A Genome-Scale Insight into the Effect of Shear Stress During the Fed-Batch Production of Clavulanic Acid by Streptomyces Clavuligerus
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of a New Genome Scale Model of S. Clavuligerus
2.2. Flux Balance Analysis
2.3. Microorganism, Cultivation Media and Experimental Conditions
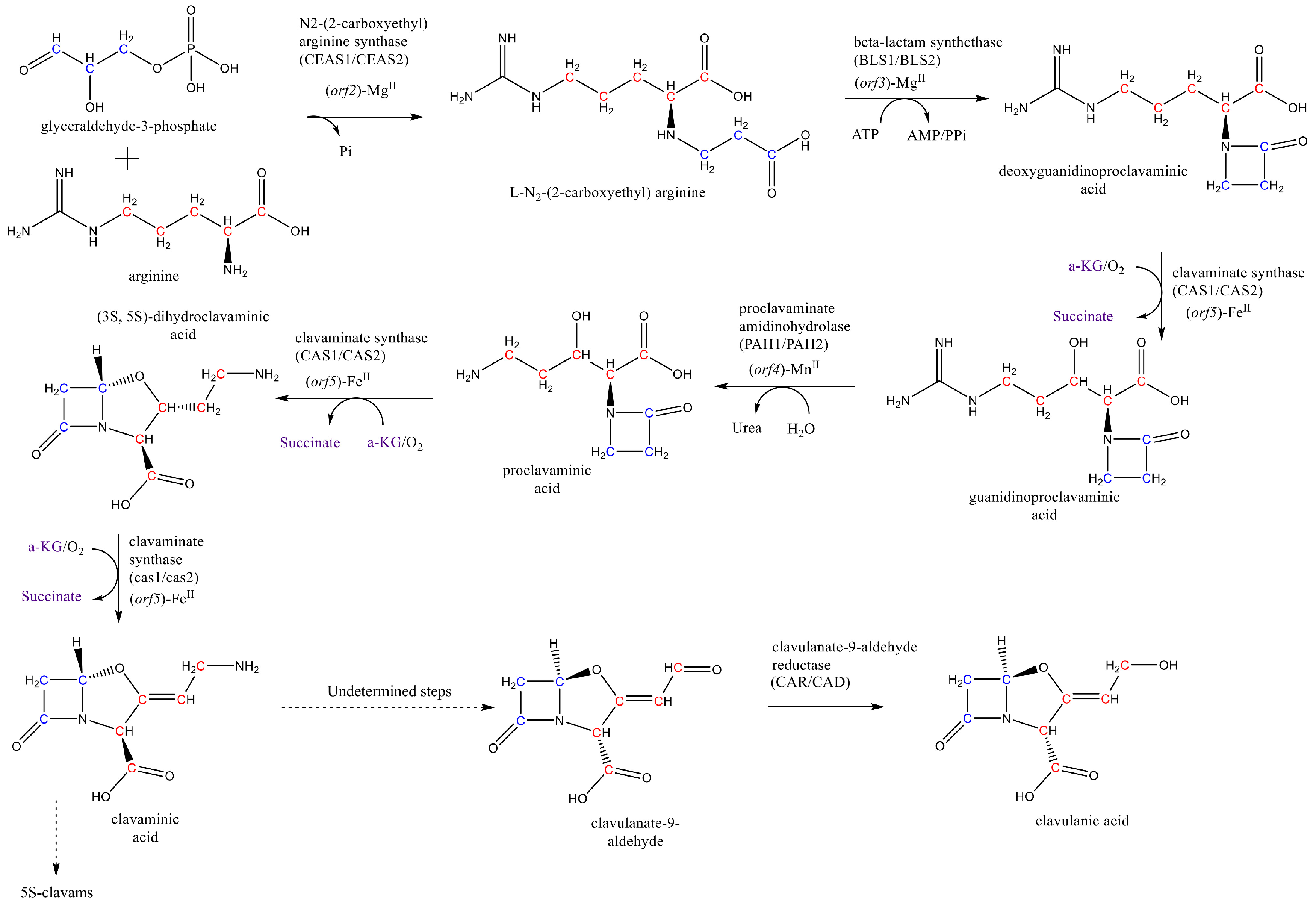
3. Results and Discussion
3.1. Development of a New and Improved Genome Scale Model of S. Clavuligerus
3.2. Model Validation
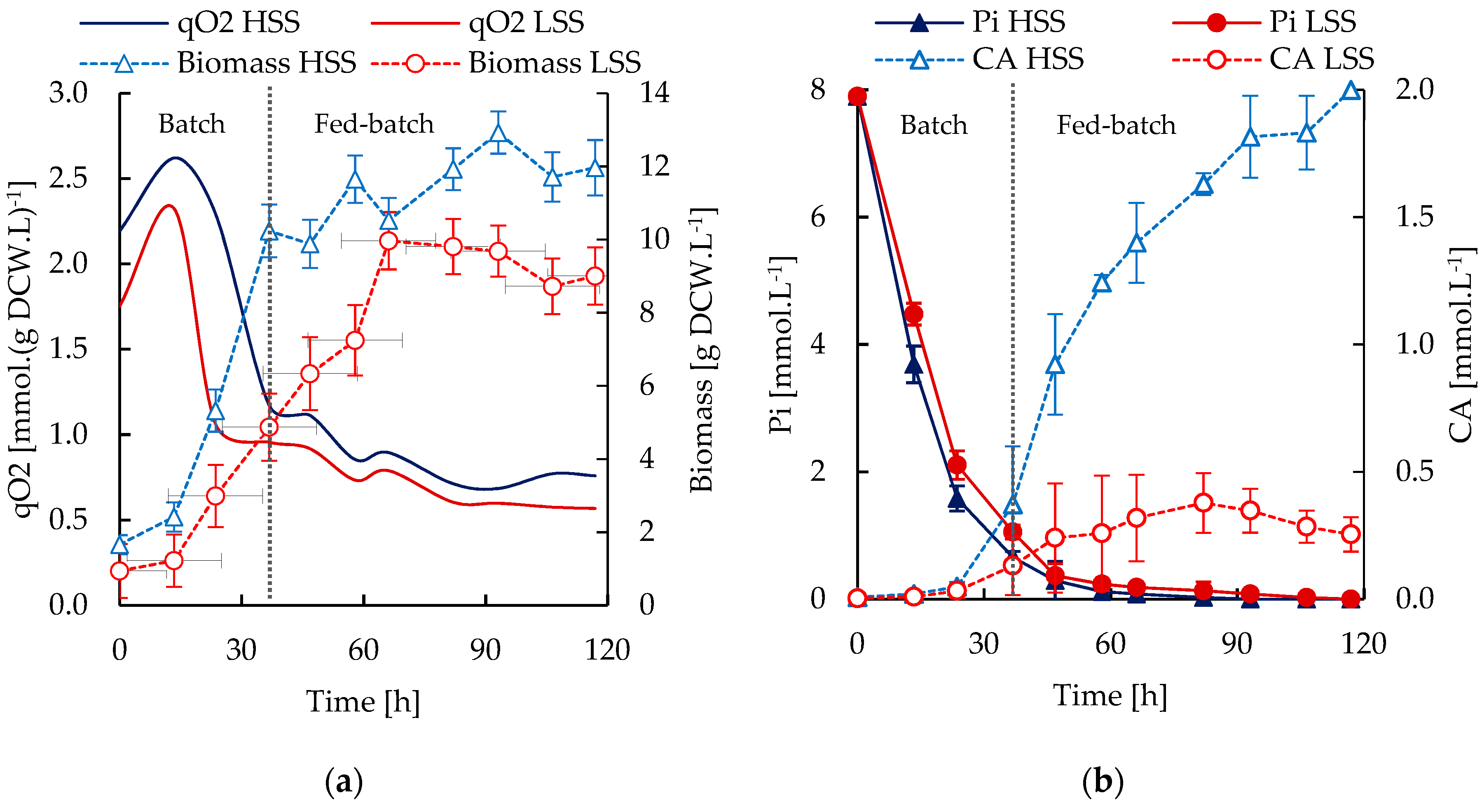
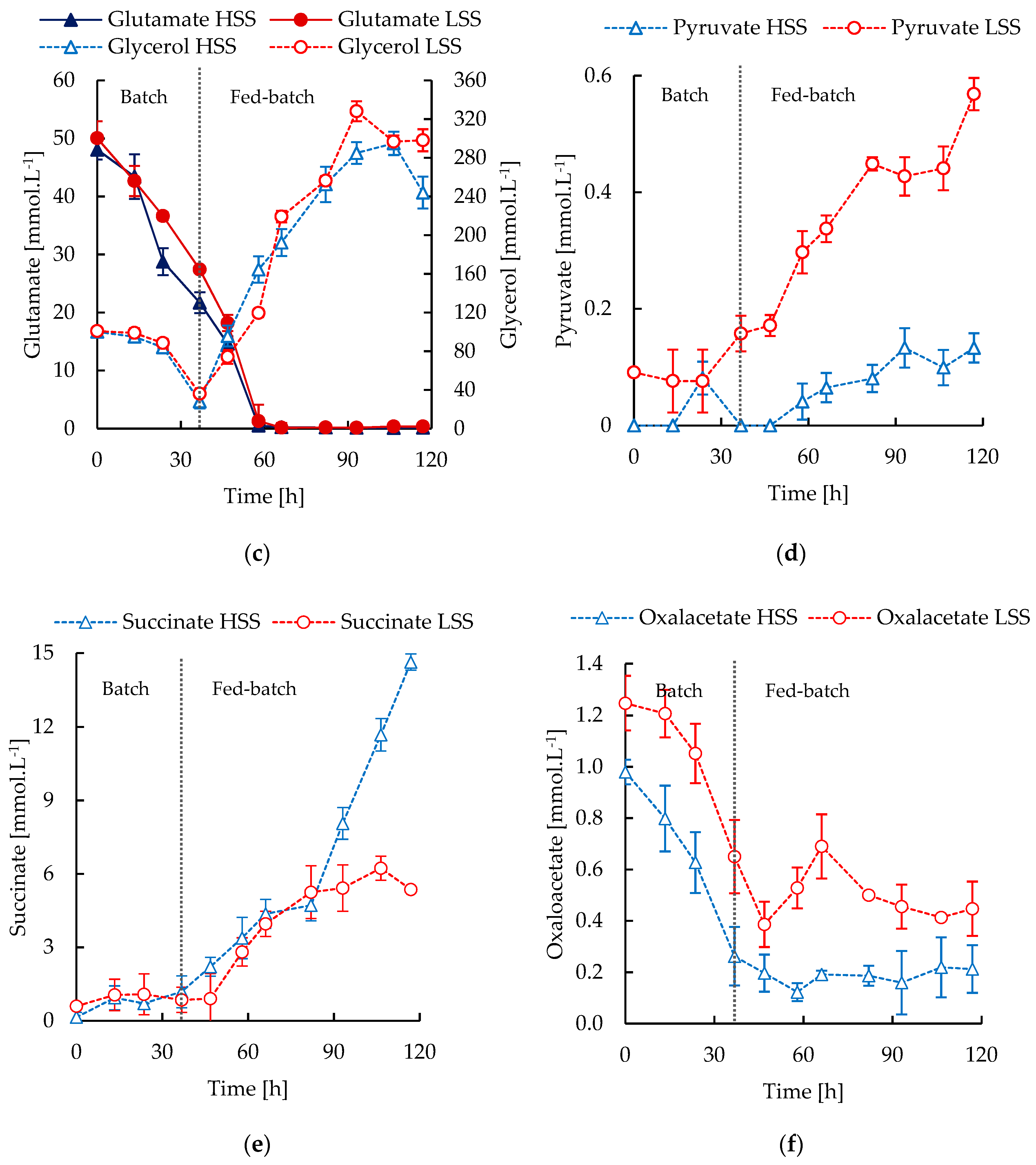
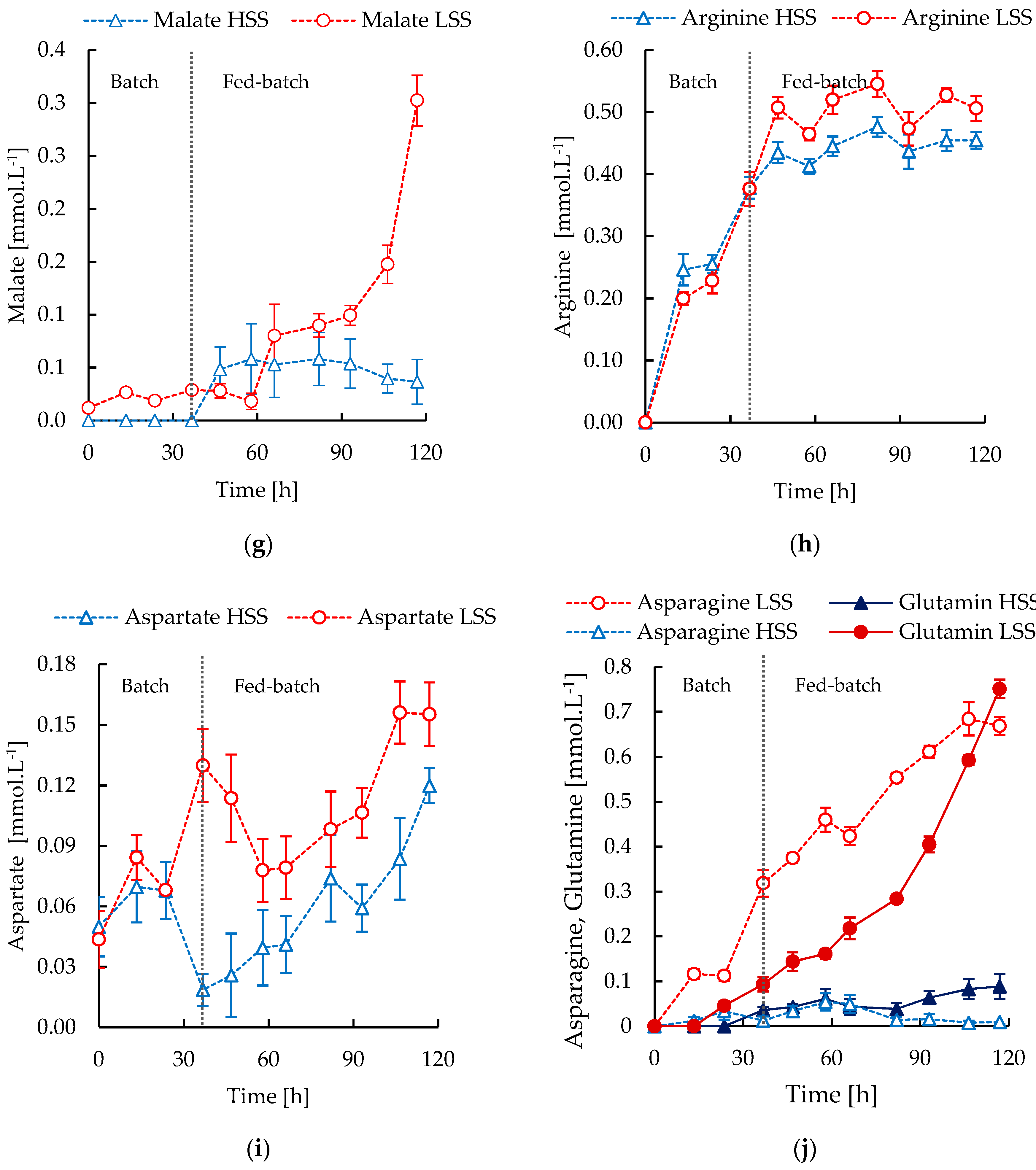
3.3. S. Clavuligerus Cultivation under High and Low Shear Stress Conditions
3.4. In Silico Metabolic Correlations of Environmental Cultivation Conditions and CA Biosynthesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gómez-Ríos, D.; Ramírez-Malule, H. Bibliometric analysis of recent research on multidrug and antibiotics resistance (2017–2018). J. Appl. Pharm. Sci. 2019, 9, 112–116. [Google Scholar] [CrossRef]
- Gómez-Ríos, D.; Ramírez-Malule, H.; Neubauer, P.; Junne, S.; Ríos-Estepa, R. Data of clavulanic acid and clavulanate-imidazole stability at low temperatures. Data Br. 2019, 23, 103775. [Google Scholar] [CrossRef]
- Gómez-Ríos, D.; Ramírez-Malule, H.; Neubauer, P.; Junne, S.; Ríos-Estepa, R. Degradation Kinetics of Clavulanic Acid in Fermentation Broths at Low Temperatures. Antibiotics 2019, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.-L.; Law, J.W.-F.; Chaiyakunapruk, N.; Jacob, S.A.; Palanisamy, U.D.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Fermentation Conditions that Affect Clavulanic Acid Production in Streptomyces clavuligerus: A Systematic Review. Front. Microbiol. 2016, 7, 522. [Google Scholar] [CrossRef] [PubMed]
- Saudagar, P.S.; Singhal, R.S. Optimization of nutritional requirements and feeding strategies for clavulanic acid production by Streptomyces clavuligerus. Bioresour. Technol. 2007, 98, 2010–2017. [Google Scholar] [CrossRef] [PubMed]
- Rosa, J.C.; Baptista Neto, A.; Hokka, C.O.; Badino, A.C. Influence of dissolved oxygen and shear conditions on clavulanic acid production by Streptomyces clavuligerus. Bioprocess Biosyst. Eng. 2005, 99–104. [Google Scholar] [CrossRef]
- Gómez-Ríos, D.; Junne, S.; Neubauer, P.; Ochoa, S.; Ríos-Estepa, R.; Ramírez-Malule, H. Characterization of the Metabolic Response of Streptomyces clavuligerus to Shear Stress in Stirred Tanks and Single-Use 2D Rocking Motion Bioreactors for Clavulanic Acid Production. Antibiotics 2019, 8, 168. [Google Scholar] [CrossRef]
- Jensen, S.E. Biosynthesis of clavam metabolites. J. Ind. Microbiol. Biotechnol. 2012, 39, 1407–1419. [Google Scholar] [CrossRef]
- Hamed, R.B.; Gomez-Castellanos, J.R.; Henry, L.; Ducho, C.; McDonough, M.A.; Schofield, C.J. The enzymes of β-lactam biosynthesis. Nat. Prod. Rep. 2013, 30, 21–107. [Google Scholar] [CrossRef]
- Ramirez-malule, H.; Junne, S.; Cruz-bournazou, M.N.; Neubauer, P. Streptomyces clavuligerus shows a strong association between TCA cycle intermediate accumulation and clavulanic acid biosynthesis. Appl. Microbiol. Biotechnol. 2018, 102, 4009–4402. [Google Scholar] [CrossRef]
- Bachmann, B.O.; Townsend, C.A. Kinetic mechanism of the β-lactam synthetase of streptomyces clavuligerus. Biochemistry 2000, 39, 11187–11193. [Google Scholar] [CrossRef] [PubMed]
- Tahlan, K.; Park, H.U.; Wong, A.; Beatty, P.H.; Jensen, S.E. Two Sets of Paralogous Genes Encode the Enzymes Involved in the Early Stages of Clavulanic Acid and Clavam Metabolite Biosynthesis in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 2004, 48, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ren, J.S.; Harlos, K.; McKinnon, C.H.; Clifton, I.J.; Schofield, C.J. Crystal structure of a clavaminate synthase-Fe(II)-2-oxoglutarate-substrate-NO complex: Evidence for metal centred rearrangements. FEBS Lett. 2002, 517, 7–12. [Google Scholar] [CrossRef]
- Caines, M.E.C.; Elkins, J.M.; Hewitson, K.S.; Schofield, C.J. Crystal Structure and Mechanistic Implications of N 2-(2-Carboxyethyl)arginine Synthase, the First Enzyme in the Clavulanic Acid Biosynthesis Pathway. J. Biol. Chem. 2004, 279, 5685–5692. [Google Scholar] [CrossRef]
- Wu, T.K.; Busby, R.W.; Houston, T.A.; Mcilwaine, D.B.; Egan, L.A.; Townsend, C.A.; Wu, T.; Busby, R.W.; Houston, T.A.; Ilwaine, D.B.M.C.; et al. Identification, Cloning, Sequencing, and overexpression of the gene encoding proclavaminate amidino hydrolase and characterization of protein function in clavulanic acid biosynthesis. J. Bacteriol. 1995, 177, 3714–3720. [Google Scholar]
- Shrestha, B.; Dhakal, D.; Darsandhari, S.; Pandey, R.P.; Pokhrel, A.R.; Jnawali, H.N.; Sohng, J.K. Heterologous production of clavulanic acid intermediates in Streptomyces venezuelae. Biotechnol. Bioprocess Eng. 2017, 22, 359–365. [Google Scholar] [CrossRef]
- Ramirez-Malule, H.; Restrepo, A.; Cardona, W.; Junne, S.; Neubauer, P.; Rios-Estepa, R. Inversion of the stereochemical configuration (3S, 5S)-clavaminic acid into (3R, 5R)-clavulanic acid: A computationally-assisted approach based on experimental evidence. J. Theor. Biol. 2016, 395, 40–50. [Google Scholar] [CrossRef]
- MacKenzie, A.K.; Kershaw, N.J.; Hernandez, H.; Robinson, C.V.; Schofield, C.J.; Andersson, I. Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the β-Lactamase Inhibitor Clavulanic Acid. Biochemistry 2007, 46, 1523–1533. [Google Scholar] [CrossRef]
- Kurt-Kizildoğan, A.; Vanli-Jaccard, G.; Mutlu, A.; Sertdemir, I.; Özcengiz, G. Genetic engineering of an industrial strain of Streptomyces clavuligerusfor further enhancement of clavulanic acid production. Turkish J. Biol. 2017, 41, 342–353. [Google Scholar] [CrossRef]
- Arulanantham, H.; Kershaw, N.J.; Hewitson, K.S.; Hughes, C.E.; Thirkettle, J.E.; Schofield, C.J. ORF17 from the clavulanic acid biosynthesis gene cluster catalyzes the ATP-dependent formation of N-glycyl-clavaminic acid. J. Biol. Chem. 2006, 281, 279–287. [Google Scholar] [CrossRef]
- López-Agudelo, V.A.; Baena, A.; Ramirez-Malule, H.; Ochoa, S.; Barrera, L.F.; Ríos-Estepa, R. Metabolic adaptation of two in silico mutants of Mycobacterium tuberculosis during infection. BMC Syst. Biol. 2017, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Alam, M.T.; Heijne, W.H.M.; Van Den Berg, M.A.; Müller, U.; Trefzer, A.; Bovenberg, R.A.L.; Breitling, R.; Takano, E. Genome-wide gene expression changes in an industrial clavulanic acid overproduction strain of Streptomyces clavuligerus. Microb. Biotechnol. 2011, 4, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Toro, L.; Pinilla, L.; Avignone-Rossa, C.; Ríos-Estepa, R. An enhanced genome-scale metabolic reconstruction of Streptomyces clavuligerus identifies novel strain improvement strategies. Bioprocess Biosyst. Eng. 2018, 41, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Cerón, S.; Galindo-Betancur, D.; Ramírez-Malule, H. Data set of in silico simulation for the production of clavulanic acid and cephamycin C by Streptomyces clavuligerus using a genome scale metabolic model. Data Br. 2019, 24, 103992. [Google Scholar] [CrossRef] [PubMed]
- Machado, D.; Andrejev, S.; Tramontano, M.; Patil, K.R. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018, 46, 7542–7553. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Zhong, C.; Zong, G.; Fu, J.; Liu, Z.; Zhang, G.; Qin, R. Complete Genome Sequence of Streptomyces clavuligerus F613-1, an Industrial Producer of Clavulanic Acid. Genome Announc. 2016, 4, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Bushell, M.E.; Kirk, S.; Zhao, H.; Avignone-rossa, C.A. Manipulation of the physiology of clavulanic acid biosynthesis with the aid of metabolic flux analysis. Enzyme Microb. Technol. 2006, 39, 149–157. [Google Scholar] [CrossRef]
- Cavallieri, A.P.; Baptista, A.S.; Leite, C.A.; Araujo, M.L.G. da C. A case study in flux balance analysis: Lysine, a cephamycin C precursor, can also increase clavulanic acid production. Biochem. Eng. J. 2016, 112, 42–53. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59. [Google Scholar] [CrossRef]
- Schellenberger, J.; Lewis, N.E.; Palsson, B. Elimination of thermodynamically infeasible loops in steady-state metabolic models. Biophys. J. 2011, 100, 544–553. [Google Scholar] [CrossRef]
- Henry, C.S.; Broadbelt, L.J.; Hatzimanikatis, V. Thermodynamics-based metabolic flux analysis. Biophys. J. 2007, 92, 1792–1805. [Google Scholar] [CrossRef] [PubMed]
- Noor, E. Removing both Internal and Unrealistic Energy-Generating Cycles in Flux Balance Analysis. arXiv 2018, arXiv:1803.04999v1. [Google Scholar]
- Desouki, A.A.; Jarre, F.; Gelius-Dietrich, G.; Lercher, M.J. CycleFreeFlux: Efficient removal of thermodynamically infeasible loops from flux distributions. Bioinformatics 2015, 31, 2159–2165. [Google Scholar] [CrossRef] [PubMed]
- Martínez, V.S.; Nielsen, L.K. NExT: Integration of Thermodynamic Constraints and Metabolomics Data into a Metabolic Network. In Metabolic Flux Analysis. Methods in Molecular Biology (Methods and Protocols); Krömer, J., Nielsen, L., Blank, L., Eds.; Humana Press: New York, NY, USA, 2012; Volume 1191, pp. 65–78. ISBN 9781617796173. [Google Scholar]
- Yousofshahi, M.; Ullah, E.; Stern, R.; Hassoun, S. MC3: A steady-state model and constraint consistency checker for biochemical networks. BMC Syst. Biol. 2013, 7, 129. [Google Scholar] [CrossRef]
- Lieven, C.; Beber, M.E.; Olivier, B.G.; Bergmann, F.T.; Ataman, M.; Babaei, P.; Bartell, J.A.; Blank, L.M.; Chauhan, S.; Correia, K.; et al. MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol. 2020, 38, 272–276. [Google Scholar] [CrossRef]
- Orth, J.D.; Thiele, I.; Palsson, B.O. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef]
- Holzhütter, H.G. The principle of flux minimization and its application to estimate stationary fluxes in metabolic networks. Eur. J. Biochem. 2004, 271, 2905–2922. [Google Scholar] [CrossRef]
- Roubos, J.A.; Krabben, P.; De Laat, W.; Heijnen, J.J. Clavulanic Acid Degradation in Streptomyces clavuligerus Fed-Batch Cultivations. Biotechnol. Prog. 2002, 18, 451–457. [Google Scholar] [CrossRef]
- Ramirez-Malule, H.; Junne, S.; López, C.; Zapata, J.; Sáez, A.; Neubauer, P.; Rios-Estepa, R. An improved HPLC-DAD method for clavulanic acid quantification in fermentation broths of Streptomyces clavuligerus. J. Pharm. Biomed. Anal. 2016, 120, 241–247. [Google Scholar] [CrossRef]
- Junne, S.; Klingner, A.; Kabisch, J.; Schweder, T.; Neubauer, P. A two-compartment bioreactor system made of commercial parts for bioprocess scale-down studies: Impact of oscillations on Bacillus subtilis fed-batch cultivations. Biotechnol. J. 2011, 6, 1009–1017. [Google Scholar] [CrossRef]
- Lemoine, A.; Martınez-Iturralde, N.M.; Spann, R.; Neubauer, P. Response of Corynebacterium glutamicum Exposed to Oscillating Cultivation Conditions in a Two- and a Novel Three-Compartment Scale- Down Bioreactor. Biotechnol. Bioeng. 2015, 112, 1220–1231. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Trefzer, A.; Kovalchuk, A.; van den Berg, M.; Müller, U.; Heijne, W.; Wu, L.; Alam, M.T.; Ronning, C.M.; Nierman, W.C.; et al. The Sequence of a 1.8-Mb Bacterial Linear Plasmid Reveals a Rich Evolutionary Reservoir of Secondary Metabolic Pathways. Genome Biol. Evol. 2010, 2, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Marashi, S.A.; Bockmayr, A. Flux coupling analysis of metabolic networks is sensitive to missing reactions. BioSystems 2011, 103, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Inoue, O.O.; Schmidell Netto, W.; Padilla, G.; Facciotti, M.C.R. Carbon catabolite repression of retamycin production by Streptomyces olindensis ICB20. Braz. J. Microbiol. 2007, 38, 58–61. [Google Scholar] [CrossRef][Green Version]
- Ciemniecki, J.A.; Newman, D.K. The Potential for Redox-Active Metabolites To Enhance or Unlock Anaerobic Survival Metabolisms in Aerobes. J. Bacteriol. 2020, 202, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Coze, F.; Gilard, F.; Tcherkez, G.; Virolle, M.-J.; Guyonvarch, A. Carbon-Flux Distribution within Streptomyces coelicolor Metabolism: A Comparison between the Actinorhodin-Producing Strain M145 and Its Non-Producing Derivative M1146. PLoS ONE 2013, 8, e84151. [Google Scholar] [CrossRef]
- Sandoval-Calderón, M.; Nguyen, D.D.; Kapono, C.A.; Herron, P.; Dorrestein, P.C.; Sohlenkamp, C. Plasticity of Streptomyces coelicolor Membrane Composition Under Different Growth Conditions and During Development. Front. Microbiol. 2015, 6, 1–13. [Google Scholar] [CrossRef]
- Gamboa-Suasnavart, R.A.; Valdez-Cruz, N.A.; Gaytan-Ortega, G.; Cereceda-Reynoso, G.I.; Cabrera-Santos, D.; López-Griego, L.; Klöckner, W.; Büchs, J.; Trujillo-Roldán, M.A. The metabolic switch can be activated in a recombinant strain of Streptomyces lividans by a low oxygen transfer rate in shake flasks. Microb. Cell Fact. 2018, 1–12. [Google Scholar] [CrossRef]
- Gallmetzer, M.; Burgstaller, W. Efflux of organic acids in Penicillium simplicissimum is an energy-spilling process, adjusting the catabolic carbon flow to the nutrient supply and the activity of catabolic pathways. Microbiology 2002, 148, 1143–1149. [Google Scholar] [CrossRef]
- Saudagar, P.S.; Survase, S.A.; Singhal, R.S. Clavulanic acid: A review. Biotechnol. Adv. 2008, 26, 335–351. [Google Scholar] [CrossRef]
- Virolle, M.-J. A Challenging View: Antibiotics Play a Role in the Regulation of the Energetic Metabolism of the Producing Bacteria. Antibiotics 2020, 9, 83. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, C.; Martínez-Castro, M. Regulation of the phosphate metabolism in Streptomyces genus: Impact on the secondary metabolites. Appl. Microbiol. Biotechnol. 2019, 103, 1643–1658. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-García, A.; Barreiro, C.; Santos-Beneit, F.; Sola-Landa, A.; Martín, J.F. Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ΔphoP mutant. Proteomics 2007, 7, 2410–2429. [Google Scholar] [CrossRef] [PubMed]
- Esnault, C.; Dulermo, T.; Smirnov, A.; Askora, A.; David, M.; Deniset-Besseau, A.; Holland, I.B.; Virolle, M.J. Strong antibiotic production is correlated with highly active oxidative metabolism in Streptomyces coelicolor M145. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Millan-Oropeza, A.; Henry, C.; Lejeune, C.; David, M.; Virolle, M.-J. Expression of genes of the Pho regulon is altered in Streptomyces coelicolor. Sci. Rep. 2020, 10, 8492. [Google Scholar] [CrossRef]
- Cho, H.; Uehara, T.; Bernhardt, T.G. Beta-Lactam Antibiotics Induce a Lethal Malfunctioning of the Bacterial Cell Wall Synthesis Machinery. Cell 2014, 159, 1300–1311. [Google Scholar] [CrossRef]

| Feature | iMM865 | iLT1021 | iGG1534 | iDG1237 |
|---|---|---|---|---|
| Metabolites | 1173 | 1162 | 1199 | 1518 |
| Reactions | 1492 | 1494 | 1534 | 2177 |
| Genes | 864 | 1021 | 871 | 1237 |
| Reversible reactions | 1492 | 576 | 610 | 707 |
| Dead-end metabolites | 311 | 333 | 338 | 48 |
| Coupled reactions | 0 | 422 | 338 | 880 |
| Inconsistent coupling | 0 | 127 | 35 | 0 |
| Zero-flux reactions | 383 | 473 | 486 | 105 |
| Unbounded reactions | 1024 | 496 | 598 | 86 |
| TICs | 411 | 135 | 185 | 9 |
| Constraints | iMM865 | iLT1021 | iGG1534 | iDG1237 | μexp | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| vex,Pi | vex,Glyc | μ | vex,CA | μ | vex,CA | μ | vex,CA | μ | vex,CA | ||
| -0.20 | -0.50 | 1.60 | 0 | 0.03 | 0 | 0.92 | 0 | 0.03 | 0 | 0.04 | [27] |
| -0.20 | -0.60 | 1.61 | 0 | 0.04 | 0 | 0.93 | 0 | 0.04 | 0 | 0.05 | [27] |
| -0.20 | -0.93 | 1.67 | 0 | 0.06 | 0 | 0.97 | 0 | 0.05 | 0 | 0.07 | [27] |
| -0.20 | -2.18 | 2.44 | 0 | 0.12 | 0 | 1.55 | 0 | 0.09 | 0 | 0.09 | [27] |
| 0 | -0.72 | 2.97 | 0 | 0.05 | 0 | 1.94 | 0 | 0.05 | 0.03 | 0.05 | [10] |
| 0 | -1.11 | 2.90 | 0 | 0.05 | 0 | 1.90 | 0 | 0.05 | 0.04 | 0.05 | [10] |
| 0 | -0.97 | 2.75 | 0 | 0.05 | 0 | 1.78 | 0 | 0.05 | 0.03 | 0.04 | [10] |
| MSE | 1.506 | 0.393 | 0.700 | 0.172 | |||||||
| Time (h) | Scenario | Shear Condition | Experimental Constraints | Results | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| vex,Glyc | vex,Pi | vex,NH4 | vex,Glu | μ | vex,CA | vex,O2 | RQ * | |||
| 0–22 | SC1 | HSS & LSS | −0.5 | −0.1 | −0.2 | −0.5 | 0.06 | 0 | −1.6 | 1.01 |
| 22–37 | SC2 | HSS & LSS | −2.0 | −0.1 | −0.9 | −0.5 | 0.13 | 0 | −3.84 | 1.20 |
| 37–68 | SC3 | HSS | −0.6 | 0 | −0.2 | −0.1 | 0.02 | 0.02 | −0.53 | 0.94 |
| 37–68 | SC4 | LSS | −0.6 | 0 | −0.2 | −0.1 | 0.02 | 6 × 10−3 | −0.48 | 1.01 * |
| 68–93 | SC5 | HSS | −0.6 | 0 | −0.2 | 0 | 0.01 | 9 × 10−3 | −0.48 | 0.87 |
| 68–93 | SC6 | LSS | −0.6 | 0 | −0.2 | 0 | 0.01 | 0 | −0.42 | 1.01 * |
| MSE | 1.8 × 10−4 | 9.8 × 10−5 | 0.28 | 0.02 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Ríos, D.; López-Agudelo, V.A.; Ramírez-Malule, H.; Neubauer, P.; Junne, S.; Ochoa, S.; Ríos-Estepa, R. A Genome-Scale Insight into the Effect of Shear Stress During the Fed-Batch Production of Clavulanic Acid by Streptomyces Clavuligerus. Microorganisms 2020, 8, 1255. https://doi.org/10.3390/microorganisms8091255
Gómez-Ríos D, López-Agudelo VA, Ramírez-Malule H, Neubauer P, Junne S, Ochoa S, Ríos-Estepa R. A Genome-Scale Insight into the Effect of Shear Stress During the Fed-Batch Production of Clavulanic Acid by Streptomyces Clavuligerus. Microorganisms. 2020; 8(9):1255. https://doi.org/10.3390/microorganisms8091255
Chicago/Turabian StyleGómez-Ríos, David, Victor A. López-Agudelo, Howard Ramírez-Malule, Peter Neubauer, Stefan Junne, Silvia Ochoa, and Rigoberto Ríos-Estepa. 2020. "A Genome-Scale Insight into the Effect of Shear Stress During the Fed-Batch Production of Clavulanic Acid by Streptomyces Clavuligerus" Microorganisms 8, no. 9: 1255. https://doi.org/10.3390/microorganisms8091255
APA StyleGómez-Ríos, D., López-Agudelo, V. A., Ramírez-Malule, H., Neubauer, P., Junne, S., Ochoa, S., & Ríos-Estepa, R. (2020). A Genome-Scale Insight into the Effect of Shear Stress During the Fed-Batch Production of Clavulanic Acid by Streptomyces Clavuligerus. Microorganisms, 8(9), 1255. https://doi.org/10.3390/microorganisms8091255







