Enhancement of the Molecular and Serological Assessment of Hepatitis E Virus in Milk Samples
Abstract
:1. Introduction
2. Materials and Methods
2.1. HEV Inoculum Used in the Study
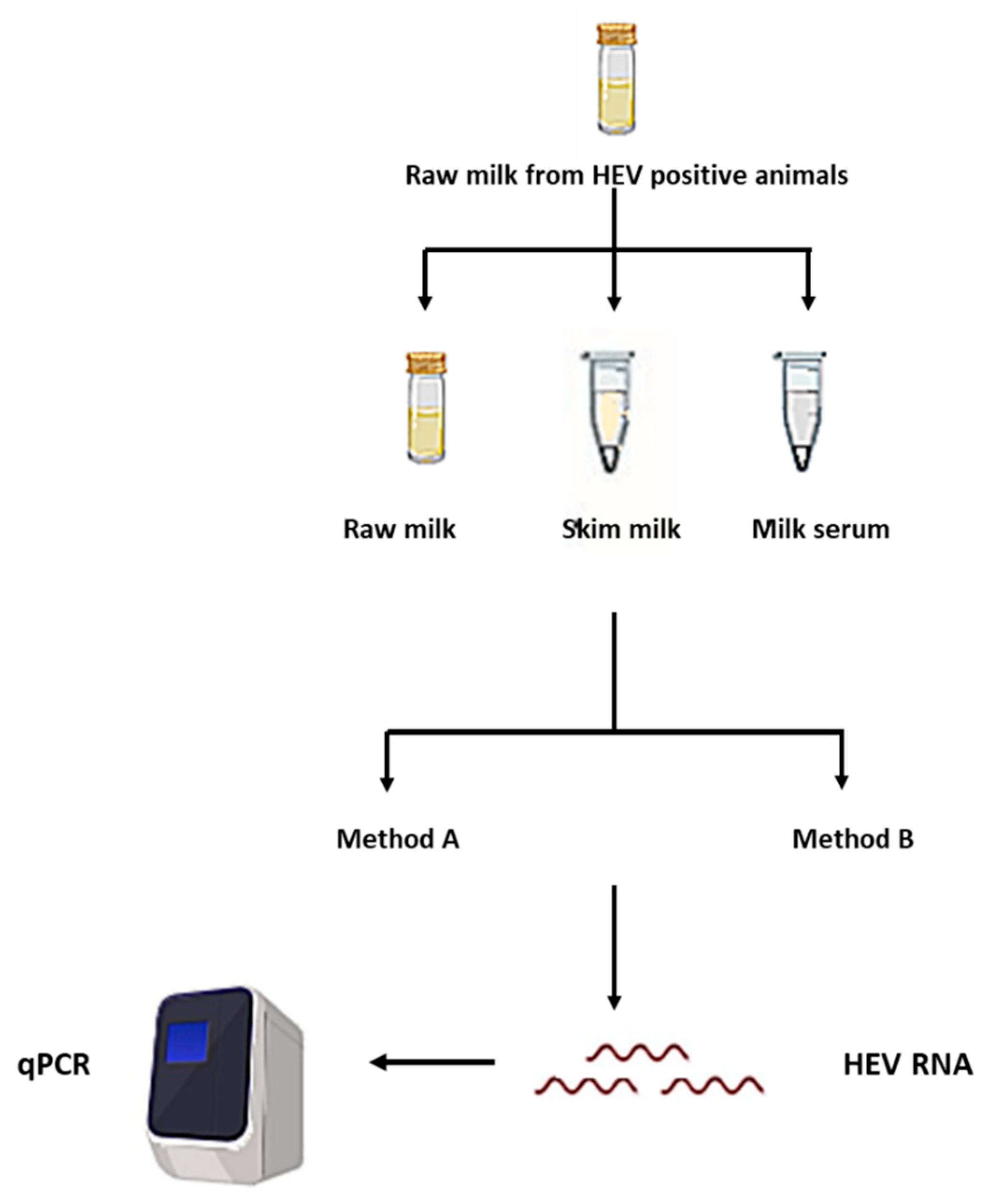
2.2. Processing of the Milk Samples and Separation of the Milk Components
2.3. RNA Extraction Methods
2.4. HEV RNA qRT-PCR
2.5. Evaluation of the Effect of Extraction Procedures and Milk Components on Potential HEV Positive Milk Samples
2.6. Detection of Anti-HEV IgG in the Milk Samples
2.7. Detection of HEV Ag in Goat and Cow Milk Samples
2.8. Statistics
3. Results
3.1. HEV Molecular Assay is affected by the Milk Matrix
3.2. Milk Components Affect the Performance of HEV Molecular Assay
3.3. Evaluation of the Effect of Extraction Procedures and Milk Components on Potential HEV Positive Milk Samples
3.4. Removal of the Fat Globules Increased the Performance of Anti-HEV IgG and HEV Ag Assays in the Milk Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rein, D.B.; Stevens, G.A.; Theaker, J.; Wittenborn, J.S.; Wiersma, S.T. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 2012, 55, 988–997. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Simmonds, P.; Izopet, J.; Oliveira-Filho, E.F.; Ulrich, R.G.; Johne, R.; Koenig, M.; Jameel, S.; Harrison, T.J.; Meng, X.-J.; et al. Proposed reference sequences for hepatitis E virus subtypes. J. Gen. Virol. 2016, 97, 537–542. [Google Scholar] [CrossRef]
- Smith, D.B.; Simmonds, P.; Jameel, S.; Emerson, S.U.; Harrison, T.J.; Meng, X.-J.; Okamoto, H.; Van Der Poel, W.H.M.; Purdy, M.A.; members of the International Committee on the Taxonomy of Viruses Hepeviridae Study Group; et al. Consensus proposals for classification of the family Hepeviridae. J. Gen. Virol. 2014, 95, 2223–2232. [Google Scholar] [CrossRef] [PubMed]
- Sayed, I.M.; Vercauteren, K.; Abdelwahab, S.F.; Meuleman, P. The emergence of hepatitis E virus in Europe. Futur. Virol. 2015, 10, 763–778. [Google Scholar] [CrossRef] [Green Version]
- Sayed, I.M.; Vercouter, A.S.; Abdelwahab, S.F.; Vercauteren, K.; Meuleman, P. Is hepatitis E virus an emerging problem in industrialized countries. Hepatology 2015, 62, 1883–1892. [Google Scholar] [CrossRef] [Green Version]
- Sayed, I.M.; Elkhawaga, A.A.; El-Mokhtar, M.A. In vivo models for studying Hepatitis E virus infection; Updates and applications. Virus Res. 2019, 274, 197765. [Google Scholar] [CrossRef]
- Sayed, I.M.; Meuleman, P. Updates in Hepatitis E virus (HEV) field; lessons learned from human liver chimeric mice. Rev. Med. Virol. 2020, 30, e2086. [Google Scholar] [CrossRef]
- Pavio, N.; Doceul, V.; Bagdassarian, E.; Johne, R. Recent knowledge on hepatitis E virus in Suidae reservoirs and transmission routes to human. Vet. Res. 2017, 48, 78. [Google Scholar] [CrossRef] [Green Version]
- Pavio, N.; Meng, X.-J.; Doceul, V. Zoonotic origin of hepatitis E. Curr. Opin. Virol. 2015, 10, 34–41. [Google Scholar] [CrossRef]
- Meng, X.J.; Purcell, R.H.; Halbur, P.G.; Lehman, J.R.; Webb, D.M.; Tsareva, T.S.; Haynes, J.S.; Thacker, B.J.; Emerson, S.U. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 1997, 94, 9860–9865. [Google Scholar] [CrossRef] [Green Version]
- Demirci, M.; Yigin, A.; Unlu, O.; Kilic, A.S. Detection of HEV RNA amounts and genotypes in raw milks obtained from different animals. Mikrobiyol. Bul. 2019, 53, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Li, Y.; Yu, W.; Jing, S.; Wang, J.; Long, F.; He, Z.; Yang, C.; Bi, Y.; Cao, W.; et al. Excretion of infectious hepatitis E virus into milk in cows imposes high risks of zoonosis. Hepatology. 2016, 64, 350–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Mokhtar, M.A.; Elkhawaga, A.A.; Sayed, I.M. Assessment of hepatitis E virus (HEV) in the edible goat products pointed out a risk for human infection in Upper Egypt. Int. J. Food Microbiol. 2020, 330, 108784. [Google Scholar] [CrossRef] [PubMed]
- Sayed, I.M.; Elkhawaga, A.A.; El-Mokhtar, M.A. Circulation of hepatitis E virus (HEV) and/or HEV-like agent in non-mixed dairy farms could represent a potential source of infection for Egyptian people. Int. J. Food Microbiol. 2020, 317, 108479. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.; Tsang, A.K.L.; Joseph, M.; Wong, E.Y.; Tang, Y.; Sivakumar, S.; Xie, J.; Bai, R.; et al. New Hepatitis E Virus Genotype in Camels, the Middle East. Emerg. Infect. Dis. 2014, 20, 1044–1048. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Teng, J.L.L.; Lau, S.K.P.; Sridhar, S.; Fu, H.; Gong, W.; Li, M.; Xu, Q.; He, Y.; Zhuang, H.; et al. Transmission of a Novel Genotype of Hepatitis E Virus from Bactrian Camels to Cynomolgus Macaques. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, G.-H.; Tan, B.-H.; Teo, E.C.-Y.; Lim, S.-G.; Dan, Y.-Y.; Wee, A.; Aw, P.P.K.; Zhu, Y.O.; Hibberd, M.; Tan, C.-K.; et al. Chronic Infection With Camelid Hepatitis E Virus in a Liver Transplant Recipient Who Regularly Consumes Camel Meat and Milk. Gastroenterology 2016, 150, 355–357.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, M.; Nishizawa, T.; Sato, H.; Sato, Y.; Jirintai; Nagashima, S.; Okamoto, H. Analysis of the full-length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J. Gen. Virol. 2011, 92, 902–908. [Google Scholar] [CrossRef]
- Butot, S.; Putallaz, T.; Sánchez, G. Procedure for Rapid Concentration and Detection of Enteric Viruses from Berries and Vegetables. Appl. Environ. Microbiol. 2007, 73, 186–192. [Google Scholar] [CrossRef] [Green Version]
- Hennechart-Collette, C.; Martin-Latil, S.; Fraisse, A.; Perelle, S. Comparison of three extraction methods to detect noroviruses in dairy products. Food Microbiol. 2017, 61, 113–119. [Google Scholar] [CrossRef]
- Summa, M.; Von Bonsdorff, C.-H.; Maunula, L. Evaluation of four virus recovery methods for detecting noroviruses on fresh lettuce, sliced ham, and frozen raspberries. J. Virol. Methods 2012, 183, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Blaise-Boisseau, S.; Hennechart-Collette, C.; Guillier, L.; Perelle, S. Duplex real-time qRT-PCR for the detection of hepatitis A virus in water and raspberries using the MS2 bacteriophage as a process control. J. Virol. Methods 2010, 166, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Suffredini, E.; Lanni, L.; Arcangeli, G.; Pepe, T.; Mazzette, R.; Ciccaglioni, G.; Croci, L. Qualitative and quantitative assessment of viral contamination in bivalve molluscs harvested in Italy. Int. J. Food Microbiol. 2014, 184, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Maunula, L.; Kaupke, A.; Vasickova, P.; Soderberg, K.; Kozyra, I.; Lazić, S.; Van Der Poel, W.H.; Bouwknegt, M.; Rutjes, S.; Willems, K.A.; et al. Tracing enteric viruses in the European berry fruit supply chain. Int. J. Food Microbiol. 2013, 167, 177–185. [Google Scholar] [CrossRef]
- Battistini, R.; Rossini, I.; Listorti, V.; Ercolini, C.; Maurella, C.; Serracca, L. HAV detection from milk-based products containing soft fruits: Comparison between four different extraction methods. Int. J. Food Microbiol. 2020, 328, 108661. [Google Scholar] [CrossRef]
- Yavarmanesh, M.; Abbaszadegan, M.; Alum, A.; Mortazavi, A.; Najafi, M.B.H.; Bassami, M.R.; Nassiri, M.R. Impact of Milk Components on Recovery of Viral RNA from MS2 Bacteriophage. Food Environ. Virol. 2013, 5, 103–109. [Google Scholar] [CrossRef]
- Yavarmanesh, M.; Alum, A.; Abbaszadegan, M. Occurrence of Noroviruses and Their Correlation with Microbial Indicators in Raw Milk. Food Environ. Virol. 2015, 7, 232–238. [Google Scholar] [CrossRef]
- Vercouter, A.-S.; Sayed, I.M.; Lipkens, Z.; De Bleecker, K.; De Vliegher, S.; Colman, R.; Koppelman, M.H.; Supré, K.; Meuleman, P. Absence of zoonotic hepatitis E virus infection in Flemish dairy cows. Int. J. Food Microbiol. 2018, 281, 54–59. [Google Scholar] [CrossRef]
- Rivero-Juárez, A.; Rodriguez-Cano, D.; Frias, M.; Cuenca-Lopez, F.; Rivero, A. Isolation of Hepatitis E Virus From Breast Milk During Acute Infection: Table 1. Clin. Infect. Dis. 2016, 62, 1464. [Google Scholar] [CrossRef] [Green Version]
- Long, F.; Yu, W.; Yang, C.; Wang, J.; Li, Y.; Li, Y.; Huang, F. High prevalence of hepatitis E virus infection in goats. J. Med Virol. 2017, 89, 1981–1987. [Google Scholar] [CrossRef]
- Sayed, I.M.; Seddik, M.I.; Gaber, M.A.; Saber, S.H.; Mandour, S.A.; El-Mokhtar, M.A. Replication of Hepatitis E Virus (HEV) in Primary Human-Derived Monocytes and Macrophages In Vitro. Vaccines 2020, 8, 239. [Google Scholar] [CrossRef] [PubMed]
- El-Mokhtar, M.A.; Othman, E.R.; Khashbah, M.Y.; Ismael, A.; Ghaliony, M.A.; Seddik, M.I.; Sayed, I.M. Evidence of the Extrahepatic Replication of Hepatitis E Virus in Human Endometrial Stromal Cells. Pathogens 2020, 9, 295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- A Baylis, S.; Blümel, J.; Mizusawa, S.; Matsubayashi, K.; Sakata, H.; Okada, Y.; Nübling, C.M.; Hanschmann, K.-M.O. World Health Organization International Standard to Harmonize Assays for Detection of Hepatitis E Virus RNA. Emerg. Infect. Dis. 2013, 19, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Blans, K.; Hansen, M.S.; Sørensen, L.V.; Hvam, M.L.; Howard, K.A.; Moeller, A.; Wiking, L.; Larsen, L.B.; Rasmussen, J.T. Pellet-free isolation of human and bovine milk extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2017, 6, 1294340. [Google Scholar] [CrossRef]
- Bohren, H.; Wenner, V. Natural State of Milk Proteins. I. Composition of the Micellar and Soluble Casein of Milk After Ultracentrifugal Sedimentation. J. Dairy Sci. 1961, 44, 1213–1223. [Google Scholar] [CrossRef]
- Sayed, I.M.; Verhoye, L.; Cocquerel, L.; Abravanel, F.; Foquet, L.; Montpellier, C.; Debing, Y.; Farhoudi, A.; Wychowski, C.; Dubuisson, J.; et al. Study of hepatitis E virus infection of genotype 1 and 3 in mice with humanised liver. Gut 2017, 66, 920–929. [Google Scholar] [CrossRef]
- Sayed, I.M.; Foquet, L.; Verhoye, L.; Abravanel, F.; Farhoudi, A.; Leroux-Roels, G.; Izopet, J.; Meuleman, P. Transmission of hepatitis E virus infection to human-liver chimeric FRG mice using patient plasma. Antivir. Res. 2017, 141, 150–154. [Google Scholar] [CrossRef]
- Sayed, I.M.; Meuleman, P. Murine Tissues of Human Liver Chimeric Mice Are Not Susceptible to Hepatitis E Virus Genotypes 1 and 3. J. Infect. Dis. 2017, 216, 919–920. [Google Scholar] [CrossRef]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.; Hill, V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef]
- Wilrich, C.; Wilrich, P.-T. Estimation of the POD Function and the LOD of a Qualitative Microbiological Measurement Method. J. AOAC Int. 2009, 92, 1763–1772. [Google Scholar] [CrossRef] [Green Version]
- Sayed, I.M.; Verhoye, L.; Montpellier, C.; Abravanel, F.; Izopet, J.; Cocquerel, L.; Meuleman, P. Hepatitis E Virus (HEV) Open Reading Frame 2 Antigen Kinetics in Human-Liver Chimeric Mice and Its Impact on HEV Diagnosis. J. Infect. Dis. 2019, 220, 811–819. [Google Scholar] [CrossRef]
- Bronzwaer, S.; Kass, G.; Robinson, T.; Tarazona, J.; Verhagen, H.; Verloo, D.; Vrbos, D.; Hugas, M. Food Safety Regulatory Research Needs 2030. EFSA J. 2019, 17, e170622. [Google Scholar] [PubMed]
- Abravanel, F.; Lhomme, S.; El Costa, H.; Schvartz, B.; Peron, J.-M.; Kamar, N.; Izopet, J. Rabbit Hepatitis E Virus Infections in Humans, France. Emerg. Infect. Dis. 2017, 23, 1191–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colson, P.; Borentain, P.; Queyriaux, B.; Kaba, M.; Moal, V.; Gallian, P.; Heyries, L.; Raoult, D.; Gerolami, R. Pig Liver Sausage as a Source of Hepatitis E Virus Transmission to Humans. J. Infect. Dis. 2010, 202, 825–834. [Google Scholar] [CrossRef] [Green Version]
- Said, B.; Ijaz, S.; Kafatos, G.; Booth, L.; Thomas, H.L.; Walsh, A.; Ramsay, M.; Morgan, D. Hepatitis E Outbreak on Cruise Ship. Emerg. Infect. Dis. 2009, 15, 1738–1744. [Google Scholar] [CrossRef]
- Mesquita, J.R.; Santos-Ferreira, N.; Ferreira, A.S.; Albuquerque, C.; Nóbrega, C.; Esteves, F.; Cruz, R.; Vala, H.; Nascimento, M.S.J. Increased risk of hepatitis E virus infection in workers occupationally exposed to sheep. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.B.; Kelton, D.F.; Hietala, S.K.; Duffield, T.F. Evaluation of enzyme-linked immunosorbent assays performed on milk and serum samples for detection of neosporosis and leukosis in lactating dairy cows. Can. Vet. J. 2013, 54, 347–352. [Google Scholar] [PubMed]
- Cuevas-Ferrando, E.; Martínez-Murcia, A.; Pérez-Cataluña, A.; Sánchez, G.; Randazzo, W. Assessment of ISO Method 15216 to Quantify Hepatitis E Virus in Bottled Water. Microorganisms 2020, 8, 730. [Google Scholar] [CrossRef]




| Virus Inoculum (IU/mL) | Method A (QIAamp Viral RNA Mini) | Method A (High Pure Viral Nucleic Acid, Roche) | Method B | ||||
|---|---|---|---|---|---|---|---|
| PBS | Milk | PBS | Milk | PBS | Milk | ||
| Pos/Total a | Pos/Total a | Pos/Total a | Pos/Total a | Pos/Total a | Pos/Total a | ||
| Positive control stool | 3 × 105 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| 3 × 104 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | |
| 3 × 103 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 2/3 | |
| 3 × 102 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 1/3 | |
| 3 × 101 | 2/3 | 0/3 | 2/3 | 0/3 | 3/3 | 0/3 | |
| 3 | 0/3 | 0/3 | 0/3 | 0/3 | 1/3 | 0/3 | |
| LoD95% of HEV RNA (IU/mL) b | 96.6 | 300 | 96.6 | 300 | 19.4 | 6.127 × 103 | |
| LoD95% of IPC (copies/mL) | 64.2 | 651.8 | 64.2 | 651.8 | 12.9 | 1412.3 | |
| Virus Stock | Undiluted RNA | 1/10 Diluted RNA | ||||
|---|---|---|---|---|---|---|
| Pos/Total a | Ct b | Mean HEV Recovery | Pos/Total a | Ct b | Mean HEV Recovery | |
| 3 × 105 | 3/3 | 26.12 ± 0.35 | 3.75 | 3/3 | 29.34 ± 0.43 | 2.28% |
| 3 × 104 | 3/3 | 29.67 ± 0.21 | 1.71 | 1/3 | 32.31 | 1% |
| 3 × 103 | 2/3 | 32.76 ± 0.51 | 1.16 | 0/3 | NA | NA |
| Method | LoD95% a | Whole Milk Matrix b | Skim Milk Matrix | Milk Serum |
|---|---|---|---|---|
| Method A (manual extraction) | LoD95% HEV (IU/mL) | 300 | 300 | 300 |
| LoD95% IPC (copies/mL) | 651.8 | 200 | 140.22 | |
| Method B (automated extraction) | LoD95% HEV (IU/mL) | 6.127 × 103 | 977 IU/mL | 96.34 IU/mL |
| LoD95% IPC (copies/mL) | 1412.3 | 140.22 | 24.98 |
| Sample | Reference | HEV Markers in Milk (Data from Previous Reports) | Detection of HEV RNA | |||||
|---|---|---|---|---|---|---|---|---|
| Method A | Method B | |||||||
| Whole Milk | Skim Milk | Milk Serum | Whole Milk | Skim Milk | Milk Serum | |||
| Cow milk (n = 8) | [14] | Sample (n = 1) (+ve for anti-HEV IgG, HEV Ag, and HEV RNA) | + a 1/1 | + 1/1 | + 1/1 | − b 0/1 | + 1/1 | + 1/1 |
| Samples (n = 7) positive for anti-HEV IgG and negative for HEV RNA and HEV Ag | − 0/7 | − 0/7 | − 0/7 | − 0/7 | − 0/7 | + (1 out of 7) Titer 2.5 × 102 IU/mL | ||
| Goat milk (n = 20) | [13] | Samples (n = 2) (+ve for anti-HEV IgG, HEV Ag, and HEV RNA) | + 2/2 | + 2/2 | + 2/2 | - 0/2 | + 1/2 | + 2/2 |
| Samples (n = 3) (+ve for anti-HEV IgG and HEV Ag and negative for HEV RNA | − 0/3 | − 0/3 | − 0/3 | − 0/3 | − 0/3 | + (1 out of 3) Titer 1.87 × 102 IU/mL | ||
| Samples (n = 15) positive for anti-HEV IgG only | − 0/15 | − 0/15 | − 0/15 | − 0/15 | − 0/15 | + (1 out of 15) Titer 1.68 × 102 IU/mL | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sayed, I.M.; Hammam, A.R.A.; Elfaruk, M.S.; Alsaleem, K.A.; Gaber, M.A.; Ezzat, A.A.; Salama, E.H.; Elkhawaga, A.A.; El-Mokhtar, M.A. Enhancement of the Molecular and Serological Assessment of Hepatitis E Virus in Milk Samples. Microorganisms 2020, 8, 1231. https://doi.org/10.3390/microorganisms8081231
Sayed IM, Hammam ARA, Elfaruk MS, Alsaleem KA, Gaber MA, Ezzat AA, Salama EH, Elkhawaga AA, El-Mokhtar MA. Enhancement of the Molecular and Serological Assessment of Hepatitis E Virus in Milk Samples. Microorganisms. 2020; 8(8):1231. https://doi.org/10.3390/microorganisms8081231
Chicago/Turabian StyleSayed, Ibrahim M., Ahmed R. A. Hammam, Mohamed Salem Elfaruk, Khalid A. Alsaleem, Marwa A. Gaber, Amgad A. Ezzat, Eman H. Salama, Amal A. Elkhawaga, and Mohamed A. El-Mokhtar. 2020. "Enhancement of the Molecular and Serological Assessment of Hepatitis E Virus in Milk Samples" Microorganisms 8, no. 8: 1231. https://doi.org/10.3390/microorganisms8081231
APA StyleSayed, I. M., Hammam, A. R. A., Elfaruk, M. S., Alsaleem, K. A., Gaber, M. A., Ezzat, A. A., Salama, E. H., Elkhawaga, A. A., & El-Mokhtar, M. A. (2020). Enhancement of the Molecular and Serological Assessment of Hepatitis E Virus in Milk Samples. Microorganisms, 8(8), 1231. https://doi.org/10.3390/microorganisms8081231








