16S and 23S rRNA Gene Mutation Independent Multidrug Resistance of Non-Tuberculous Mycobacteria Isolated from South Korean Soil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Isolation of NTM from Soil Samples
2.3. Extraction of Mycobacterial DNA
2.4. Sequence-Based Identification of Environmental NTM Isolates
2.5. Phylogenetic Tree Analysis
2.6. Antibiotic Resistance Test
2.7. PCR and Sequence Analysis Associated with Antibiotic Resistance
3. Results
3.1. Isolation and Identification of NTM from Soil Samples
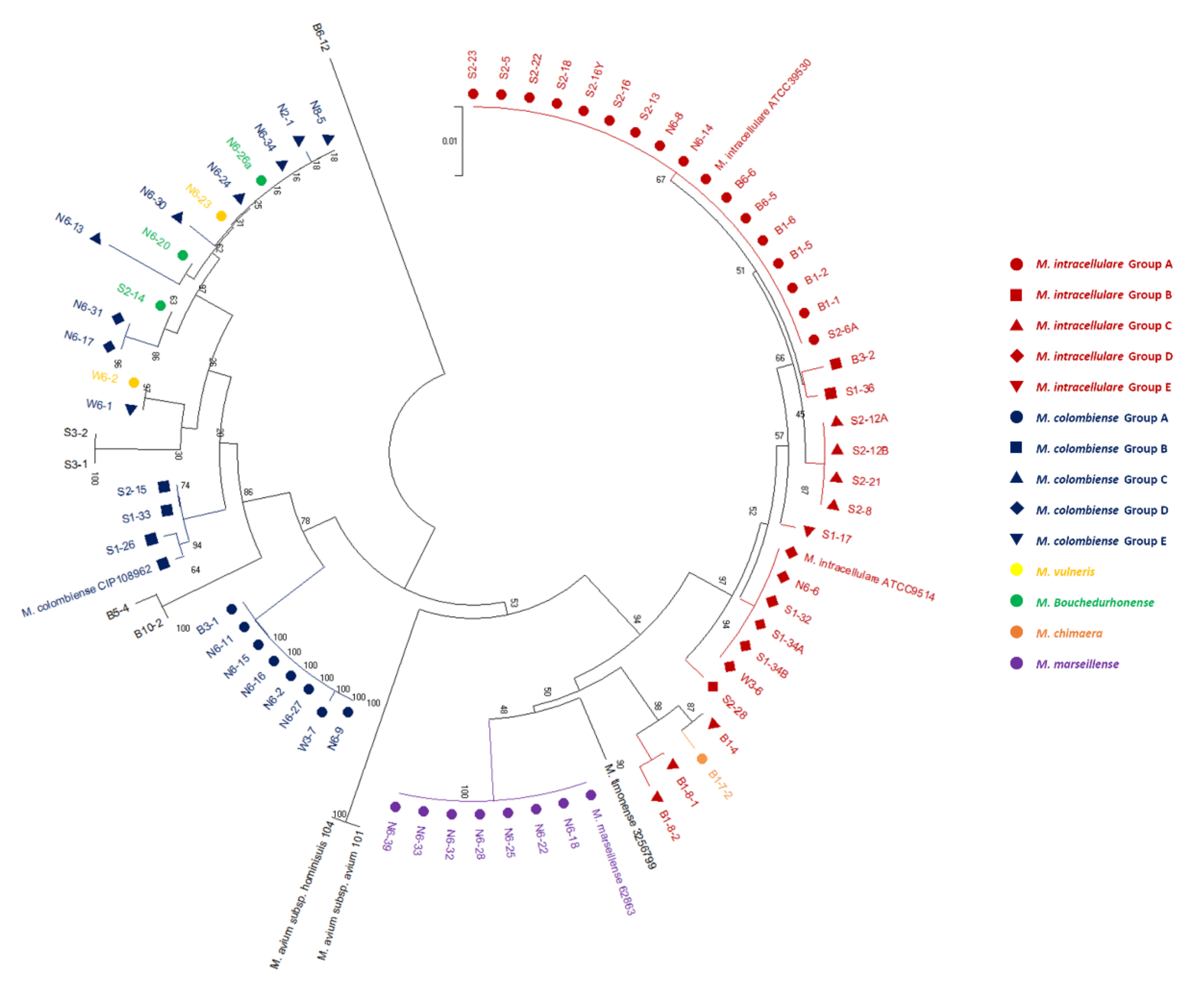
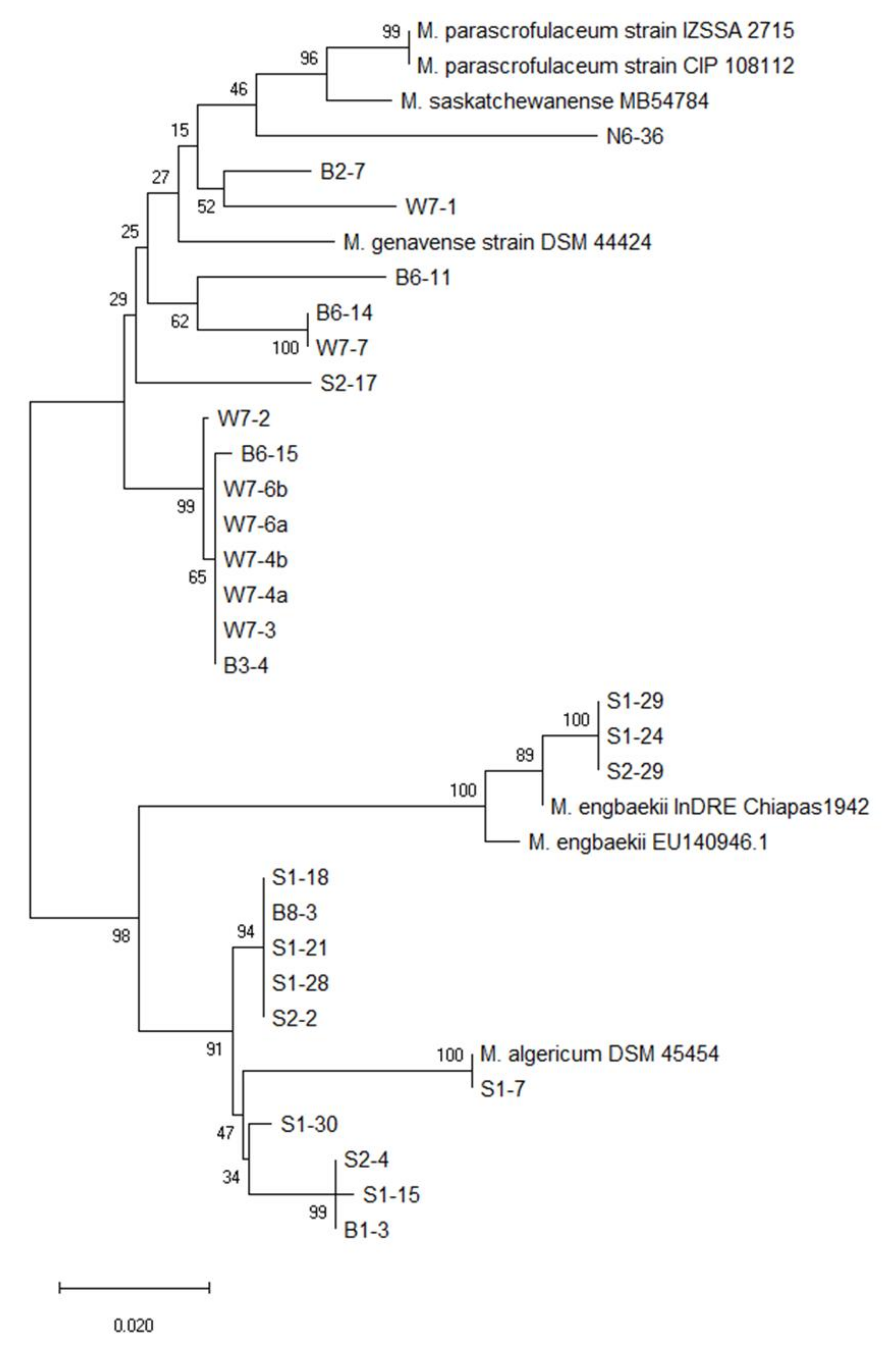
3.2. Phylogenetic Tree Analysis
3.3. Antibiotic Resistance Tests
3.4. PCR and Sequence Analysis Associated with Antibiotic Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Primm, T.P.; Lucero, C.A.; Falkinham, J.O., 3rd. Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 2004, 17, 98–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, J.R.; Virdi, R.; Chan, E.D. Global environmental nontuberculous mycobacteria and their contemporaneous man-made and natural niches. Front. Microbiol. 2018, 9, 2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falkinham, J.O., 3rd. Environmental sources of nontuberculous mycobacteria. Clin. Chest Med. 2015, 36, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Wassilew, N.; Hoffmann, H.; Andrejak, C.; Lange, C. Pulmonary disease caused by non-tuberculous mycobacteria. Respiration 2016, 91, 386–402. [Google Scholar] [CrossRef]
- Koh, W.J. Nontuberculous mycobacteria-overview. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Johnson, M.M.; Odell, J.A. Nontuberculous mycobacterial pulmonary infections. J. Thorac. Dis. 2014, 6, 210–220. [Google Scholar]
- Wu, U.I.; Holland, S.M. Host susceptibility to non-tuberculous mycobacterial infections. Lancet Infect. Dis. 2015, 15, 968–980. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, K.; Ito, Y.; Hirai, T.; Kubo, T.; Imai, S.; Tatsumi, S.; Fujita, K.; Takakura, S.; Niimi, A.; Iinuma, Y.; et al. Environmental risk factors for pulmonary Mycobacterium avium-intracellulare complex disease. Chest 2011, 140, 723–729. [Google Scholar] [CrossRef] [Green Version]
- Pereira, S.G.; Alarico, S.; Tiago, I.; Reis, D.; Nunes-Costa, D.; Cardoso, O.; Maranha, A.; Empadinhas, N. Studies of antimicrobial resistance in rare mycobacteria from a nosocomial environment. BMC Microbiol. 2019, 19, 62. [Google Scholar] [CrossRef] [Green Version]
- Gebert, M.J.; Delgado-Baquerizo, M.; Oliverio, A.M.; Webster, T.M.; Nichols, L.M.; Honda, J.R.; Chan, E.D.; Adjemian, J.; Dunn, R.R.; Fierer, N. Ecological analyses of mycobacteria in showerhead biofilms and their relevance to human health. mBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Brennan, P.J.; Nikaido, H. The envelope of mycobacteria. Annu. Rev. Biochem. 1995, 64, 29–63. [Google Scholar] [CrossRef] [PubMed]
- Jarlier, V.; Nikaido, H. Mycobacterial cell wall: Structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 1994, 123, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, N.; Frehel, C.; Ryter, A.; Ohayon, H.; Lesourd, M.; David, H.L. Multiple drug resistance in Mycobacterium avium: Is the wall architecture responsible for exclusion of antimicrobial agents? Antimicrob. Agents Chemother. 1981, 20, 666–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steed, K.A.; Falkinham, J.O., 3rd. Effect of growth in biofilms on chlorine susceptibility of Mycobacterium avium and Mycobacterium intracellulare. Appl. Environ. Microbiol. 2006, 72, 4007–4011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henkle, E.; Winthrop, K.L. Nontuberculous mycobacteria infections in immunosuppressed hosts. Clin. Chest Med. 2015, 36, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Ryu, Y.J.; Koh, W.J.; Daley, C.L. Diagnosis and treatment of nontuberculous mycobacterial lung disease: Clinicians’ perspectives. Tuberc. Respir. Dis. 2016, 79, 74–84. [Google Scholar] [CrossRef]
- Cowman, S.; Burns, K.; Benson, S.; Wilson, R.; Loebinger, M.R. The antimicrobial susceptibility of non-tuberculous mycobacteria. J. Infect. 2016, 72, 324–331. [Google Scholar] [CrossRef]
- Sweetline Anne, N.; Ronald, B.S.M.; Senthil Kumar, T.M.A.; Thangavelu, A. Conventional and molecular determination of drug resistance in Mycobacterium tuberculosis and Mycobacterium bovis isolates in cattle. Tuberculosis 2018, 114, 113–118. [Google Scholar] [CrossRef]
- Huh, H.J.; Kim, S.Y.; Shim, H.J.; Kim, D.H.; Yoo, I.Y.; Kang, O.K.; Ki, C.S.; Shin, S.Y.; Jhun, B.W.; Shin, S.J.; et al. GenoType NTM-DR performance evaluation for identification of Mycobacterium avium complex and Mycobacterium abscessus and determination of clarithromycin and amikacin resistance. J. Clin. Microbiol. 2019, 57, e00516-19. [Google Scholar] [CrossRef] [Green Version]
- Maningi, N.E.; Daum, L.T.; Rodriguez, J.D.; Said, H.M.; Peters, R.P.H.; Sekyere, J.O.; Fischer, G.W.; Chambers, J.P.; Fourie, P.B. Multi- and extensively drug resistant Mycobacterium tuberculosis in South Africa: A molecular analysis of historical isolates. J. Clin. Microbiol. 2018, 56, e01214-17. [Google Scholar] [CrossRef] [Green Version]
- Jo, K.W.; Lee, S.; Kang, M.R.; Sung, H.; Kim, M.N.; Shim, T.S. Frequency and type of disputed rpoB mutations in Mycobacterium tuberculosis isolates from South Korea. Tuberc. Respir. Dis. 2017, 80, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Sreevatsan, S.; Pan, X.; Stockbauer, K.E.; Williams, D.L.; Kreiswirth, B.N.; Musser, J.M. Characterization of rpsL. and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob. Agents Chemother. 1996, 40, 1024–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, J.R.; Hess, T.; Malcolm, K.C.; Ovrutsky, A.R.; Bai, X.; Irani, V.R.; Dobos, K.M.; Chan, E.D.; Flores, S.C. Pathogenic nontuberculous mycobacteria resist and inactivate cathelicidin: Implication of a novel role for polar mycobacterial lipids. PLoS ONE 2015, 10, e0126994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.; Russell, C.; Soll, B.; Chow, D.; Bamrah, S.; Brostrom, R.; Kim, W.; Scott, J.; Bankowski, M.J. Increasing prevalence of nontuberculous mycobacteria in respiratory specimens from US-affiliated pacific island jurisdictions. Emerg. Infect. Dis. 2018, 24, 485–491. [Google Scholar] [CrossRef] [Green Version]
- Simons, S.; van Ingen, J.; Hsueh, P.R.; van Hung, N.; Dekhuijzen, P.N.; Boeree, M.J.; van Soolingen, D. Nontuberculous mycobacteria in respiratory tract infections, eastern Asia. Emerg. Infect. Dis. 2011, 17, 343–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.C.; Kang, M.J.; Han, C.H.; Lee, S.M.; Kim, C.J.; Lee, J.M.; Kang, Y.A. Prevalence, incidence, and mortality of nontuberculous mycobacterial infection in Korea: A nationwide population-based study. BMC Pulm. Med. 2019, 19, 140. [Google Scholar] [CrossRef] [Green Version]
- Izumi, K.; Morimoto, K.; Hasegawa, N.; Uchimura, K.; Kawatsu, L.; Ato, M.; Mitarai, S. Epidemiology of adults and children treated for nontuberculous mycobacterial pulmonary disease in Japan. Ann. Am. Thorac. Soc. 2019, 16, 341–347. [Google Scholar] [CrossRef]
- Blanc, P.; Dutronc, H.; Peuchant, O.; Dauchy, F.A.; Cazanave, C.; Neau, D.; Wirth, G.; Pellegrin, J.L.; Morlat, P.; Mercié, P.; et al. Nontuberculous mycobacterial infections in a French hospital: A 12-year retrospective study. PLoS ONE 2016, 11, e0168290. [Google Scholar] [CrossRef]
- Ding, L.W.; Lai, C.C.; Lee, L.N.; Hsueh, P.R. Disease caused by non-tuberculous mycobacteria in a university hospital in Taiwan, 1997–2003. Epidemiol. Infect. 2006, 134, 1060–1067. [Google Scholar] [CrossRef]
- Lai, C.C.; Tan, C.K.; Chou, C.H.; Hsu, H.L.; Liao, C.H.; Huang, Y.T.; Yang, P.C.; Luh, K.T.; Hsueh, P.R. Increasing incidence of nontuberculous mycobacteria, Taiwan, 2000–2008. Emerg. Infect. Dis. 2010, 16, 294–296. [Google Scholar] [CrossRef]
- Hoefsloot, W.; van Ingen, J.; Andrejak, C.; Angeby, K.; Bauriaud, R.; Bemer, P.; Beylis, N.; Boeree, M.J.; Cacho, J.; Chihota, V.; et al. Nontuberculous mycobacteria network European trials group. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: An NTM-NET collaborative study. Eur. Respir. J. 2013, 42, 1604–1613. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Myung, W.; Koh, W.J.; Moon, S.M.; Jhun, B.W. Epidemiology of nontuberculous mycobacterial infection, South Korea, 2007–2016. Emerg. Infect. Dis. 2019, 25, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Zweijpfenning, S.M.H.; Ingen, J.V.; Hoefsloot, W. Geographic distribution of nontuberculous mycobacteria isolated from clinical specimens: A systematic review. Semin. Respir. Crit. Care Med. 2018, 39, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.W.; Saito, H.; Yoshii, Z. Environmental mycobacteria in Korea. I. Distribution of the organisms. Microbiol. Immunol. 1984, 28, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Park, H.T.; Shin, M.K.; Sung, K.Y.; Park, H.E.; Cho, Y.I.; Yoo, H.S. Effective DNA extraction method to improve detection of Mycobacterium avium subsp. paratuberculosis in bovine feces. Korean J. Vet. Res. 2014, 54, 55–57. [Google Scholar] [CrossRef] [Green Version]
- Telenti, A.; Marchesi, F.; Balz, M.; Bally, F.; Böttger, E.C.; Bodmer, T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 1993, 31, 175–178. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.J.; Lee, B.S.; Koh, W.J.; Manning, E.J.; Anklam, K.; Sreevatsan, S.; Lambrecht, R.S.; Collins, M.T. Efficient differentiation of Mycobacterium avium complex species and subspecies by use of five-target multiplex PCR. J. Clin. Microbiol. 2010, 48, 4057–4062. [Google Scholar] [CrossRef] [Green Version]
- Adékambi, T.; Colson, P.; Drancourt, M. rpoB-Based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 2003, 41, 5699–5708. [Google Scholar] [CrossRef] [Green Version]
- Nessar, R.; Reyrat, J.M.; Murray, A.; Gicquel, B. Genetic analysis of new 16S rRNA mutations conferring aminoglycoside resistance in Mycobacterium abscessus. J. Antimicrob. Chemother. 2011, 66, 1719–1724. [Google Scholar] [CrossRef]
- Inagaki, T.; Yagi, T.; Ichikawa, K.; Nakagawa, T.; Moriyama, M.; Uchiya, K.; Nikai, T.; Ogawa, K. Evaluation of a rapid detection method of clarithromycin resistance genes in Mycobacterium avium complex isolates. J. Antimicrob. Chemother. 2011, 66, 722–729. [Google Scholar] [CrossRef] [Green Version]
- Nash, K.A.; Andini, N.; Zhang, Y.; Brown-Elliott, B.A.; Wallace, R.J., Jr. Intrinsic macrolide resistance in rapidly growing mycobacteria. Antimicrob. Agents Chemother. 2006, 50, 3476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical Laboratory Standards Institute. Susceptibility Testing of Mycobacteria, 201 Nocardiae, and Other Aerobic Actinomycetes; Approved Standard, 2nd ed.; CLSI 202 document No. M24-A2; Clinical Laboratory Standards Institute: Wayne, PA, USA, 2011. [Google Scholar]
- Van Ingen, J.; Kuijper, E.J. Drug susceptibility testing of nontuberculous mycobacteria. Future Microbiol. 2014, 9, 1095–1110. [Google Scholar] [CrossRef] [PubMed]
- Adjemian, J.; Olivier, K.N.; Seitz, A.E.; Holland, S.M.; Prevots, D.R. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am. J. Respir. Crit. Care Med. 2012, 185, 881–886. [Google Scholar] [CrossRef] [Green Version]
- Vinnard, C.; Longworth, S.; Mezochow, A.; Patrawalla, A.; Kreiswirth, B.N.; Hamilton, K. Deaths related to nontuberculous mycobacterial infections in the United States, 1999–2014. Ann. Am. Thorac. Soc. 2016, 13, 1951–1955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, H.J.; Choi, H.Y.; Ki, M. Nontuberculosis mycobacterial infections at a specialized tuberculosis treatment centre in the Republic of Korea. BMC Infect. Dis. 2017, 17, 432. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, K.; Iwai, K.; Uchimura, K.; Okumura, M.; Yoshiyama, T.; Yoshimori, K.; Ogata, H.; Kurashima, A.; Gemma, A.; Kudoh, S. A steady increase in nontuberculous mycobacteriosis mortality and estimated prevalence in Japan. Ann. Am. Thorac. Soc. 2014, 11, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ringshausen, F.C.; Wagner, D.; de Roux, A.; Diel, R.; Hohmann, D.; Hickstein, L.; Welte, T.; Rademacher, J. Prevalence of nontuberculous mycobacterial pulmonary disease, Germany, 2009–2014. Emerg. Infect. Dis. 2016, 22, 1102–1105. [Google Scholar] [CrossRef]
- Cowman, S.A.; James, P.; Wilson, R.; Cookson, W.O.C.; Moffatt, M.F.; Loebinger, M.R. Profiling mycobacterial communities in pulmonary nontuberculous mycobacterial disease. PLoS ONE 2018, 13, e0208018. [Google Scholar] [CrossRef]
- Jang, M.A.; Koh, W.J.; Huh, H.J.; Kim, S.Y.; Jeon, K.; Ki, C.S.; Lee, N.Y. Distribution of nontuberculous mycobacteria by multigene sequence-based typing and clinical significance of isolated strains. J. Clin. Microbiol. 2014, 52, 1207–1212. [Google Scholar] [CrossRef] [Green Version]
- Walsh, C.M.; Gebert, M.J.; Delgado-Baquerizo, M.; Maestre, F.T.; Fierer, N. A global survey of mycobacterial diversity in soil. Appl. Environ. Microbiol. 2019, 85, e01180-19. [Google Scholar] [CrossRef] [Green Version]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [Green Version]
- Churgin, D.S.; Tran, K.D.; Gregori, N.Z.; Young, R.C.; Alabiad, C.; Flynn, H.W., Jr. Multi-Drug resistant Mycobacterium chelonae scleral buckle infection. Am. J. Ophthalmol. Case Rep. 2018, 10, 276–278. [Google Scholar] [CrossRef]
- Hurst-Hess, K.; Rudra, P.; Ghosh, P. Mycobacterium abscessus WhiB7 regulates a species-specific repertoire of genes to confer extreme antibiotic resistance. Antimicrob. Agents Chemother. 2017, 61, e01347-17. [Google Scholar] [CrossRef] [Green Version]
- Rudra, P.; Hurst-Hess, K.; Lappierre, P.; Ghosh, P. High levels of intrinsic tetracycline resistance in Mycobacterium abscessus are conferred by a tetracycline-modifying monooxygenase. Antimicrob. Agents Chemother. 2018, 62, e00119-18. [Google Scholar] [CrossRef] [Green Version]
- Rominski, A.; Roditscheff, A.; Selchow, P.; Böttger, E.C.; Sander, P. Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J. Antimicrob. Chemother. 2017, 72, 376–384. [Google Scholar] [CrossRef] [Green Version]
- Falkinham, J.O., 3rd. Nontuberculous mycobacteria from household plumbing of patients with nontuberculous mycobacteria disease. Emerg. Infect. Dis. 2011, 17, 419–424. [Google Scholar] [CrossRef]
- Marks, J.; Jenkins, P.A.; Schaefer, W.B. Thin-layer chromatography of mycobacterial lipids as an aid to classification: Technical improvements: Mycobacterium avium, M. intracellulare (Battey bacilli). Tubercle 1971, 52, 219–225. [Google Scholar] [CrossRef]
- Ripoll, F.; Deshayes, C.; Pasek, S.; Laval, F.; Beretti, J.L.; Biet, F.; Risler, J.L.; Daffé, M.; Etienne, G.; Gaillard, J.L.; et al. Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. BMC Genom. 2007, 8, 114. [Google Scholar] [CrossRef] [Green Version]
- Schorey, J.S.; Sweet, L. The mycobacterial glycopeptidolipids: Structure, function, and their role in pathogenesis. Glycobiology 2008, 18, 832–841. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Lian, L.L.; Wan, L.; Zhang, J.; Zhao, X.; Jiang, Y.; Zhao, L.L.; Liu, H.; Wan, K. Antimicrobial susceptibility of standard strains of nontuberculous mycobacteria by microplate Alamar Blue assay. PLoS ONE 2013, 8, e84065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.C.; Wu, M.F.; Chen, H.C.; Huang, W.C. In vitro activity of aminoglycosides, clofazimine, d-cycloserine and dapsone against 83 Mycobacterium avium complex clinical isolates. J. Microbiol. Immunol. Infect. 2018, 51, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Heifets, L.; Lindholm-Levy, P. Comparison of bactericidal activities of streptomycin, amikacin, kanamycin, and capreomycin against Mycobacterium avium and M. tuberculosis. Antimicrob. Agents Chemother. 1989, 33, 1298–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurer, F.P.; Rüegger, V.; Ritter, C.; Bloemberg, G.V.; Böttger, E.C. Acquisition of clarithromycin resistance mutations in the 23S rRNA gene of Mycobacterium abscessus in the presence of inducible erm (41). J. Antimicrob. Chemother. 2012, 67, 2606–2611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arthur, M.; Courvalin, P. Contribution of two different mechanisms to erythromycin resistance in Escherichia coli. Antimicrob. Agents Chemother. 1986, 30, 694–700. [Google Scholar] [CrossRef] [Green Version]
- O’Hara, K.; Kanda, T.; Ohmiya, K.; Ebisu, T.; Kono, M. Purification and characterization of macrolide 2’-phosphotransferase from a strain of Escherichia coli that is highly resistant to erythromycin. Antimicrob. Agents Chemother. 1989, 33, 1354–1357. [Google Scholar]
- Sugimoto, Y.; Suzuki, S.; Nonaka, L.; Boonla, C.; Sukpanyatham, N.; Chou, H.Y.; Wu, J.H. The novel mef (C)–mph (G) macrolide resistance genes are conveyed in the environment on various vectors. J. Glob. Antimicrob. Resist. 2017, 10, 47–53. [Google Scholar] [CrossRef]
- Tiberi, S.; Scardigli, A.; Centis, R.; D’Ambrosio, L.; Muñoz-Torrico, M.; Salazar-Lezama, M.Á.; Spanevello, A.; Visca, D.; Zumla, A.; Migliori, G.B.; et al. Classifying new anti-tuberculosis drugs: Rationale and future perspectives. Int. J. Infect. Dis. 2017, 56, 181–184. [Google Scholar] [CrossRef] [Green Version]
- Otchere, I.D.; Asante-Poku, A.; Osei-Wusu, S.; Aboagye, S.Y.; Yeboah-Manu, D. Isolation and characterization of nontuberculous mycobacteria from patients with pulmonary tuberculosis in Ghana. Int. J. Mycobacteriol. 2017, 6, 70–75. [Google Scholar] [CrossRef]
- Wu, M.L.; Aziz, D.B.; Dartois, V.; Dick, T. NTM drug discovery: Status, gaps and the way forward. Drug Discov. Today 2018, 23, 1502–1519. [Google Scholar] [CrossRef]
- Wang, D.M.; Liao, Y.; Li, Q.F.; Zhu, M.; Wu, G.H.; Xu, Y.H.; Zhong, J.; Luo, J.; Li, Y.J. Drug resistance and pathogenic spectrum of patients coinfected with nontuberculous mycobacteria and human-immunodeficiency virus in Chengdu, China. Chin. Med. J. 2019, 132, 1293–1297. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.S.; Donkor, E.S. Carbapenem resistance: A review. Med. Sci. 2017, 6, E1. [Google Scholar] [CrossRef] [Green Version]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [Green Version]
- Griffith, D.E.; Aksamit, T.; Brown-Elliott, B.A.; Catanzaro, A.; Daley, C.; Gordin, F.; Holland, S.M.; Horsburgh, R.; Huitt, G.; Iademarco, M.F.; et al. An official ATS/IDSA statement: Diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 2007, 175, 367–416. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Yoo, H.K.; Kim, S.H.; Koh, W.J.; Kim, C.K.; Park, Y.K.; Kim, H.J. The drug resistance profile of Mycobacterium abscessus group strains from Korea. Ann. Lab. Med. 2014, 34, 31–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.H.; Huh, H.J.; Song, D.J.; Moon, S.M.; Lee, S.H.; Shin, S.Y.; Kim, C.K.; Ki, C.S.; Koh, W.J.; Lee, N.Y. Differences in drug susceptibility pattern between Mycobacterium avium and Mycobacterium intracellulare isolated in respiratory specimens. J. Infect. Chemother. 2018, 24, 315–318. [Google Scholar] [CrossRef]





| Target Gene | Primer Sequence | Product Size (bp) | Reference | |
|---|---|---|---|---|
| Identification of non-tuberculosis mycobacteria | ||||
| 16s rRNA | F | ATAAGCCTGGGAAACTGGGT | 484 | [37] |
| R | CACGCTCACAGTTAAGCCGT | |||
| hsp65 | F | ACCAACGATGGTGTGTCCAT | 439 | [36] |
| R | CTTGTCGAACCGCATACCCT | |||
| rpoB | F | GGCAAGGTCACCCCGAAGGG | 723 | [38] |
| R | AGCGGCTGCTGGGTGATCATC | |||
| Identification of antibiotic resistance genes | ||||
| rrs | F | ATGACGTCAAGTCATCATGCC | 341 | [39] |
| R | AGGTGATCCAGCCGCACCTTC | |||
| rrl | F | TTTAAGCCCCAGTAAACGGC | 420 | [40] |
| R | GTCCAGGTTGAGGGAACCTT | |||
| erm | F | ACGTGGTGGTGGGCAAYCTG | 175 | [41] |
| R | AATTCGAACCACGGCCACCACT | |||
| Antibiotics | MIC Breakpoints | |||||
|---|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | ||||
| SGM | RGM | SGM | RGM | SGM | RGM | |
| Rifampicin | ≤0.5 | <1 | 1–4 | N/A | ≥8 | ≥1 |
| Streptomycin | <5 | <5 | N/A | N/A | ≥5 | ≥5 |
| Amikacin | ≤16 | ≤16 | 32 | 32 | ≥64 | ≥64 |
| Azithromycin | ≤8 | ≤2 | 16 | 4 | ≥32 | ≥8 |
| Ethambutol | ≤2 | <5 | 4 | N/A | ≥8 | ≥5 |
| Isoniazid | ≤0.5 | <1 | N/A | N/A | ≥1 | ≥1 |
| Moxifloxacin | ≤1 | ≤1 | 2 | 2 | ≥4 | ≥4 |
| Imipenem | ≤4 | ≤4 | 8–16 | 8–16 | ≥32 | ≥32 |
| Species | Strain No. | Presence of erm Gene | Sequencing Results | MIC Value (μg/mL) | |||
|---|---|---|---|---|---|---|---|
| rrs | rrl | STR | AMK | AZI | |||
| M.intracellulare | S2-16Y | ND | G1190A | WT | 0.5 | 1 | 0.25 |
| M.intracellulare | B1-8-1 | ND | G1446T | WT | 8 | 64 | 32 |
| M.intracellulare | B1-4 | ND | G1446T | WT | 2 | 16 | 1 |
| M.colombiense | S1-33 | ND | C1520G G1513A | WT | 0.25 | 2 | 1 |
| M.peregrinum | S1-3 | ND | C1235T | WT | 8 | 2 | 0.5 |
| M.sinense | S2-4 | ND | T1191G | WT | 128 | 2 | 4 |
| M.intracellulare | B1-1 | ND | WT | T2419C | 8 | 128 | 32 |
| M.intracellulare | B1-6 | ND | WT | T2419C | 16 | 128 | 32 |
| M.intracellulare | S2-16 | ND | WT | T2419C | 8 | 128 | 64 |
| M.intracellulare | S2-18 | ND | WT | T2419C | 4 | 64 | 16 |
| M.intracellulare | S2-22 | ND | WT | T2419C | 16 | 128 | 64 |
| M.intracellulare | S2-23 | ND | WT | T2419C | 4 | 64 | 32 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.-E.; Kim, S.; Shim, S.; Park, H.-T.; Park, W.B.; Im, Y.B.; Yoo, H.S. 16S and 23S rRNA Gene Mutation Independent Multidrug Resistance of Non-Tuberculous Mycobacteria Isolated from South Korean Soil. Microorganisms 2020, 8, 1114. https://doi.org/10.3390/microorganisms8081114
Park H-E, Kim S, Shim S, Park H-T, Park WB, Im YB, Yoo HS. 16S and 23S rRNA Gene Mutation Independent Multidrug Resistance of Non-Tuberculous Mycobacteria Isolated from South Korean Soil. Microorganisms. 2020; 8(8):1114. https://doi.org/10.3390/microorganisms8081114
Chicago/Turabian StylePark, Hyun-Eui, Suji Kim, Soojin Shim, Hong-Tae Park, Woo Bin Park, Young Bin Im, and Han Sang Yoo. 2020. "16S and 23S rRNA Gene Mutation Independent Multidrug Resistance of Non-Tuberculous Mycobacteria Isolated from South Korean Soil" Microorganisms 8, no. 8: 1114. https://doi.org/10.3390/microorganisms8081114
APA StylePark, H.-E., Kim, S., Shim, S., Park, H.-T., Park, W. B., Im, Y. B., & Yoo, H. S. (2020). 16S and 23S rRNA Gene Mutation Independent Multidrug Resistance of Non-Tuberculous Mycobacteria Isolated from South Korean Soil. Microorganisms, 8(8), 1114. https://doi.org/10.3390/microorganisms8081114





