Refining the Galleria mellonella Model by Using Stress Marker Genes to Assess Clostridioides difficile Infection and Recuperation during Phage Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates and Inocula Preparation
2.2. Preparation of Phage Cocktail
2.3. Preparation of G mellonella Larvae and Model Set Up
2.4. Bacterial Infection and Phage Therapy Regimens in G. mellonella Larvae
2.5. Survival Rates, Bacterial Enumeration and Toxin Assay from the Treated Insects
2.6. RNA Extraction, cDNA Synthesis and qPCR
3. Results
3.1. Impact of Phage Therapy Regimens on G. mellonella Survival
3.2. Impact of Phage Therapy Regimens on C. difficile Colonisation on G. mellonella
3.3. Impact of Phage Therapy on C. difficile Toxins Levels in G. mellonella
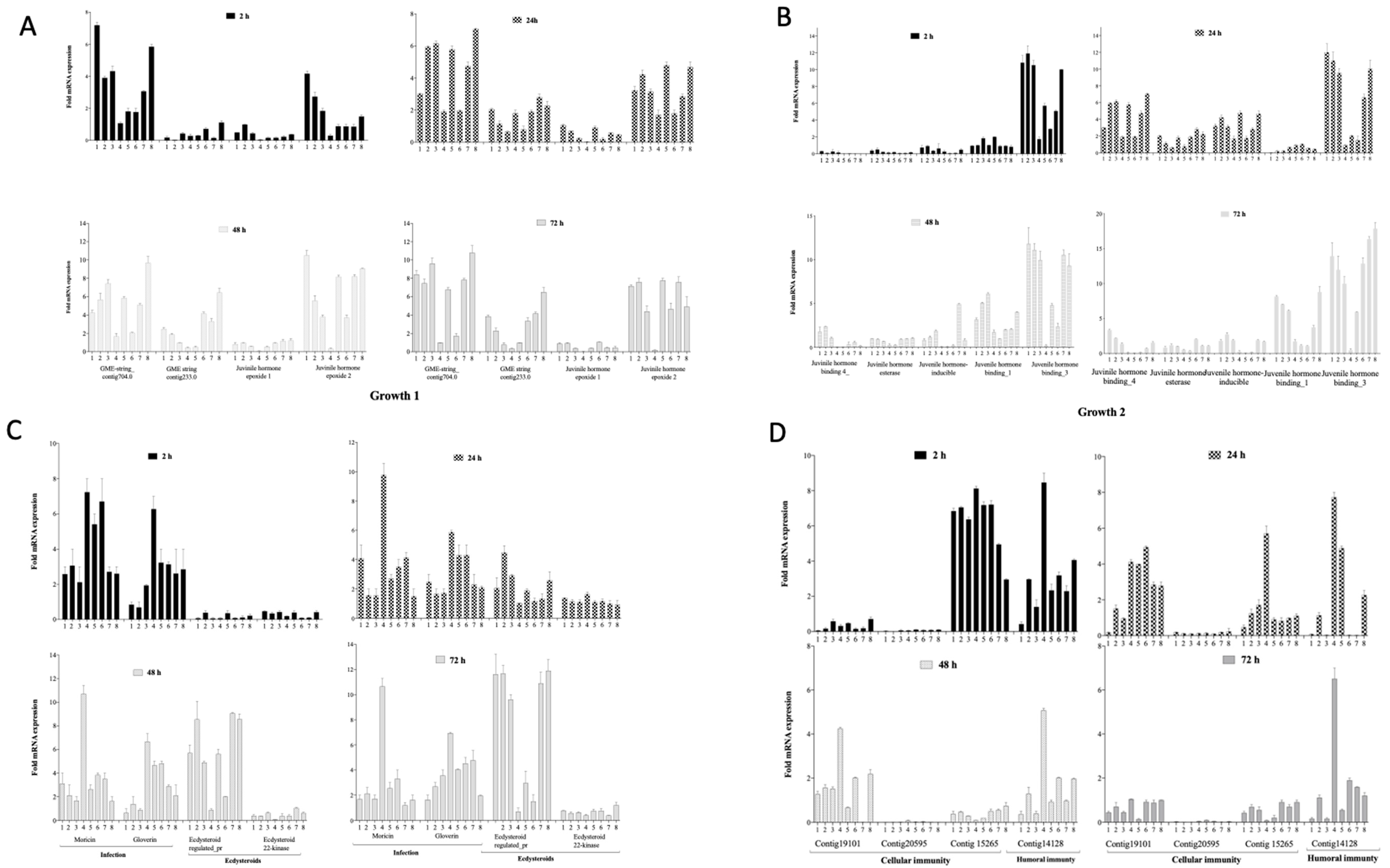
3.4. Gene Regulation during Infection and Phage Therapy in G. mellonella
3.4.1. Impact on Growth Markers
3.4.2. Impact on Markers of Infection, Molting and Reproduction
3.4.3. Impact on Cellular and Humoral Immunity Markers
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Olsen, M.A.; Young-Xu, Y.; Stwalley, D.; Kelly, C.P.; Gerding, D.N.; Saeed, M.J.; Mahé, C.; Dubberke, E.R. The burden of Clostridium difficile infection: Estimates of the incidence of CDI from U.S. Administrative databases. BMC Infect. Dis. 2016, 16, 177. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.H.; Olsen, M.A.; Dubberke, E.R. The Morbidity, Mortality, and Costs Associated with Clostridium difficile Infection. Infect. Dis. Clin. N. Am. 2015, 29, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Bacci, S.; Molbak, K.; Kjeldsen, M.K.; Olsen, K.E. Binary toxin and death after Clostridium difficile infection. Emerg. Infect. Dis. 2011, 17, 976–982. [Google Scholar] [CrossRef] [PubMed]
- Lyerly, D.M.; Krivan, H.C.; Wilkins, T.D. Clostridium difficile: Its disease and toxins. Clin. Microbiol. Rev. 1988, 1, 1–18. [Google Scholar] [CrossRef]
- Legenza, L.; Barnett, S.; Rose, W.; Bianchini, M.; Safdar, N.; Coetzee, R. Epidemiology and outcomes of Clostridium difficile infection among hospitalised patients: Results of a multicentre retrospective study in South Africa. BMJ Glob. Health 2018, 3. [Google Scholar] [CrossRef]
- Lessa, F.C.; Mu, Y.; Bamberg, W.M.; Beldavs, Z.G.; Dumyati, G.K.; Dunn, J.R.; Farley, M.M.; Holzbauer, S.M.; Meek, J.I.; Phipps, E.C.; et al. Burden of Clostridium difficile Infection in the United States. N. Engl. J. Med. 2015, 372, 825–834. [Google Scholar] [CrossRef]
- Lessa, F.C.; Gould, C.V.; McDonald, L.C. Current Status of Clostridium difficile Infection Epidemiology. Clin. Infect. Dis. 2012, 55, S65–S70. [Google Scholar] [CrossRef]
- Kuijper, E.J.; Barbut, F.; Brazier, J.S.; Kleinkauf, N.; Eckmanns, T.; Lambert, M.L.; Drudy, D.; Fitzpatrick, F.; Wiuff, C.; Brown, D.J.; et al. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Eurosurveillance 2008, 13, 18942. [Google Scholar]
- Akerlund, T.; Persson, I.; Unemo, M.; Noren, T.; Svenungsson, B.; Wullt, M.; Burman, L.G. Increased sporulation rate of epidemic Clostridium difficile Type 027/NAP1. J. Clin. Microbiol. 2008, 46, 1530–1533. [Google Scholar] [CrossRef]
- Vonberg, R.P.; Kuijper, E.J.; Wilcox, M.H.; Barbut, F.; Tull, P.; Gastmeier, P.; The European C. difficile-Infection Control Group; The European Centre for Disease Prevention and Control (ECDC); van den Broek, P.J.; Colville, A.; et al. Infection control measures to limit the spread of Clostridium difficile. Clin. Microbiol. Infect. 2008, 14, 2–20. [Google Scholar] [CrossRef]
- Peng, Y.Z.; Huang, G.T. Phage therapy for bacterial infection of burn. Zhonghua Shao Shang Za Zhi 2016, 32, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Dang, M.D.; Hasbun, R.; Koo, H.L.; Jiang, Z.D.; DuPont, H.L.; Garey, K.W. Clostridium difficile infection: Update on emerging antibiotic treatment options and antibiotic resistance. Expert Rev. Anti-Infect. Ther. 2010, 8, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Debast, S.B.; Bauer, M.P.; Kuijper, E.J. European Society of Clinical Microbiology and Infectious Diseases: Update of the Treatment Guidance Document for Clostridium difficile Infection. Clin. Microbiol. Infect. 2014, 20, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Koss, C.A.; Baras, D.C.; Lane, S.D.; Aubry, R.; Marcus, M.; Markowitz, L.E.; Koumans, E.H. Investigation of Metronidazole Use during Pregnancy and Adverse Birth Outcomes. Antimicrob. Agents Chemother. 2012, 56, 4800–4805. [Google Scholar] [CrossRef]
- Nelson, R.L.; Suda, K.J.; Evans, C.T. Antibiotic treatment for Clostridium difficile-associated diarrhoea in adults. Cochrane Database Syst. Rev. 2017. [Google Scholar] [CrossRef]
- Baines, S.D.; Wilcox, M.H. Antimicrobial Resistance and Reduced Susceptibility in Clostridium difficile: Potential Consequences for Induction, Treatment, and Recurrence of C. difficile Infection. Antibiotics 2015, 4, 267–298. [Google Scholar] [CrossRef]
- Zucca, M.; Scutera, S.; Savoia, D. Novel avenues for Clostridium difficile infection drug discovery. Expert Opin. Drug Discov. 2013, 8, 459–477. [Google Scholar] [CrossRef]
- Giau, V.V.; Lee, H.; An, S.S.A.; Hulme, J. Recent advances in the treatment of C. difficile using biotherapeutic agents. Infect. Drug Resist. 2019, 12, 1597–1615. [Google Scholar] [CrossRef]
- Slopek, S.; Weber-Dabrowska, B.; Dabrowski, M.; Kucharewicz-Krukowska, A. Results of bacteriophage treatment of suppurative bacterial infections in the years 1981-1986. Arch. Immunol. Ther. Exp. 1987, 35, 569–583. [Google Scholar]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef]
- Brüssow, H. What is needed for phage therapy to become a reality in Western medicine? Virology 2012, 434, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, K.R.; Clokie, M.R.J. Clostridium difficile phages: Still difficult? Front. Microbiol. 2014, 5, 184. [Google Scholar] [CrossRef] [PubMed]
- Nale, J.Y.; Spencer, J.; Hargreaves, K.R.; Buckley, A.M.; Trzepiński, P.; Douce, G.R.; Clokie, M.R.J. Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrob. Agents Chemother. 2016, 60, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Nale, J.Y.; Chutia, M.; Carr, P.; Hickenbotham, P.; Clokie, M.R.J. ‘Get in early’; biofilm and wax moth (Galleria mellonella) models reveal new insights into the therapeutic potential of Clostridium difficile bacteriophages. Front. Microbiol. 2016, 7, 1383. [Google Scholar] [CrossRef] [PubMed]
- Nale, J.; Redgwell, T.A.; Millard, A.; Clokie, M.R.J. Efficacy of an Optimised Bacteriophage Cocktail to Clear Clostridium difficile in a Batch Fermentation Model. Antibiotics 2018, 7, 13. [Google Scholar] [CrossRef]
- Best, E.L.; Freeman, J.; Wilcox, M.H. Models for the study of Clostridium difficile infection. Gut Microbes 2012, 3, 145–167. [Google Scholar] [CrossRef]
- Mukherjee, K.; Hain, T.; Fischer, R.; Chakraborty, T.; Vilcinskas, A. Brain infection and activation of neuronal repair mechanisms by the human pathogen Listeria monocytogenes in the lepidopteran model host Galleria mellonella. Virulence 2013, 4, 324–332. [Google Scholar] [CrossRef]
- Mukherjee, K.; Altincicek, B.; Hain, T.; Domann, E.; Vilcinskas, A.; Chakraborty, T. Galleria mellonella as a Model System for Studying Listeria Pathogenesis. Appl. Environ. Microbiol. 2010, 76, 310–317. [Google Scholar] [CrossRef]
- Beeton, M.L.; Alves, D.R.; Enright, M.C.; Jenkins, A.T.A. Assessing phage therapy against Pseudomonas aeruginosa using a Galleria mellonella infection model. Int. J. Antimicrob. Agents 2015, 46, 196–200. [Google Scholar] [CrossRef]
- Viegas, S.; Mil-Homens, D.; Fialho, A.; Arraiano, C. The Virulence of Salmonella enterica Serovar Typhimurium in the Insect Model Galleria mellonella Is Impaired by Mutations in RNase E and RNase III. Appl. Environ. Microbiol. 2013, 79, 6124–6133. [Google Scholar] [CrossRef] [PubMed]
- Vogel, H.; Altincicek, B.; Glöckner, G.; Vilcinskas, A. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genom. 2011, 12, 308. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, K.R.; Kropinski, A.M.; Clokie, M.R.J. What Does the Talking?: Quorum Sensing Signalling Genes Discovered in a Bacteriophage Genome. PLoS ONE 2014, 9, e85131. [Google Scholar] [CrossRef] [PubMed]
- Weld, R.J.; Butts, C.; Heinemann, J.A. Models of phage growth and their applicability to phage therapy. J. Theor. Biol. 2004, 227, 1–11. [Google Scholar] [CrossRef]
- O‘Sullivan, L.; Buttimer, C.; McAuliffe, O.; Bolton, D.; Coffey, A. Bacteriophage-based tools: Recent advances and novel applications. F1000Res 2016, 5, 2782. [Google Scholar] [CrossRef]
- Shan, J.; Ramachandran, A.; Thanki, A.M.; Vukusic, F.B.I.; Barylski, J.; Clokie, M.R.J. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci. Rep. 2018, 8, 5091. [Google Scholar] [CrossRef]
- Flecknell, P. Replacement, reduction and refinement. Altex 2002, 19, 73–78. [Google Scholar]
- Meyers, J.R. Zebrafish: Development of a Vertebrate Model Organism. Curr. Protoc. Essent. Lab. Tech. 2018, 16, e19. [Google Scholar] [CrossRef]
- Ramarao, N.; Nielsen-Leroux, C.; Lereclus, D. The Insect Galleria mellonella as a Powerful Infection Model to Investigate Bacterial Pathogenesis. J. Vis. Exp. 2012, e4392. [Google Scholar] [CrossRef]
- Mukherjee, K.; Abu Mraheil, M.; Silva, S.; Müller, D.; Cemic, F.; Hemberger, J.; Hain, T.; Vilcinskas, A.; Chakraborty, T. Anti-Listeria activities of Galleria mellonella hemolymph proteins. Appl. Environ. Microbiol. 2011, 77, 4237–4240. [Google Scholar] [CrossRef]
- Kay, S.; Edwards, J.; Brown, J.; Dixon, R. Galleria mellonella Infection Model Identifies Both High and Low Lethality of Clostridium perfringens Toxigenic Strains and Their Response to Antimicrobials. Front. Microbiol. 2019, 10, 1281. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, V.; Fralick, J.A.; Rolfe, R.D. Prevention of Clostridium difficile-induced ileocecitis with Bacteriophage. Anaerobe 1999, 5, 69–78. [Google Scholar] [CrossRef]
- Govind, R.; Fralick, J.A.; Rolfe, R.D. In vivo lysogenization of a Clostridium difficile bacteriophage ΦCD119. Anaerobe 2011, 17, 125–129. [Google Scholar] [CrossRef]
- Buckley, A.M.; Spencer, J.; Maclellan, L.M.; Candlish, D.; Irvine, J.J.; Douce, G.R. Susceptibility of Hamsters to Clostridium difficile Isolates of Differing Toxinotype. PLoS ONE 2013, 8, e64121. [Google Scholar] [CrossRef]
- Lange, A.; Schäfer, A.; Bender, A.; Steimle, A.; Beier, S.; Parusel, R.; Frick, J.-S. Galleria mellonella: A Novel Invertebrate Model to Distinguish Intestinal Symbionts From Pathobionts. Front. Immunol. 2018, 9, 2114. [Google Scholar] [CrossRef]
- Coates, C.J.; Lim, J.; Harman, K.; Rowley, A.F.; Griffiths, D.J.; Emery, H.; Layton, W. The insect, Galleria mellonella, is a compatible model for evaluating the toxicology of okadaic acid. Cell Biol. Toxicol. 2019, 35, 219–232. [Google Scholar] [CrossRef]
- Chippendale, G.M.; Reddy, G.P.V. Dietary carbohydrates: Role in feeding behaviour and growth of the Southwestern corn borer, Diatraea grandiosella. J. Insect Physiol. 1974, 20, 751–759. [Google Scholar] [CrossRef]
- Ge, L.; Jiang, Y.; Xia, T.; Song, Q.; Stanley, D.; Kuai, P.; Lu, X.; Yang, G.; Wu, J. Silencing a sugar transporter gene reduces growth and fecundity in the brown planthopper, Nilaparvata lugens (Stål) (Hemiptera: Delphacidae). Sci. Rep. 2015, 5, 12194. [Google Scholar] [CrossRef]
- Mukherjee, K.; Fischer, R.; Vilcinskas, A. Histone acetylation mediates epigenetic regulation of transcriptional reprogramming in insects during metamorphosis, wounding and infection. Front. Zool. 2012, 9, 1–12. [Google Scholar] [CrossRef]
- Farhadian, S.F.; Suárez-Fariñas, M.; Cho, C.E.; Pellegrino, M.; Vosshall, L.B. Post-fasting olfactory, transcriptional, and feeding responses in Drosophila. Physiol. Behav. 2012, 105, 544–553. [Google Scholar] [CrossRef]
- Browne, N.; Heelan, M.; Kavanagh, K. An analysis of the structural and functional similarities of insect hemocytes and mammalian phagocytes. Virulence 2013, 4, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Mowlds, P.; Kavanagh, K. Effect of pre-incubation temperature on susceptibility of Galleria mellonella larvae to infection by Candida albicans. Mycopathologia 2008, 165, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Grizanova, E.V.; Dubovskiy, I.M.; Whitten, M.M.A.; Glupov, V.V. Contributions of cellular and humoral immunity of Galleria mellonella larvae in defence against oral infection by Bacillus thuringiensis. J. Invertebr. Pathol. 2014, 119, 40–46. [Google Scholar] [CrossRef] [PubMed]
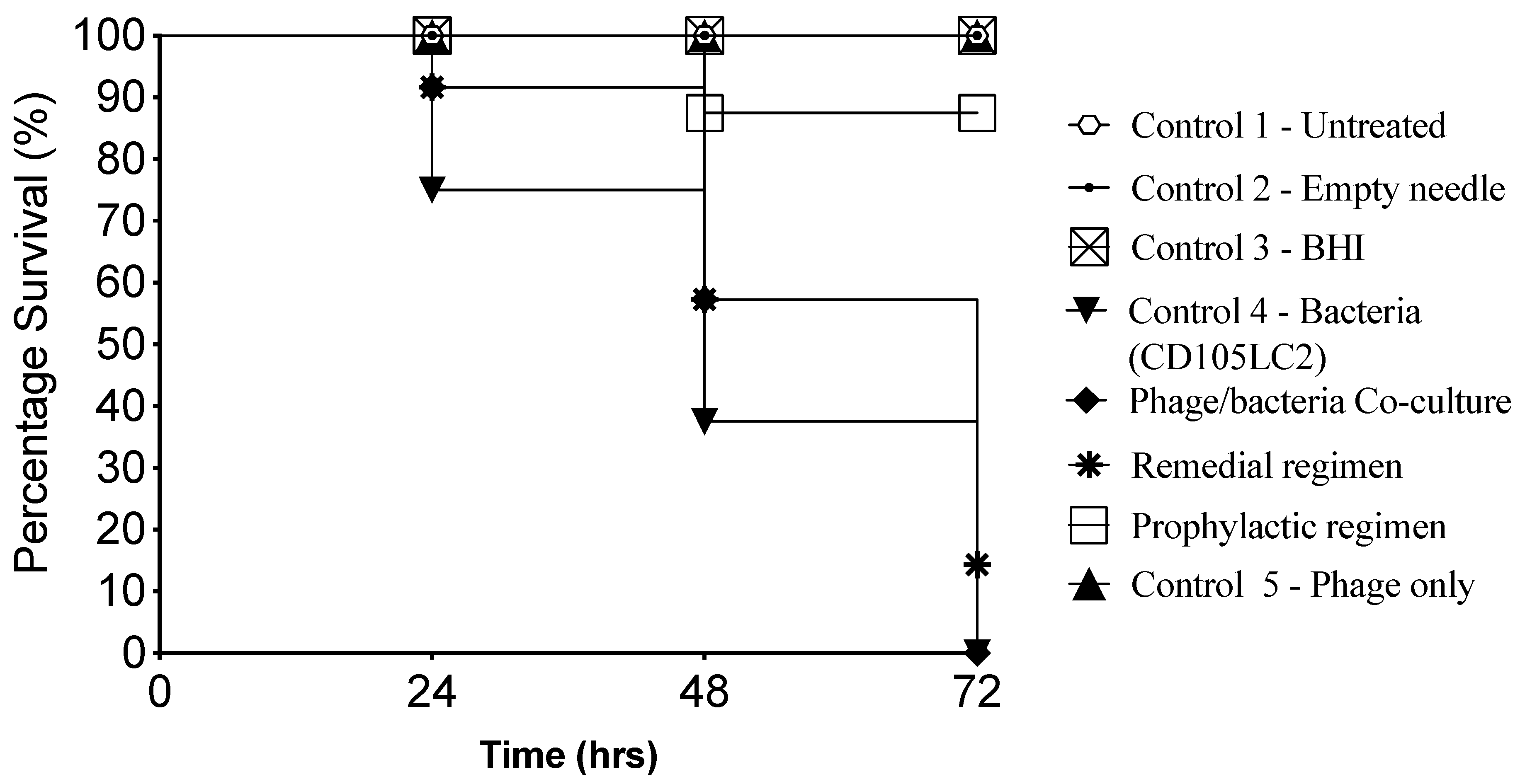
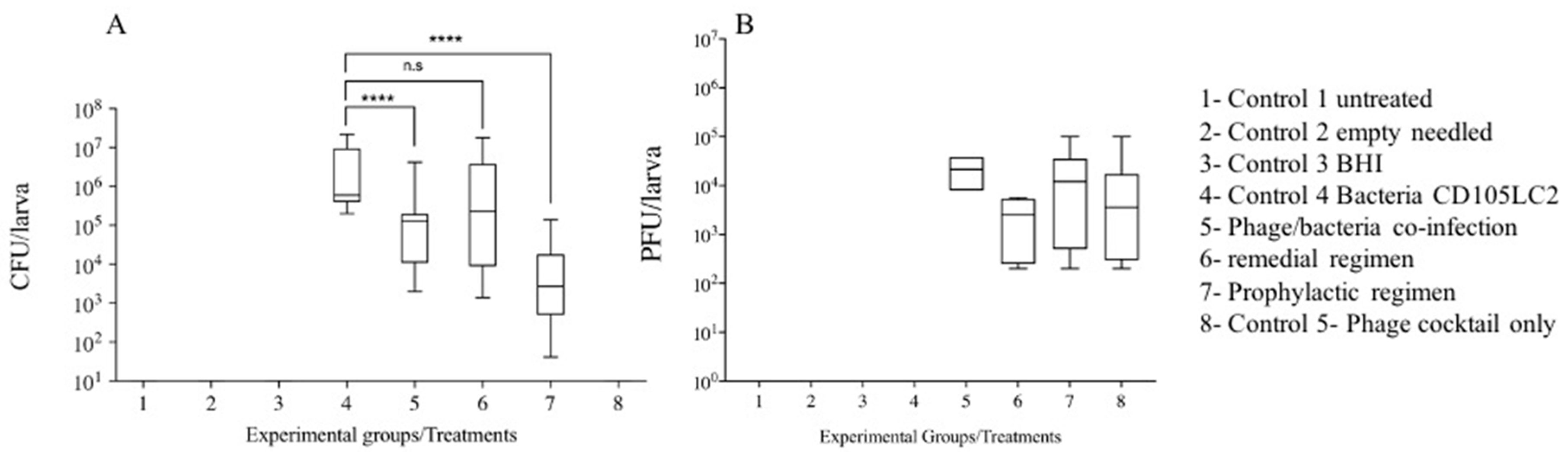
| Experimental Groups | Treatments | Time (h) | ||||
|---|---|---|---|---|---|---|
| 0 | 2 | 24 | 48 | 72 | ||
| 1 | Control 1—Untreated | - | - | - | - | - |
| 2 | Control 2—Empty needle | N | N | - | - | - |
| 3 | Control 3—BHI | BHI | BHI | - | - | - |
| 4 | Control 4—Bacteria (CD105LC2) | B | BHI | - | - | - |
| 5 | Phage/bacteria co-infection | P+B | BHI | - | - | - |
| 6 | Remedial regimen | B | P | - | - | - |
| 7 | Prophylactic regimen | P | B | - | - | - |
| 8 | Control 5—Phage only | P | BHI | - | - | - |
| Gene Function | Number of Genes | Gene Name/Locus Tag | Reference |
|---|---|---|---|
| Growth | 9 | GME-string_contig704.0, GME-string_contig233.0 Juvenile hormone epoxide hydrolase_1 Juvenile hormone epoxide hydrolase _2 Juvenile hormone binding_1 Juvenile hormone esterase Juvenile hormone inducible Juvenile hormone binding_3 Juvenile hormone binding_4 | [28] |
| Cellular immunity | 3 | Contig19101_1 Contig20595_1 Contig15265_1 | |
| Humoral immunity | 1 | Contig14128_1 | |
| Infection | 2 | Moricin, Gloverin | |
| Reproduction | 2 | Ecdysteroid_regulated_pr Ecdysteroid_22-kinase |
| Time (h) | Experimental Treatment Groups (OD450 Measurements) | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Untreated | Needle | BHI | CD105LC2 | Co-Infection | Remedial | Prophylaxis | Phage | |
| 2 | 0.063 | 0.064 | 0.067 | 0.072 | 0.111 | 0.108 | 0.127 | 0.061 |
| 24 | 0.068 | 0.058 | 0.060 | 0.192 | 0.201 | 0.293 | 0.201 | 0.054 |
| 48 | 0.067 | 0.059 | 0.057 | 0.210 | 0.239 | 0.223 | 0.108 | 0.060 |
| 72 | 0.065 | 0.065 | 0.091 | 0.234 | 0.193 | 0.260 | 0.129 | 0.059 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nale, J.Y.; Chutia, M.; Cheng, J.K.J.; Clokie, M.R.J. Refining the Galleria mellonella Model by Using Stress Marker Genes to Assess Clostridioides difficile Infection and Recuperation during Phage Therapy. Microorganisms 2020, 8, 1306. https://doi.org/10.3390/microorganisms8091306
Nale JY, Chutia M, Cheng JKJ, Clokie MRJ. Refining the Galleria mellonella Model by Using Stress Marker Genes to Assess Clostridioides difficile Infection and Recuperation during Phage Therapy. Microorganisms. 2020; 8(9):1306. https://doi.org/10.3390/microorganisms8091306
Chicago/Turabian StyleNale, Janet Y., Mahananda Chutia, Jeffrey K. J. Cheng, and Martha R. J. Clokie. 2020. "Refining the Galleria mellonella Model by Using Stress Marker Genes to Assess Clostridioides difficile Infection and Recuperation during Phage Therapy" Microorganisms 8, no. 9: 1306. https://doi.org/10.3390/microorganisms8091306
APA StyleNale, J. Y., Chutia, M., Cheng, J. K. J., & Clokie, M. R. J. (2020). Refining the Galleria mellonella Model by Using Stress Marker Genes to Assess Clostridioides difficile Infection and Recuperation during Phage Therapy. Microorganisms, 8(9), 1306. https://doi.org/10.3390/microorganisms8091306







