Isolation, Characterization, and Pathogenicity of Two Pseudomonas syringae Pathovars from Populus trichocarpa Seeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Sources for Bacteria, Plants, Seeds, and Chemicals
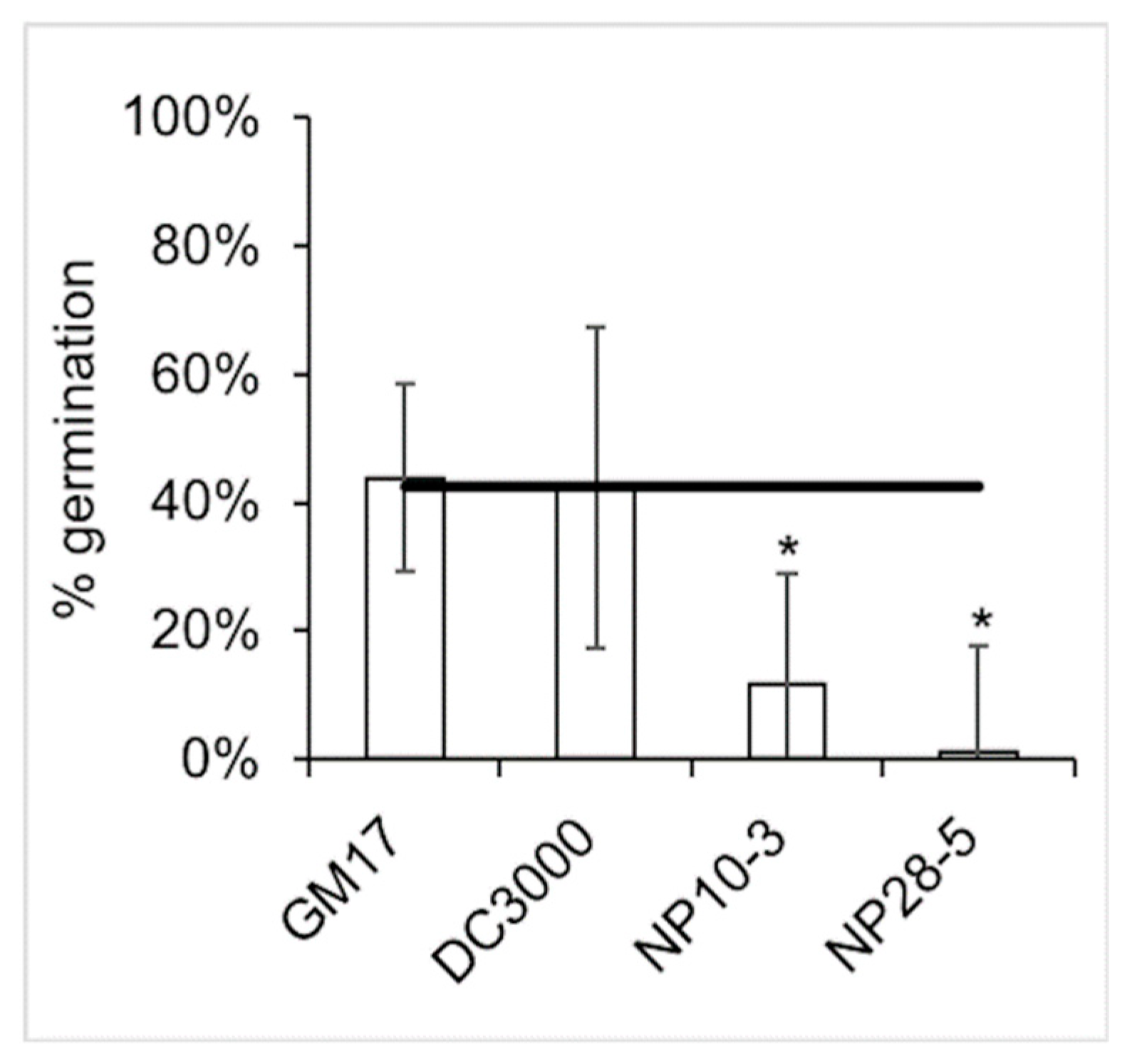
2.2. Seed Germination Assay
2.3. P. trichocarpa Seed Microbial Isolations and Growth Conditions
2.4. DNA Isolation and PCR
2.5. Preparation of Sterile Filtered Culture Supernatants
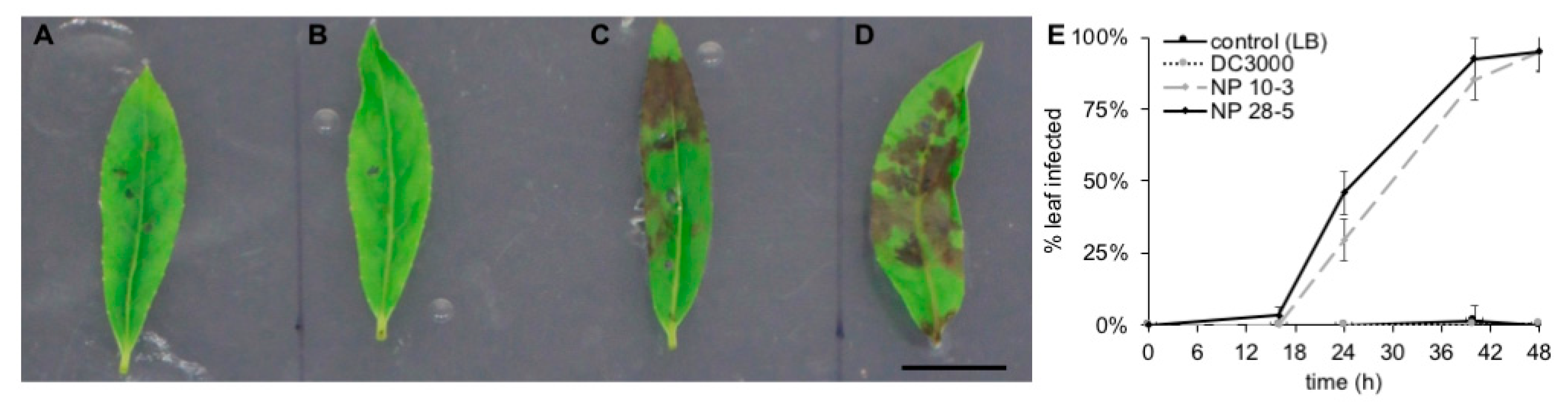
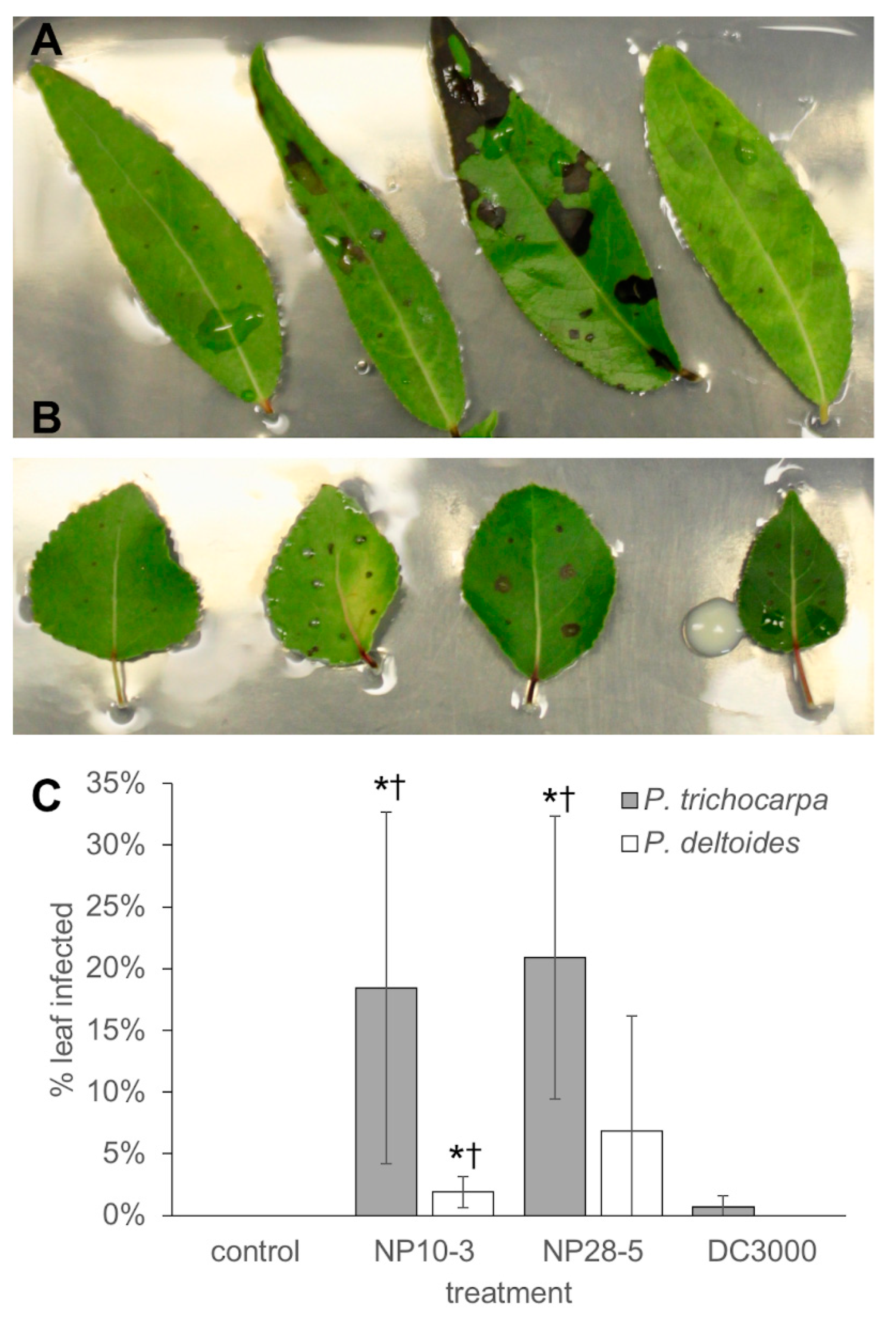
2.6. Leaf Infection Assays
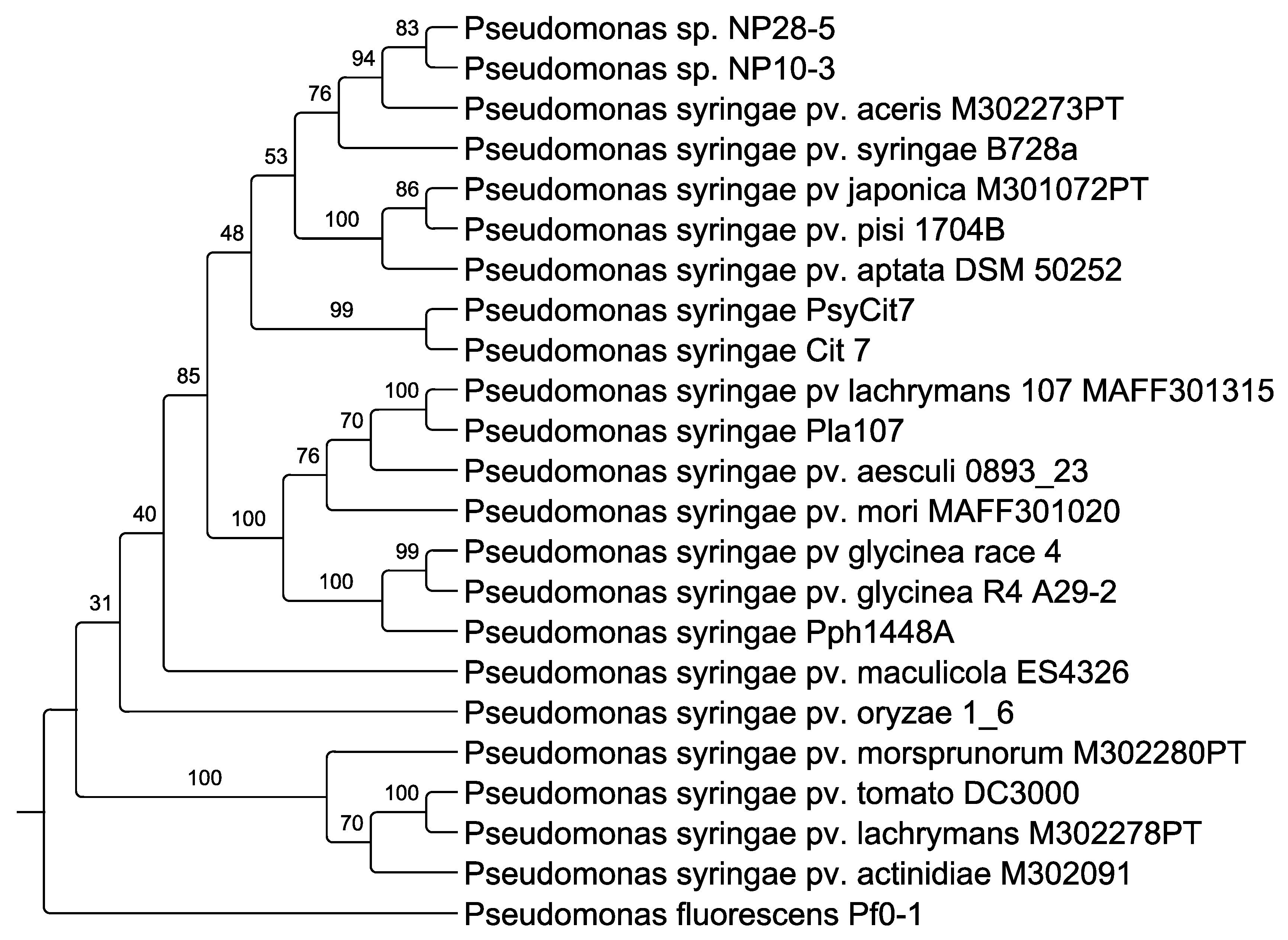
2.7. Phylogenetic Tree
2.8. Antibiotic Resistance Assay
2.9. Aromatic Compound Degradation Assay
2.10. IAA Production Assay
2.11. Biofilm Formation Assay
2.12. Motility Assay
2.13. Ice Nucleation Activity Assay
2.14. P. trichocarpa Metabolomics
3. Results
3.1. Isolation of P. syringae from P. trichocarpa
3.2. Effects on Germination of P. trichocarpa Seeds
3.3. Genome Analysis
3.4. Biosynthetic Potential of P. syringae Isolates
3.5. Pathogenicity of P. syringae Isolates
3.6. IAA and Motility Assays
3.7. Metabolomics of Populus Trichocarpa in Response to P. syringae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability Statement
References
- Xin, X.F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Hwang, M.S.H.; Morgan, R.L.; Sarkar, S.F.; Wang, P.W.; Guttman, D.S. Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl. Environ. Microbiol. 2005, 71, 5182–5191. [Google Scholar] [CrossRef]
- Hirano, S.S.; Upper, C.D. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—A pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 2000, 64, 624–653. [Google Scholar] [CrossRef] [PubMed]
- Block, A.; Li, G.; Fu, Z.Q.; Alfano, J.R. Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 2008, 11, 396–403. [Google Scholar] [CrossRef]
- Galán, J.E.; Collmer, A. Type III secretion machines: Bacterial devices for protein delivery into host cells. Science 1999, 284, 1322–1328. [Google Scholar]
- Collmer, A.; Badel, J.L.; Charkowski, A.O.; Deng, W.L.; Fouts, D.E.; Ramos, A.R.; Rehm, A.H.; Anderson, D.M.; Schneewind, O.; van Dijk, K.; et al. Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 8770. [Google Scholar] [CrossRef] [PubMed]
- Morel, J.B.; Dangl, J.L. The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 1997, 4, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Nurnberger, T.; Nennstiel, D.; Jabs, T.; Sacks, W.R.; Hahlbrock, K.; Scheel, D. High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 1994, 78, 449–460. [Google Scholar] [CrossRef]
- Buell, C.R.; Joardar, V.; Lindeberg, M.; Selengut, J.; Paulsen, I.T.; Gwinn, M.L.; Dodson, R.J.; Deboy, R.T.; Durkin, A.S.; Kolonay, J.F.; et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2003, 100, 10181–10186. [Google Scholar] [CrossRef]
- Ward, J.L.; Forcat, S.; Beckmann, M.; Bennett, M.; Miller, S.J.; Baker, J.M.; Hawkins, N.D.; Vermeer, C.P.; Lu, C.; Lin, W.; et al. The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 2010, 63, 443–457. [Google Scholar] [CrossRef]
- Rico, A.; McCraw, S.L.; Preston, G.M. The metabolic interface between Pseudomonas syringae and plant cells. Curr. Opin. Microbiol. 2011, 14, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Lindeberg, M.; Cunnac, S.; Collmer, A. The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol. Plant Pathol. 2009, 10, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Helmann, T.C.; Deutschbauer, A.M.; Lindow, S.E. Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc. Natl. Acad. Sci. USA 2019, 116, 18900–18910. [Google Scholar] [CrossRef]
- Rohmer, L.; Guttman, D.S.; Dangl, J.L. Diverse Evolutionary Mechanisms Shape the Type III Effector Virulence Factor Repertoire in the Plant Pathogen Pseudomonas syringae. Genetics 2004, 167, 1341–1360. [Google Scholar] [CrossRef][Green Version]
- Bender, C.L.; Alarcón-Chaidez, F.; Gross, D.C. Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 1999, 63, 266–292. [Google Scholar] [CrossRef]
- Block, A.; Schmelz, E.; Jones, J.B.; Klee, H.R. Coronatine and salicylic acid: The battle between Arabidopsis and Pseudomonas for phytohormone control. Mol. Plant Pathol. 2005, 6, 79–83. [Google Scholar] [CrossRef]
- Uppalapati, S.R.; Ishiga, Y.; Wangdi, T.; Urbanczyk-Wochniak, E.; Ishiga, T.; Mysore, K.S.; Bender, C.L. Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: Phenotypic and gene expression analyses of the virulence function of coronatine. Mol. Plant. Microbe Interact. 2008, 21, 383–395. [Google Scholar] [CrossRef]
- Ramos, C.; Matas, I.M.; Bardaji, L.; Aragon, I.M.; Murillo, J. Pseudomonas savastanoi pv. savastanoi: Some like it knot. Mol. Plant Pathol. 2012, 13, 998–1009. [Google Scholar] [CrossRef]
- Green, S.; Studholme, D.J.; Laue, B.E.; Dorati, F.; Lovell, H.; Arnold, D.; Cottrell, J.E.; Bridgett, S.; Blaxter, M.; Huitema, E.; et al. Comparative genome analysis provides insights into the evolution and adaptation of Pseudomonas syringae pv. aesculi on Aesculus hippocastanum. PLoS ONE 2010, 5, e10224. [Google Scholar] [CrossRef]
- Sarkar, S.F.; Gordon, J.S.; Martin, G.B.; Guttman, D.S. Comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics 2006, 174, 1041–1056. [Google Scholar] [CrossRef] [PubMed]
- Caballo-Ponce, E.; van Dillewijn, P.; Wittich, R.M.; Ramos, C. WHOP, a genomic region associated with woody hosts in the Pseudomonas syringae complex contributes to the virulence and fitness of Pseudomonas savastanoi pv. savastanoi in olive plants. Mol. Plant Microbe Interact. 2017, 30, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Otto, M.; Hammerbacher, A.; Petersen, Y.; Pierneef, R.; Coutinho, T.A. Phenolic compound degradation by Pseudomonas syringae phylogroup 2 strains. J. Plant Pathol. 2018, 100, 279–286. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Varvaro, L.; Parisi, L.; Audergon, J.M.; Morris, C.E. Chapter Four. Disease and Frost Damage of Woody Plants Caused by Pseudomonas syringae: Seeing the Forest for the Trees. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 235–295. [Google Scholar]
- Hulin, M.T.; Armitage, A.D.; Vicente, J.G.; Holub, E.B.; Baxtger, L.; Bates, H.J.; Mansfield, J.W.; Jackson, R.W.; Harrison, R.J. Comparative genomics of Pseudomonas syringae reveals convergent gene gain and loss associated with specialization onto cherry (Prunus avium). New Phytol. 2018, 219, 672–696. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa. Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Ramstedt, M.; Årström, B.; Fircks, H.A. Dieback of poplar and willow caused by Pseudomonas syringae in combination with freezing stress. Eur. J. Forest Pathol. 1994, 24, 305–315. [Google Scholar] [CrossRef]
- Haworth, R.H.; Spiers, A.G. Characterisation of bacteria from poplars and willows exhibiting leaf spotting and stem cankering in New Zealand. Eur. J. Forest Pathol. 1988, 18, 426–436. [Google Scholar] [CrossRef]
- Morris, C.E.; Sands, D.C.; Vinatzer, B.A.; Glaux, C.; Guilbaud, C.; Buffière, A.; Yan, S.; Dominguez, H.; Thompson, B.M. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2008, 2, 321–334. [Google Scholar] [CrossRef]
- Canfield, M.L.; Baca, S.; Moore, L.W. Isolation of Pseudomonas syringae from 40 cultivars of diseased woody plants with tip dieback in Pacific Northwest nurseries. Plant Dis. 1986, 70, 647–650. [Google Scholar] [CrossRef]
- Abraham, P.E.; Garcia, B.J.; Gunter, L.E.; Jawdy, S.S.; Engle, N.L.; Yang, X.; Jacobson, D.A.; Hettich, R.L.; Tuskan, G.A.; Tschapliknski, T.J. Quantitative proteome profile of water deficit stress responses in eastern cottonwood (Populus deltoides) leaves. PLoS ONE 2018, 13, e0190019. [Google Scholar] [CrossRef]
- Timm, C.M.; Pelletier, D.A.; Jawdy, S.S.; Gunter, L.E.; Henning, J.A.; Engle, N.L.; Aufrecht, J.; Gee, E.; Nookaew, I.; Yang, Z.; et al. Two poplar-associated bacterial isolates induce additive favorable responses in a constructed plant-microbiome system. Front. Plant Sci. 2016, 7, 497. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.; Utturkar, S.M.; Klingeman, D.M.; Johnson, C.M.; Martin, S.L.; Land, M.L.; Lu, T.Y.; Schadt, C.W.; Doktycz, M.J.; Pelletier, D.A. Twenty-one genome sequences from Pseudomonas species and 19 genome sequences from diverse bacteria isolated from the rhizosphere and endosphere of Populus deltoides. J. Bacteriol. 2012, 194, 5991–5993. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Mavromatis, K.; Ivanova, N.N.; Chen, I.M.; Chu, K.; Kyrpides, N.C. IMG ER: A system for microbial genome annotation expert review and curation. Bioinformatics 2009, 25, 2271–2278. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Chen, I.; Min, A.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; et al. IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012, 40, D115–D122. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T.J.; Kauffman, K.T.; Amrine, K.C.H.; Carper, D.L.; Lee, R.S.; Becich, P.J.; Canales, C.J.; Ardell, D.H. FAST: FAST Analysis of Sequences Toolbox. Front. Genet. 2015, 6, 172. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. A space-time process model for the evolution of DNA sequences. Genetics 1995, 139, 993–1005. [Google Scholar] [PubMed]
- Yang, Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 1994, 39, 306–314. [Google Scholar] [CrossRef]
- Soubrier, J.; Steel, M.; Lee, M.S.Y.; Der Sarkissian, C.; Guindon, S.; Ho, S.Y.W.; Cooper, A. The Influence of Rate Heterogeneity among Sites on the Time Dependence of Molecular Rates. Mol. Biol. Evol. 2012, 29, 3345–3358. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2017, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.J.; Berg, H.C. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. USA 1989, 86, 6973–6977. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, e2437. [Google Scholar] [CrossRef]
- Tschaplinski, T.J.; Plett, J.M.; Engle, N.L.; Deveau, A.; Cushman, K.C.; Martin, M.Z.; Doktycz, M.J.; Tuskan, G.A.; Brun, A.; Kohler, A.; et al. Populus trichocarpa and Populus deltoides exhibit different metabolomic responses to colonization by the symbiotic fungus Laccaria bicolor. Mol. Plant Microbe Interact. 2014, 27, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Tschaplinski, T.J.; Standaert, R.F.; Engle, N.L.; Martin, M.Z.; Sangha, A.K.; Parks, J.M.; Smith, J.C.; Samuel, R.; Jiang, N.; Pu, Y.; et al. Down-regulation of the caffeic acid O-methyltransferase gene in switchgrass reveals a novel monolignol analog. Biotechnol. Biofuels 2012, 5, 71. [Google Scholar] [CrossRef]
- Labbé, J.L.; Weston, D.J.; Dunkirk, N.; Pelletier, D.A.; Tuskan, G.A. Newly identified helper bacteria stimulate ectomycorrhizal formation in Populus. Front. Plant Sci. 2014, 5, 579. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Dillon, M.M.; Thakur, S.; Almeida, R.N.D.; Wang, P.W.; Weir, B.S.; Guttman, D.S. Recombination of ecologically and evolutionarily significant loci maintains genetic cohesion in the Pseudomonas syringae species complex. Genome Biol. 2019, 20, 3. [Google Scholar] [CrossRef]
- Baltrus, D.A.; Nishimura, M.R.; ROmanchuk, A.; Chang, J.H.; Mukhtar, M.S.; Cherkis, K.; Roach, J.; Grant, S.R.; Jones, C.D.; Dangl, J.L. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 2011, 7, e1002132. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, B.; Ramel, C.; Dudler, R. Pseudomonas syringae virulence factor syringolin A counteracts stomatal immunity by proteasome inhibition. Mol. Plant Microbe Interact. 2010, 23, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, M.L.; Tester, M.A.; Gross, D.C. Role of biosurfactant and ion channel-forming activities of syringomycin in transmembrane ion flux: A model for the mechanism of action in the plant-pathogen interaction. Mol. Plant Microbe Interact. 1995, 8, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Quinones, B.; Dulla, G.; Lindow, S.E. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. 2005, 18, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Braun, S.D.; Voksch, B.; Nuske, J.; Spiteller, D. 3-Methylarginine from Pseudomonas syringae pv. syringae 22d/93 suppresses the bacterial blight caused by its close relative Pseudomonas syringae pv. glycinea. ChemBioChem 2008, 9, 1913–1920. [Google Scholar] [CrossRef]
- Arrebola, E.; Cazorla, F.M.; Durán, V.E.; Rivera, E.; Olea, F.; Codina, J.C.; Pérez-García, A.; de Vicente, A. Mangotoxin: A novel antimetabolite toxin produced by Pseudomonas syringae inhibiting ornithine/arginine biosynthesis. Physiol. Mol. Plant Pathol. 2003, 63, 117–127. [Google Scholar] [CrossRef]
- Kuhlmann, A.U.; Hoffmann, T.; Bursy, J.; Jebbar, M.; Bremer, E. Ectoine and hydroxyectoine as protectants against osmotic and cold stress: Uptake through the SigB-controlled betaine-choline- carnitine transporter-type carrier EctT from Virgibacillus pantothenticus. J. Bacteriol. 2011, 193, 4699–4708. [Google Scholar] [CrossRef]
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek 2014, 106, 85–125. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef]
- Weston, D.J.; Pelletier, D.A.; Morrell-Falvey, J.L.; Tschaplinski, T.J.; Jawdy, S.S.; Lu, T.Y.; Allen, S.M.; Melton, S.J.; Martin, M.Z.; Shadt, C.W.; et al. Pseudomonas fluorescens induces strain-dependent and strain-independent host plant responses in defense networks, primary metabolism, photosynthesis, and fitness. Mol. Plant Microbe Interact. 2012, 25, 765–778. [Google Scholar] [CrossRef]
- Timm, C.M.; Campbell, A.G.; Utturkar, S.M.; Jun, S.-R.; Parales, R.E.; Tan, W.A.; Robeson, M.S.; Lu, T.S.; Jawdy, S.S.; Brown, S.D.; et al. Metabolic functions of Pseudomonas fluorescens strains from Populus deltoides depend on rhizosphere or endosphere isolation compartment. Front. Microbiol. 2015, 6, 1118. [Google Scholar] [CrossRef] [PubMed]
- Ghods, S.; Sims, I.M.; Moradali, M.F.; Rehm, B.H. Bactericidal compounds controlling growth of the plant pathogen Pseudomonas syringae pv. actinidiae, which forms biofilms composed of a novel exopolysaccharide. Appl. Environ. Microbiol. 2015, 81, 4026–4036. [Google Scholar] [CrossRef]
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef] [PubMed]
- Delshadi, S.; Ebrahimi, M.; Shirmohammadi, E. Influence of plant-growth-promoting bacteria on germination, growth and nutrients’ uptake of Onobrychis sativa L. under drought stress. J. Plant Interact. 2017, 12, 200–208. [Google Scholar] [CrossRef]
- Arnold, D.L.; Preston, G.M. Pseudomonas syringae: Enterprising epiphyte and stealthy parasite. Microbiology 2019, 165, 251–253. [Google Scholar] [CrossRef]
- Nowell, R.W.; Laue, B.E.; Sharp, P.M.; Green, S. Comparative genomics reveals genes significantly associated with woody hosts in the plant pathogen Pseudomonas syringae. Mol. Plant Pathol. 2016, 17, 1409–1424. [Google Scholar] [CrossRef]
- Macho, A.P.; Zumaquero, A.; Gonzalez-Plaza, J.J.; Ortiz-Martín, I.; Rufián, J.S.; Beuzón, C.R. Genetic analysis of the individual contribution to virulence of the type III effector inventory of Pseudomonas syringae pv. phaseolicola. PLoS ONE 2012, 7, e35871. [Google Scholar] [CrossRef]
- Washington, E.J.; Mukhtar, M.S.; Finkel, O.M.; Wan, L.; Banfield, M.J.; Kieber, J.J.; Dangl, J.L. Pseudomonas syringae type III effector HopAF1 suppresses plant immunity by targeting methionine recycling to block ethylene induction. Proc. Natl. Acad. Sci. USA 2016, 113, E3577–E3586. [Google Scholar] [CrossRef]
- Li, X.; Lin, H.; Zhang, W.; Zou, Y.; Zhang, J.; Tang, X.; Zhou, J.-M. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA 2005, 102, 12990–12995. [Google Scholar] [CrossRef]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for plant stress response. Physiol. Plant 2008, 132, 199–208. [Google Scholar] [CrossRef]
- Chen, F.; Liu, C.-J.; Tschaplinski, T.J.; Zhao, N. Genomics of secondary metabolism in Populus: Interactions with biotic and abiotic environments. Crit. Rev. Plant Sci. 2009, 28, 375–392. [Google Scholar] [CrossRef]
- Cellini, A.; Fiorentini, L.; Buriani, G.; Yu, J.; Donati, I.; Cornish, D.A.; Novak, B.; Costa, G.; Vanneste, J.L.; Spinelli, F. Elicitors of the salicylic acid pathway reduce incidence of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidae. Ann. Appl. Biol. 2014, 165, 441–453. [Google Scholar] [CrossRef]
| Species | Strain Name | Isolation Site | Host | Source | Accession |
|---|---|---|---|---|---|
| Pseudomonas syringae | NP10-3 | ID, USA | Populus trichocarpa | G Newcombe | 2757320439 |
| Pseudomonas syringae | NP28-5 | ID, USA | Populus trichocarpa | G Newcombe | 2757320523 |
| Chromosome Features a | NP10-3 | NP28-5 |
|---|---|---|
| genome size (bp) | 6045676 | 5895985 |
| plasmid (bp) | - | - |
| DNA coding sequence (%) | 5358015 (88.63%) | 5215021 (88.45%) |
| GC content (%) | 59.24% | 59.28% |
| total genes | 5331 | 5116 |
| protein coding genes | 5202 (97.6%) | 4985 (97.4%) |
| rRNA genes (5S rRNA, 16S rRNA, 23S rRNA) | 9 (7, 1, 1) | 9 (7, 1, 1) |
| tRNA | 56 | 55 |
| other RNA genes | 64 | 67 |
| total RNA genes | 129 | 131 |
| genes with assigned function | 4338 (81.37%) | 4213 (82.35%) |
| genes without assigned function | 864 (16.21%) | 772 (15.09%) |
| number of predicted enzymes | 1337 (25.08%) | 1320 (25.80%) |
| number of predicted effectors | 21 | 20 |
| biosynthetic clusters | 19 | 17 |
| genes in biosynthetic clusters | 298 (5.59%) | 301 (5.88%) |
| ANI b, P. syringae pv. syringae B728a | 98.79% | 98.78% |
| Function | Gene | Annotated Gene Product | NP10-3 Gene ID | % ID a | NP28-5 Gene ID |
|---|---|---|---|---|---|
| Catechol catabolism | catA | Catechol 1,2-dioxygenase | 2758144716 | 82 | ND b |
| catC | Muconolactone isomerase | 2758144717 | 82 | ND | |
| catB | Muconate cycloisomerase | 2758144718 | 89 | ND | |
| Anthranilate catabolism | antC | Anthranilate dioxygenase reductase component | 2758144712 | 39 | ND |
| antB | Anthranilate dioxygenase beta subunit | 2758144713 | 36 | ND | |
| antA | Anthranilate dioxygenase alpha subunit | 2758144714 | 47 | ND | |
| antR | antABC regulatory protein | 2758144715 | 25 | ND | |
| Indigo-producing oxygenase | ipoC | Involved in meta pathway of phenol degradation | ND | NA c | ND |
| ipoB | Nitrilotriacetate monooxygenase component B | ND | NA | ND | |
| ipoA | Putative oxygenase subunit | ND | NA | ND | |
| Not determined | Aerotaxis receptor | ND | NA | ND | |
| dhoB | Short chain alcohol dehydrogenase | ND | NA | ND | |
| dhoA | Dienelactone hydrolase | ND | NA | ND | |
| benR | Positive regulator of the benABCD operon | ND | NA | ND |
| Host | DC3000 | NP10-3 | NP28-5 |
|---|---|---|---|
| P. trichocarpa BESC819 | N | Y | Y |
| P. trichocarpa Nisqually-1 | N | Y | Y |
| P. deltoides WV94 | N | Y | Y |
| Arabidopsis thaliana Col-1 | Y | Y | Y |
| Avocado (Persea americana pv. Hass) | N | N | N |
| Bean (Phaseolus vulgaris pv. Tenderette) | N | N | N |
| Pea (Pisum sativum pv. Macrocarpon) | N | N | N |
| Spinach (Spinacia oleracea pv. Bloomsdale Longstanding) | N | N | N |
| Tobacco (Nicotiana tabacum pv. Little Crittenden) | N | N | N |
| Tomato (Solanum lycopersicum pv. Brandywine) | Y | N | N |
| Wheat (Triticum aestivum pv. Winter Wheat) | N | N | N |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saint-Vincent, P.M.; Ridout, M.; Engle, N.L.; Lawrence, T.J.; Yeary, M.L.; Tschaplinski, T.J.; Newcombe, G.; Pelletier, D.A. Isolation, Characterization, and Pathogenicity of Two Pseudomonas syringae Pathovars from Populus trichocarpa Seeds. Microorganisms 2020, 8, 1137. https://doi.org/10.3390/microorganisms8081137
Saint-Vincent PM, Ridout M, Engle NL, Lawrence TJ, Yeary ML, Tschaplinski TJ, Newcombe G, Pelletier DA. Isolation, Characterization, and Pathogenicity of Two Pseudomonas syringae Pathovars from Populus trichocarpa Seeds. Microorganisms. 2020; 8(8):1137. https://doi.org/10.3390/microorganisms8081137
Chicago/Turabian StyleSaint-Vincent, Patricia MB, Mary Ridout, Nancy L. Engle, Travis J. Lawrence, Meredith L. Yeary, Timothy J. Tschaplinski, George Newcombe, and Dale A. Pelletier. 2020. "Isolation, Characterization, and Pathogenicity of Two Pseudomonas syringae Pathovars from Populus trichocarpa Seeds" Microorganisms 8, no. 8: 1137. https://doi.org/10.3390/microorganisms8081137
APA StyleSaint-Vincent, P. M., Ridout, M., Engle, N. L., Lawrence, T. J., Yeary, M. L., Tschaplinski, T. J., Newcombe, G., & Pelletier, D. A. (2020). Isolation, Characterization, and Pathogenicity of Two Pseudomonas syringae Pathovars from Populus trichocarpa Seeds. Microorganisms, 8(8), 1137. https://doi.org/10.3390/microorganisms8081137






