Lauffenburger, D.; et al. IFN-γ-independent immune markers of Mycobacterium tuberculosis exposure. Nat. Med. 2019, 25, 977–987. [Google Scholar] [CrossRef] [PubMed]
- Hendon-Dunn, C.L.; Doris, K.S.; Thomas, S.R.; Allnutt, J.C.; Marriott, A.A.N.; Hatch, K.A.; Watson, R.J.; Bottley, G.; Marsh, P.D.; Taylor, S.C.; et al. A flow cytometry method for rapidly assessing M. tuberculosis responses to antibiotics with different modes of action. Antimicrob. Agents Chemother. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kamariza, M.; Shieh, P.; Ealand, C.S.; Peters, J.S.; Chu, B.; Rodriguez-Rivera, F.P.; Babu Sait, M.R.; Treuren, W.V.; Martinson, N.; Kalscheuer, R.; et al. Rapid detection of Mycobacterium tuberculosis in sputum with a solvatochromic trehalose probe. Sci. Transl. Med. 2018, 10, eaam6310. [Google Scholar] [CrossRef]
- Kim, S.H.; Shin, J.H. Identification of nontuberculous mycobacteria using multilocous sequence analysis of 16S rRNA, hsp65, and rpoB. J. Clin. Lab. Anal. 2018, 32. [Google Scholar] [CrossRef]
- de Zwaan, R.; van Ingen, J.; van Soolingen, D. Utility of rpoB gene sequencing for identification of nontuberculous mycobacteria in the Netherlands. J. Clin. Microbiol. 2014, 52, 2544–2551. [Google Scholar] [CrossRef]
- Haig, S.J.; Kotlarz, N.; LiPuma, J.J.; Raskin, L. A High-Throughput Approach for Identification of Nontuberculous Mycobacteria in Drinking Water Reveals Relationship between Water Age and Mycobacterium avium. mBio 2018, 9. [Google Scholar] [CrossRef]
- Kunze, Z.M.; Portaels, F.; McFadden, J.J. Biologically distinct subtypes of Mycobacterium avium differ in possession of insertion sequence IS901. J. Clin. Microbiol. 1992, 30, 2366. [Google Scholar] [CrossRef]
- Moss, M.T.; Malik, Z.P.; Tizard, M.L.V.; Green, E.P.; Sanderson, J.D.; Hermon-Taylor, J. IS902, an insertion element of the chronic-enteritis-causing Mycobacterium avium subsp. silvaticum. Microbiology 1992, 138, 139–145. [Google Scholar] [CrossRef][Green Version]
- Stinear, T.; Ross, B.C.; Davies, J.K.; Marino, L.; Robins-Browne, R.M.; Oppedisano, F.; Sievers, A.; Johnson, P.D.R. Identification and Characterization of Two Distinct Repeated Sequences for Detection of Mycobacterium ulcerans by PCR. J. Clin. Microbiol. 1999, 37, 1018. [Google Scholar] [CrossRef]
- Picardeau, M.; Bull, T.J.; Prod’hom, G.; Pozniak, A.L.; Shanson, D.C.; Vincent, V. Comparison of a new insertion element, IS1407, with established molecular markers for the characterization of Mycobacterium celatum. Int. J. Syst. Bacteriol. 1997, 47, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Picardeau, M.; Varnerot, A.; Rauzier, J.; Gicquel, B.; Vincent, V. Mycobacterium xenopi IS1395, a novel insertion sequence expanding the IS256 family. Microbiology 1996, 142 Pt 9, 2453–2461. [Google Scholar] [CrossRef]
- Picardeau, M.; Bull, T.J.; Vincent, V. Identification and characterization of IS-like elements in Mycobacterium gordonae. FEMS Microbiol. Lett. 1997, 154, 95–102. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Picardeau, M.; Prod’Hom, G.; Raskine, L.; LePennec, M.P.; Vincent, V. Genotypic characterization of five subspecies of Mycobacterium kansasii. J. Clin. Microbiol. 1997, 35, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Guilhot, C.; Gicquel, B.; Davies, J.; Martín, C. Isolation and analysis of IS6120, a new insertion sequence from Mycobacterium smegmatis. Mol. Microbiol. 1992, 6, 107–113. [Google Scholar] [CrossRef]
- Tortoli, E.; Mariottini, A.; Mazzarelli, G. Evaluation of INNO-LiPA MYCOBACTERIA v2: Improved reverse hybridization multiple DNA probe assay for mycobacterial identification. J. Clin. Microbiol. 2003, 41, 4418–4420. [Google Scholar] [CrossRef]
- van den Broek, T.; Janssen, N.G.; Hetem, D.J.; Bekers, W.; Kamst, M.; Fluit, A.C.; van Ingen, J.; Kusters, J.G.; Rentenaar, R.J. INNO-LiPA DNA line probe assay misidentification of M. smegmatis as Mycobacterium fortuitum complex. Diagn. Microbiol. Infect. Dis. 2019, 95, 114858. [Google Scholar] [CrossRef]
- Huh, H.J.; Kim, S.Y.; Shim, H.J.; Kim, D.H.; Yoo, I.Y.; Kang, O.K.; Ki, C.S.; Shin, S.Y.; Jhun, B.W.; Shin, S.J.; et al. GenoType NTM-DR Performance Evaluation for Identification of Mycobacterium avium Complex and Mycobacterium abscessus and Determination of Clarithromycin and Amikacin Resistance. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Mok, S.; Rogers, T.R.; Fitzgibbon, M. Evaluation of GenoType NTM-DR Assay for Identification of Mycobacterium chimaera. J. Clin. Microbiol 2017, 55, 1821–1826. [Google Scholar] [CrossRef]
- Jagielski, T.; van Ingen, J.; Rastogi, N.; Dziadek, J.; Mazur, P.K.; Bielecki, J. Current Methods in the Molecular Typing of Mycobacterium tuberculosis and Other Mycobacteria. BioMed Res. Int. 2014, 2014, 645802. [Google Scholar] [CrossRef]
- Zhang, Y.; Mann, L.B.; Wilson, R.W.; Brown-Elliott, B.A.; Vincent, V.; Iinuma, Y.; Wallace, R.J., Jr. Molecular analysis of Mycobacterium kansasii isolates from the United States. J. Clin. Microbiol. 2004, 42, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, G.H.; Hartman, S.; Zhang, Y.; Brown, B.A.; Hector, J.S.; Murphy, D.; Wallace, R.J. Large DNA restriction fragment polymorphism in the Mycobacterium avium-M. intracellulare complex: A potential epidemiologic tool. J. Clin. Microbiol. 1993, 31, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Lalande, V.; Barbut, F.; Varnerot, A.; Febvre, M.; Nesa, D.; Wadel, S.; Vincent, V.; Petit, J.C. Pseudo-outbreak of Mycobacterium gordonae associated with water from refrigerated fountains. J. Hosp. Infect. 2001, 48, 76–79. [Google Scholar] [CrossRef]
- Yakrus, M.A.; Straus, W.L. DNA polymorphisms detected in Mycobacterium haemophilum by pulsed-field gel electrophoresis. J. Clin. Microbiol. 1994, 32, 1083–1084. [Google Scholar] [CrossRef]
- Zhang, Y.; Rajagopalan, M.; Brown, B.A.; Wallace, R.J., Jr. Randomly amplified polymorphic DNA PCR for comparison of Mycobacterium abscessus strains from nosocomial outbreaks. J. Clin. Microbiol. 1997, 35, 3132–3139. [Google Scholar] [CrossRef]
- Vogiatzakis, E.; Stefanou, S.; Skroubelou, A.; Anagnostou, S.; Marinis, E.; Matsiota-Bernard, P. Molecular markers for the investigation of Mycobacterium gordonae epidemics. J. Hosp. Infect. 1998, 38, 217–222. [Google Scholar] [CrossRef]
- van Soolingen, D.; Bauer, J.; Ritacco, V.; Leão, S.C.; Pavlik, I.; Vincent, V.; Rastogi, N.; Gori, A.; Bodmer, T.; Garzelli, C.; et al. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: Proposal for standardization. J. Clin. Microbiol. 1998, 36, 3051–3054. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ma, C.; Xiao, T.; Li, M.; Liu, H.; Zhao, X.; Wan, K.; Wang, R. A New Single Gene Differential Biomarker for Mycobacterium tuberculosis Complex and Non-tuberculosis Mycobacteria. Front. Microbiol. 2019, 10, 1887. [Google Scholar] [CrossRef]
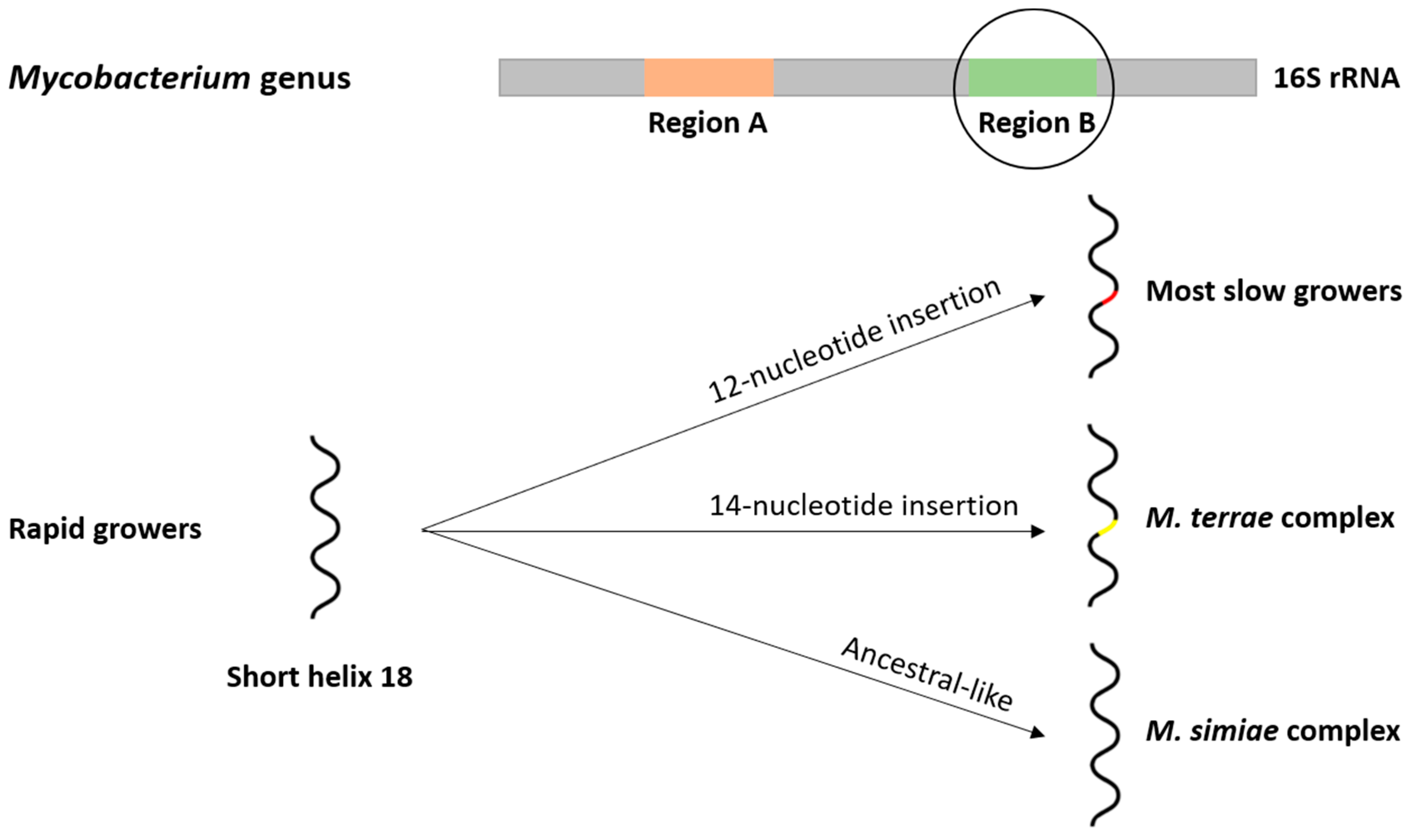
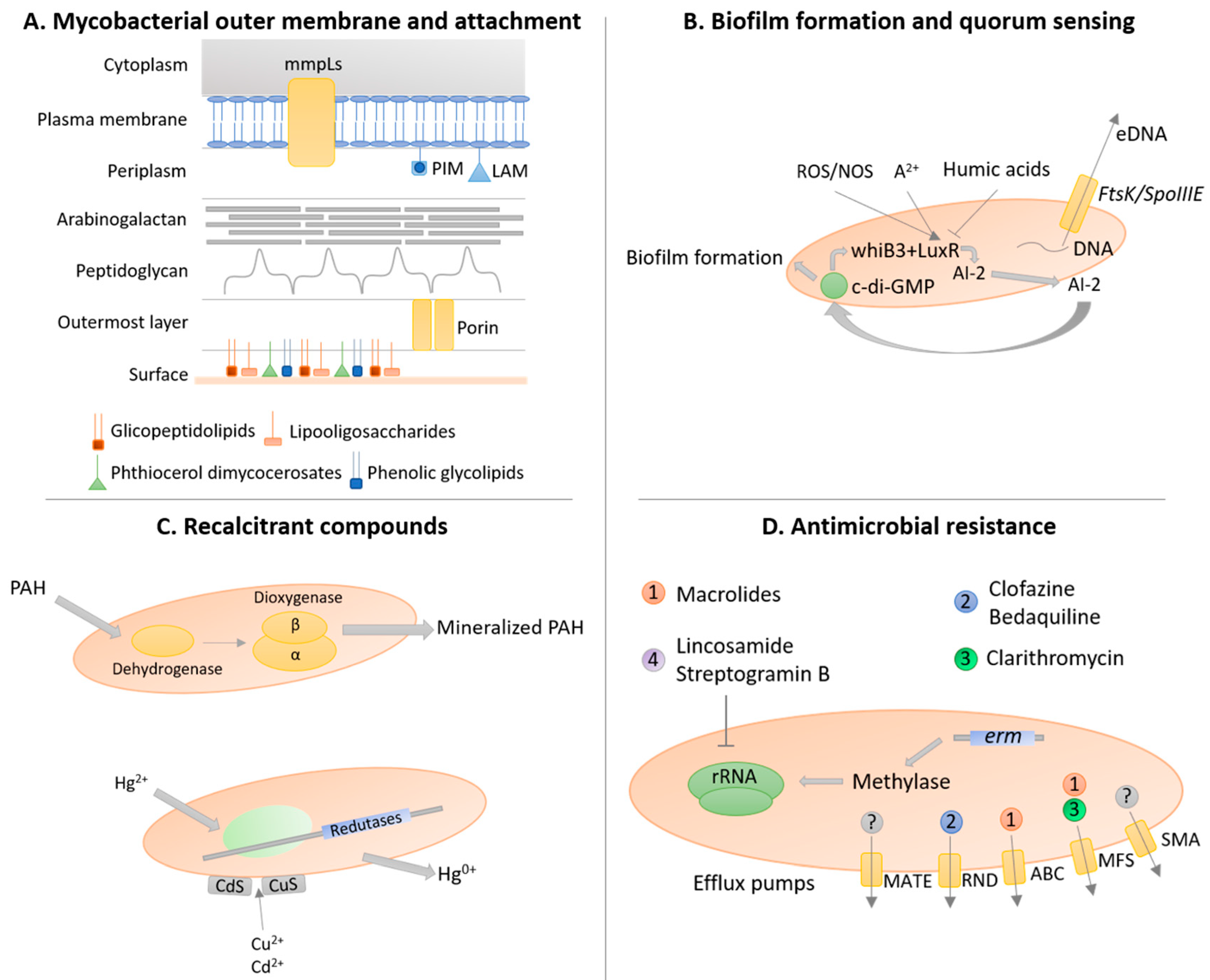
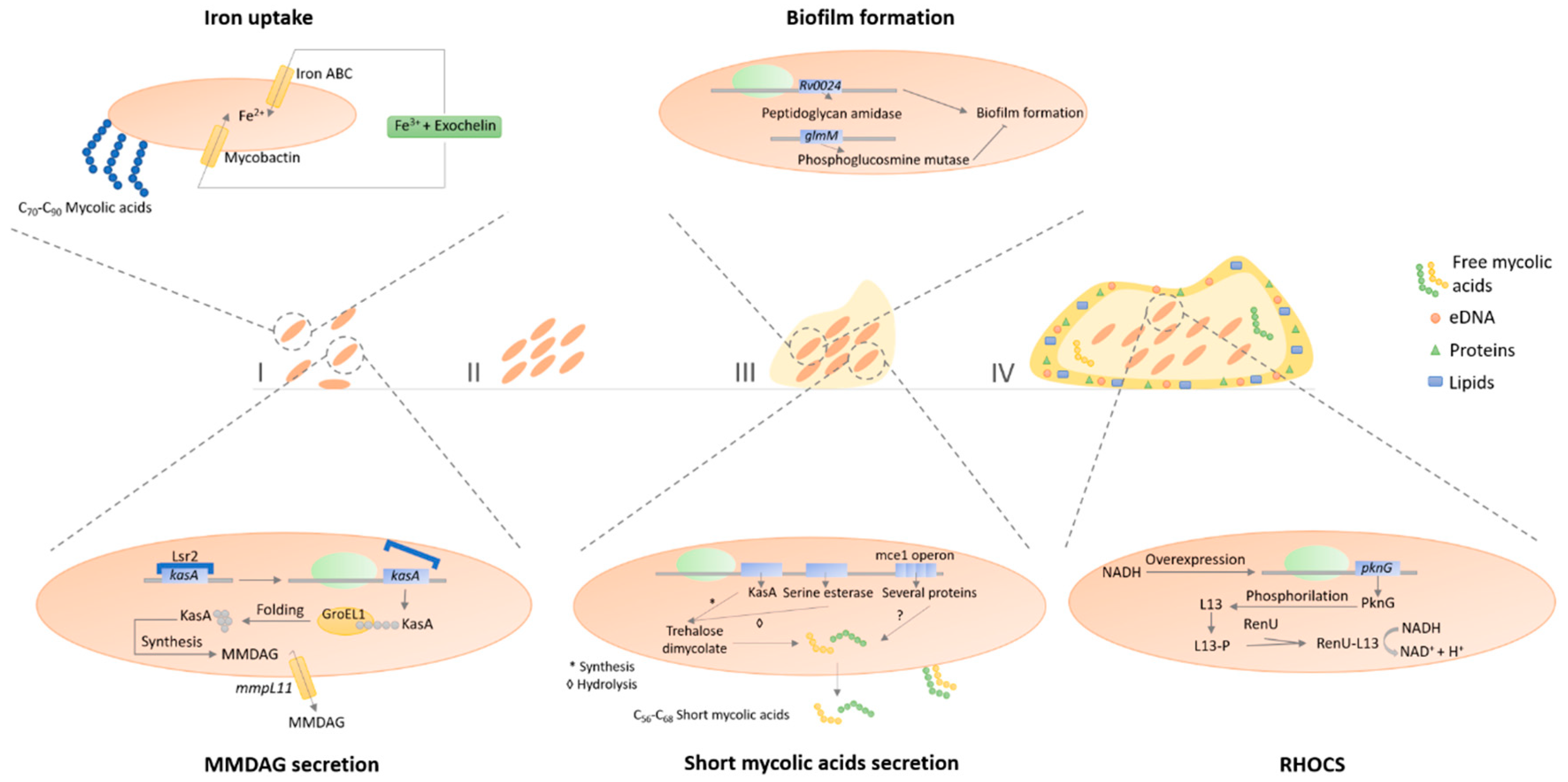
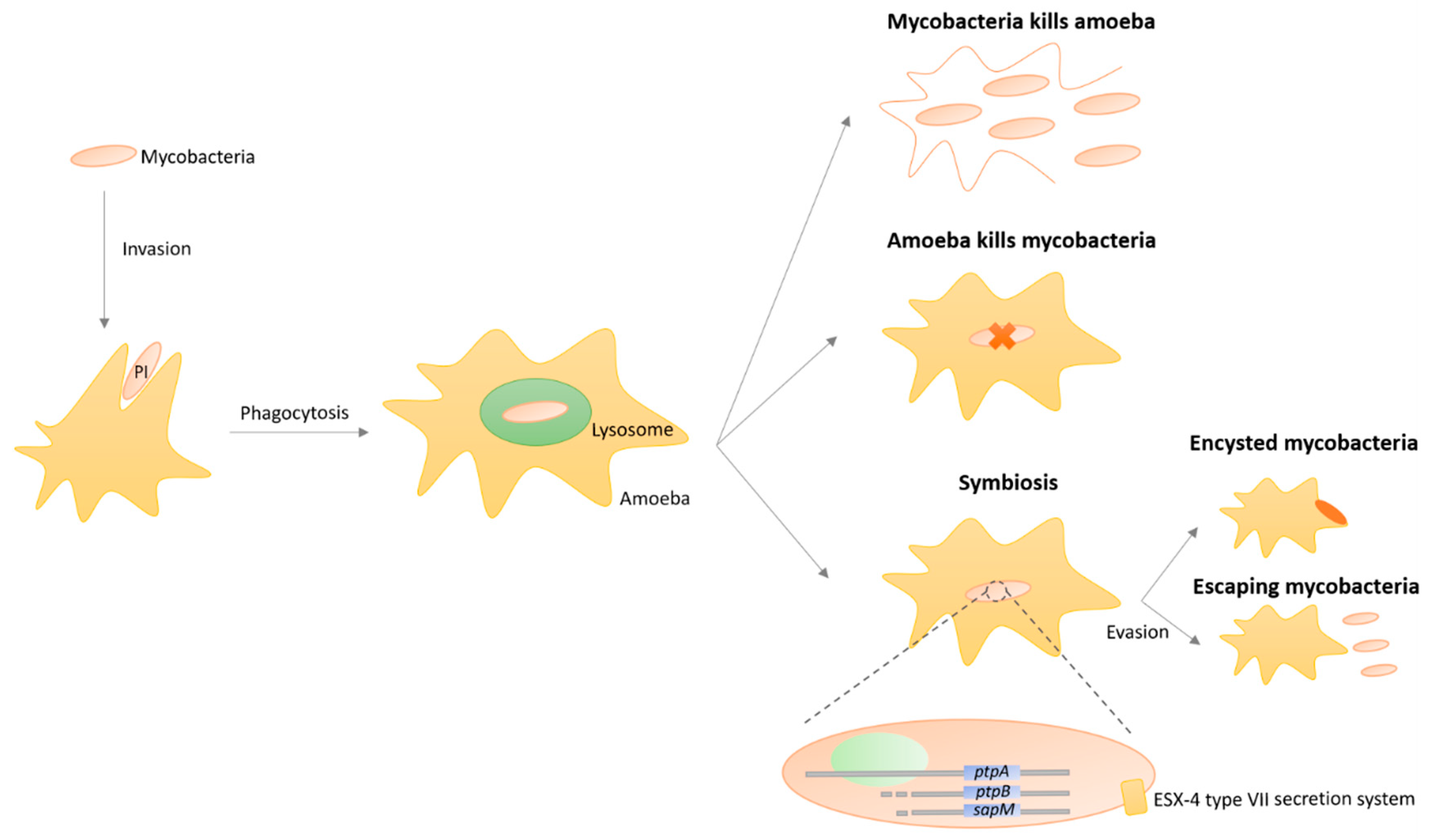
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, A.C.; Ramos, B.; Reis, A.C.; Cunha, M.V. Non-Tuberculous Mycobacteria: Molecular and Physiological Bases of Virulence and Adaptation to Ecological Niches. Microorganisms 2020, 8, 1380. https://doi.org/10.3390/microorganisms8091380
Pereira AC, Ramos B, Reis AC, Cunha MV. Non-Tuberculous Mycobacteria: Molecular and Physiological Bases of Virulence and Adaptation to Ecological Niches. Microorganisms. 2020; 8(9):1380. https://doi.org/10.3390/microorganisms8091380
Chicago/Turabian StylePereira, André C., Beatriz Ramos, Ana C. Reis, and Mónica V. Cunha. 2020. "Non-Tuberculous Mycobacteria: Molecular and Physiological Bases of Virulence and Adaptation to Ecological Niches" Microorganisms 8, no. 9: 1380. https://doi.org/10.3390/microorganisms8091380
APA StylePereira, A. C., Ramos, B., Reis, A. C., & Cunha, M. V. (2020). Non-Tuberculous Mycobacteria: Molecular and Physiological Bases of Virulence and Adaptation to Ecological Niches. Microorganisms, 8(9), 1380. https://doi.org/10.3390/microorganisms8091380




