Comprehensive Host Cell-Based Screening Assays for Identification of Anti-Virulence Drugs Targeting Pseudomonas aeruginosa and Salmonella Typhimurium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Compounds
2.2. Cell Culture
2.3. Culture Conditions of Bacteria
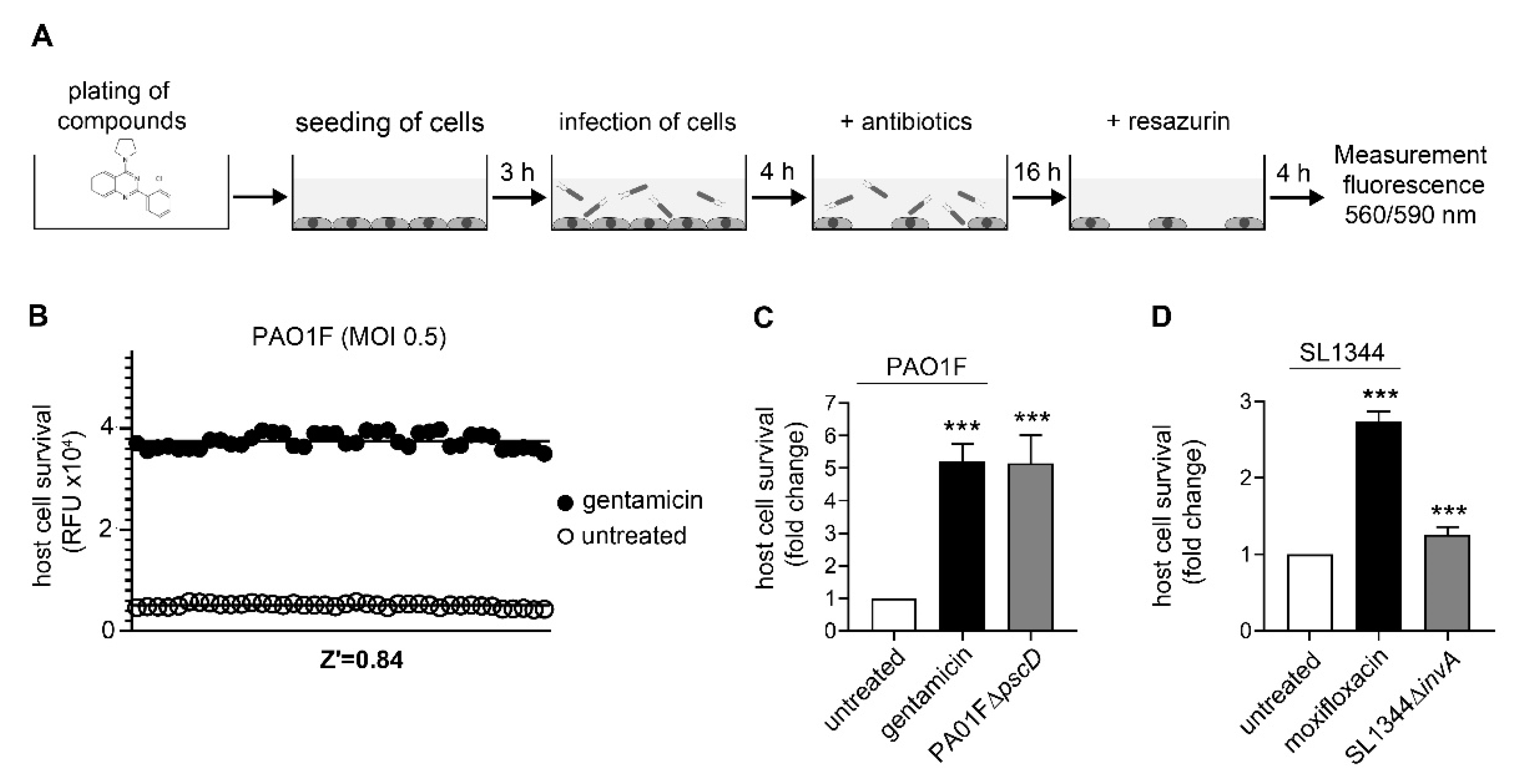
2.4. Host-Cell Survival Assays
2.5. Growth Inhibition Assays
2.6. RNA-Seq in Pseudomonas Aeruginosa
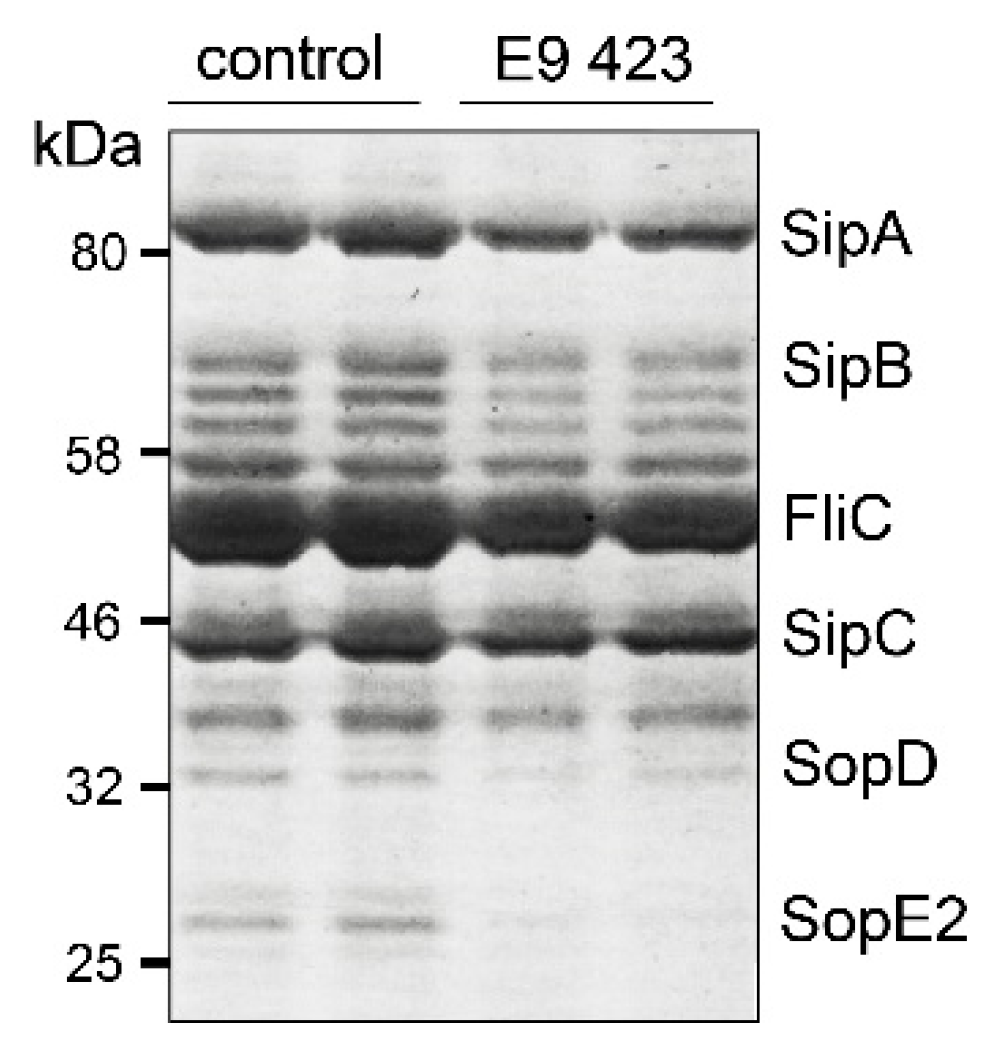
2.7. T3SS-Secretion Assay
2.8. Statistical Analysis
3. Results
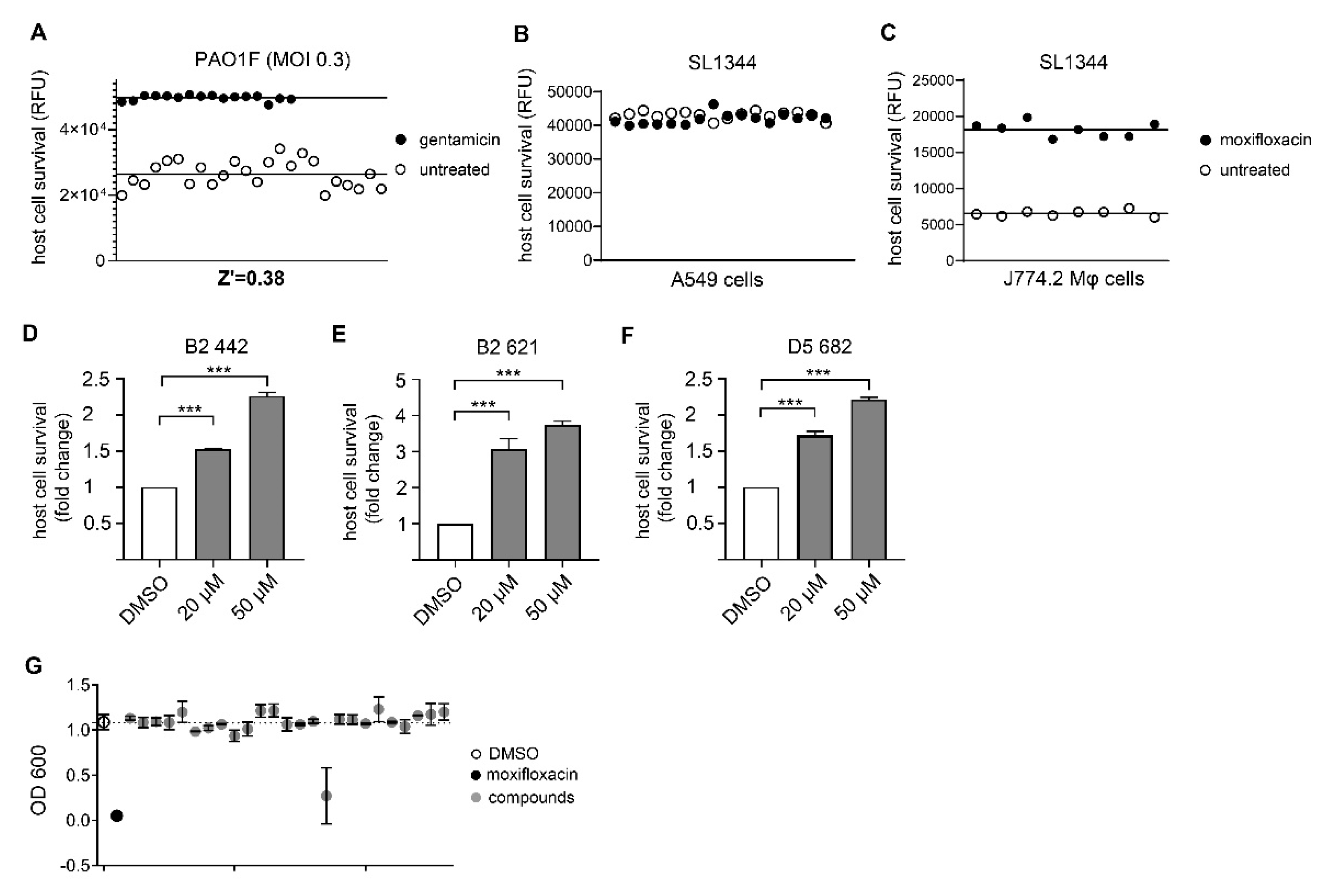
3.1. Assay Development and Validation for Pseudomonas Aeruginosa
3.2. Assay Development and Validation for Salmonella Typhimurium
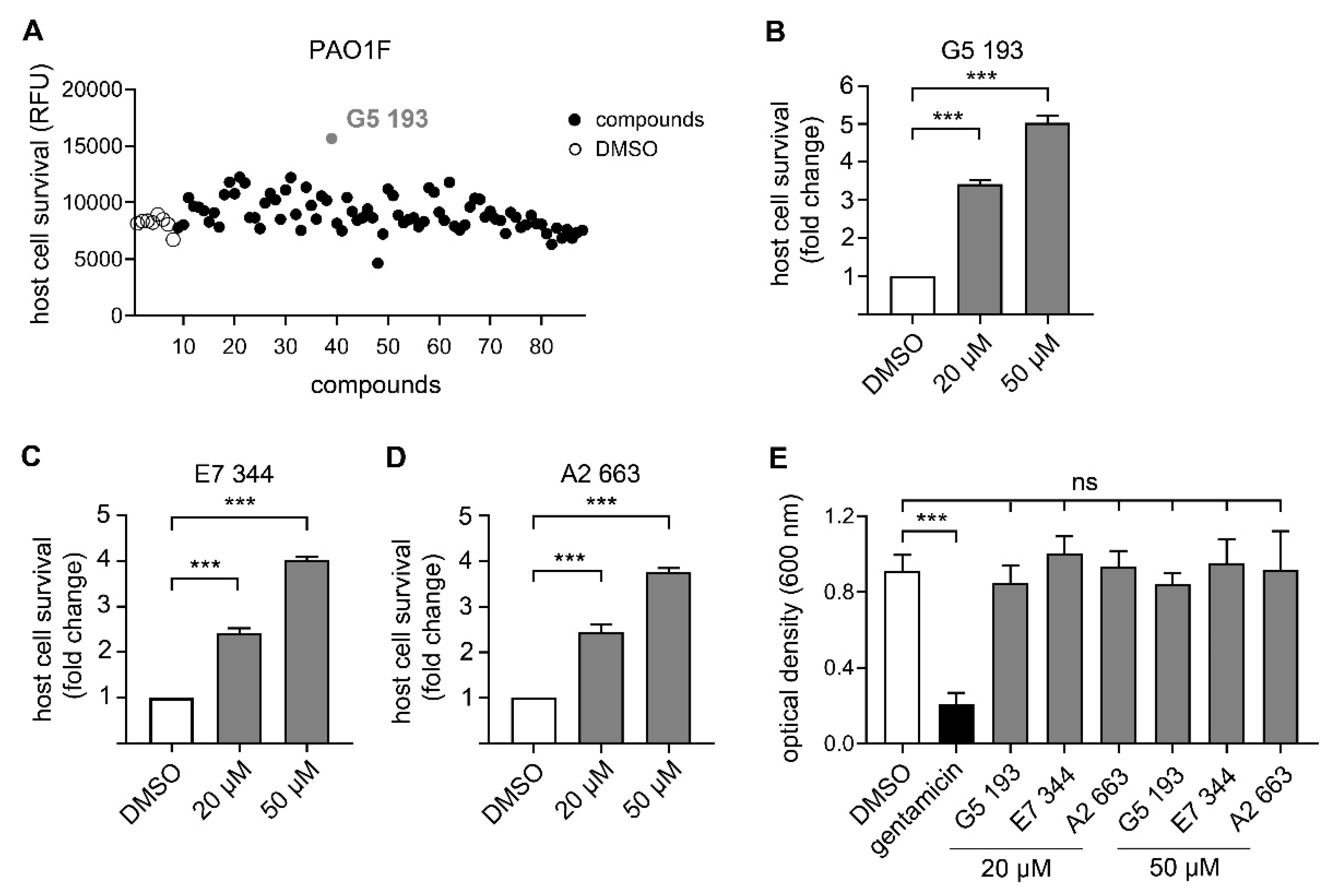
3.3. Identification of Novel Compounds with Anti-Virulence Activity against Pseudomonas Aeruginosa
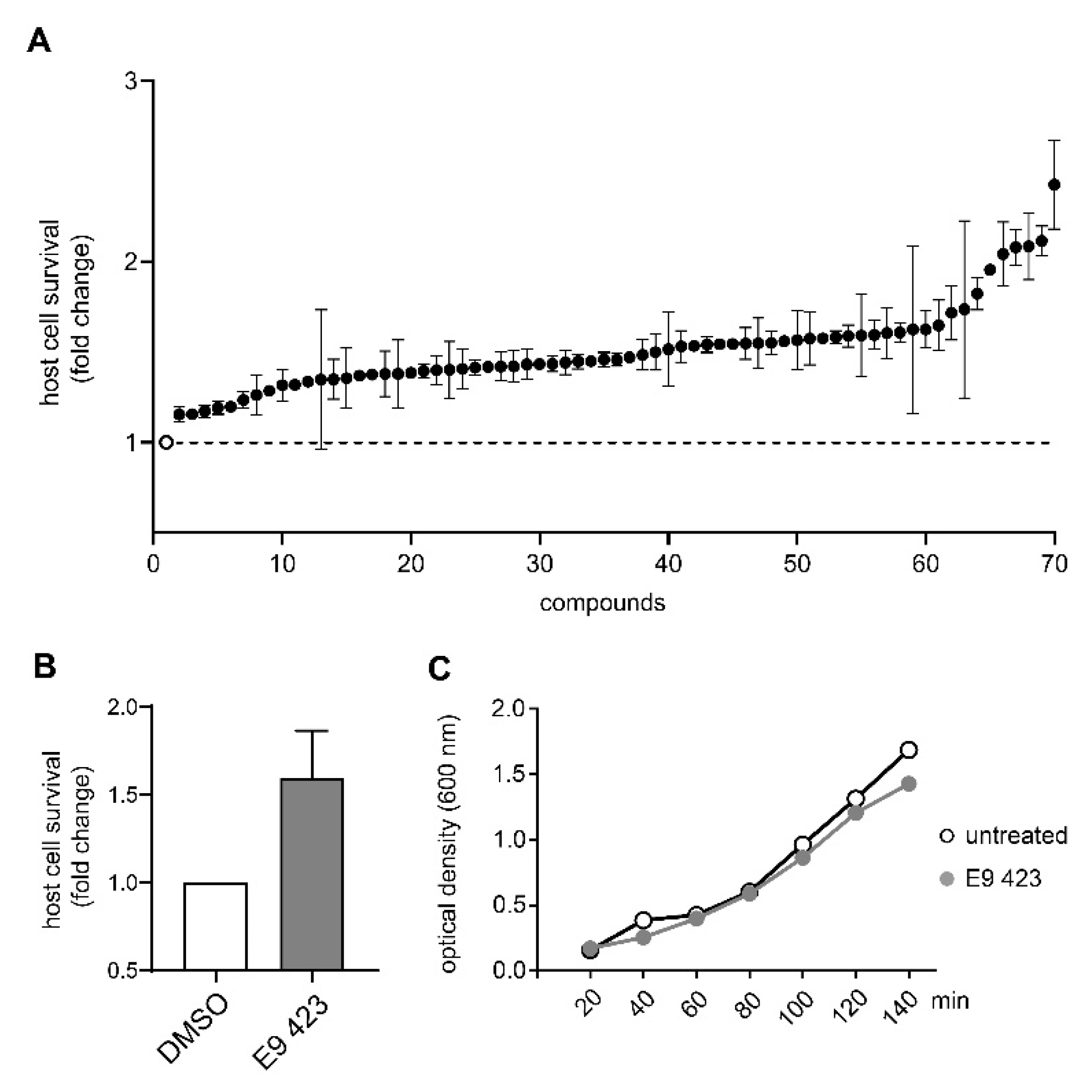
3.4. Chemical Structures of Novel Indole Compounds with Anti-Virulence Activity against Salmonella Typhimurium
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Internal Number | Specs-ID | Chemical Structure |
|---|---|---|
| D1 666 | AQ-750/42209760 | 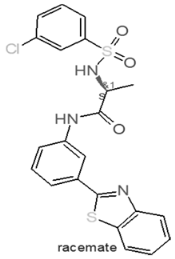 |
| A2 668 | AF-399/42309870 | 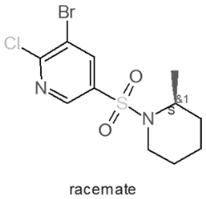 |
| G6 120 | AO-079/15259251 | 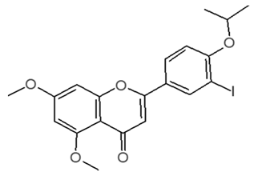 |
References
- Cassini, A.; Hogberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Bassetti, M.; Peghin, M.; Vena, A.; Giacobbe, D.R. Treatment of Infections Due to MDR Gram-Negative Bacteria. Front. Med. (Lausanne) 2019, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, E.M.; Thaden, J.T. Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics 2019, 8, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Exner, M.; Bhattacharya, S.; Christiansen, B.; Gebel, J.; Goroncy-Bermes, P.; Hartemann, P.; Heeg, P.; Ilschner, C.; Kramer, A.; Larson, E.; et al. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control. 2017, 12, Doc05. [Google Scholar] [CrossRef] [PubMed]
- Pragasam, A.K.; Veeraraghavan, B.; Nalini, E.; Anandan, S.; Kaye, K.S. An update on antimicrobial resistance and the role of newer antimicrobial agents for Pseudomonas aeruginosa. Indian J. Med. Microbiol 2018, 36, 303–316. [Google Scholar] [CrossRef]
- Uche, I.V.; MacLennan, C.A.; Saul, A. A Systematic Review of the Incidence, Risk Factors and Case Fatality Rates of Invasive Nontyphoidal Salmonella (iNTS) Disease in Africa (1966 to 2014). PLoS Negl. Trop. Dis. 2017, 11, 0005118. [Google Scholar] [CrossRef]
- Dodds, D.R. Antibiotic resistance: A current epilogue. Biochem. Pharm. 2017, 134, 139–146. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Peng, M.; Salaheen, S.; Buchanan, R.L.; Biswas, D. Alterations of Salmonella enterica Serovar Typhimurium Antibiotic Resistance under Environmental Pressure. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [Green Version]
- Tacconelli, E.; Magrini, N.; Kahlmeter, G.; Singh, N. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Anantharajah, A.; Mingeot-Leclercq, M.P.; Van Bambeke, F. Targeting the Type Three Secretion System in Pseudomonas aeruginosa. Trends Pharmacol. Sci. 2016, 37, 734–749. [Google Scholar] [CrossRef]
- Gu, L.; Zhou, S.; Zhu, L.; Liang, C.; Chen, X. Small-Molecule Inhibitors of the Type III Secretion System. Molecules 2015, 20, 17659–17674. [Google Scholar] [CrossRef] [PubMed]
- Felise, H.B.; Nguyen, H.V.; Pfuetzner, R.A.; Barry, K.C.; Jackson, S.R.; Blanc, M.P.; Bronstein, P.A.; Kline, T.; Miller, S.I. An inhibitor of gram-negative bacterial virulence protein secretion. Cell Host Microbe 2008, 4, 325–336. [Google Scholar] [CrossRef] [Green Version]
- D’Angelo, F.; Baldelli, V.; Halliday, N.; Pantalone, P.; Polticelli, F.; Fiscarelli, E.; Williams, P.; Visca, P.; Leoni, L.; Rampioni, G. Identification of FDA-Approved Drugs as Antivirulence Agents Targeting the pqs Quorum-Sensing System of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carabajal, M.A.; Asquith, C.R.M.; Laitinen, T.; Tizzard, G.J.; Yim, L.; Rial, A.; Chabalgoity, J.A.; Zuercher, W.J.; Garcia Vescovi, E. Quinazoline-Based Antivirulence Compounds Selectively Target Salmonella PhoP/PhoQ Signal Transduction System. Antimicrob. Agents Chemother. 2019, 64, e01744-19. [Google Scholar] [CrossRef] [PubMed]
- Groisman, E.A. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 2001, 183, 1835–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, S.I. PhoP/PhoQ: Macrophage-specific modulators of Salmonella virulence? Mol. Microbiol. 1991, 5, 2073–2078. [Google Scholar] [CrossRef]
- Kuijl, C.; Savage, N.D.; Marsman, M.; Tuin, A.W.; Janssen, L.; Egan, D.A.; Ketema, M.; van den Nieuwendijk, R.; van den Eeden, S.J.; Geluk, A.; et al. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature 2007, 450, 725–730. [Google Scholar] [CrossRef]
- Becker, K.A.; Riethmuller, J.; Seitz, A.P.; Gardner, A.; Boudreau, R.; Kamler, M.; Kleuser, B.; Schuchman, E.; Caldwell, C.C.; Edwards, M.J.; et al. Sphingolipids as targets for inhalation treatment of cystic fibrosis. Adv. Drug Deliv. Rev. 2018, 133, 66–75. [Google Scholar] [CrossRef]
- Grassme, H.; Jendrossek, V.; Riehle, A.; von Kurthy, G.; Berger, J.; Schwarz, H.; Weller, M.; Kolesnick, R.; Gulbins, E. Host defense against Pseudomonas aeruginosa requires ceramide-rich membrane rafts. Nat. Med. 2003, 9, 322–330. [Google Scholar] [CrossRef]
- Kornhuber, J.; Tripal, P.; Reichel, M.; Muhle, C.; Rhein, C.; Muehlbacher, M.; Groemer, T.W.; Gulbins, E. Functional Inhibitors of Acid Sphingomyelinase (FIASMAs): A novel pharmacological group of drugs with broad clinical applications. Cell Physiol. Biochem. 2010, 26, 9–20. [Google Scholar] [CrossRef]
- Rybniker, J.; Vocat, A.; Sala, C.; Busso, P.; Pojer, F.; Benjak, A.; Cole, S.T. Lansoprazole is an antituberculous prodrug targeting cytochrome bc1. Nat. Commun. 2015, 6, 7659. [Google Scholar] [CrossRef]
- SPECS Company. Available online: https://specs.net/pdf/SPECS-factsheet-world%20diversity%20set.pdf (accessed on 4 June 2020).
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Chi, E.; Mehl, T.; Nunn, D.; Lory, S. Interaction of Pseudomonas aeruginosa with A549 pneumocyte cells. Infect. Immun. 1991, 59, 822–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Karmakar, M.; Taylor, P.R.; Rietsch, A.; Pearlman, E. ExoS and ExoT ADP ribosyltransferase activities mediate Pseudomonas aeruginosa keratitis by promoting neutrophil apoptosis and bacterial survival. J. Immunol. 2012, 188, 1884–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, N.; Hulme, S.D.; Barrow, P.A. Vasoactive intestinal peptide (VIP) prevents killing of virulent and phoP mutant Salmonella typhimurium by inhibiting IFN-gamma stimulated NADPH oxidative pathways in murine macrophages. Cytokine 2006, 36, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Wemyss, M.A.; Pearson, J.S. Host Cell Death Responses to Non-typhoidal Salmonella Infection. Front. Immunol. 2019, 10, 1758. [Google Scholar] [CrossRef]
- Lee, J.; Attila, C.; Cirillo, S.L.; Cirillo, J.D.; Wood, T.K. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb. Biotechnol. 2009, 2, 75–90. [Google Scholar] [CrossRef] [Green Version]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Yuan, W.; Yu, Z.; Song, W.; Li, Y.; Fang, Z.; Zhu, B.; Li, X.; Wang, H.; Hong, W.; Sun, N. Indole-core-based novel antibacterial agent targeting FtsZ. Infect. Drug Resist. 2019, 12, 2283–2296. [Google Scholar] [CrossRef] [Green Version]
- Kohli, N.; Crisp, Z.; Riordan, R.; Li, M.; Alaniz, R.C.; Jayaraman, A. The microbiota metabolite indole inhibits Salmonella virulence: Involvement of the PhoPQ two-component system. PLoS ONE 2018, 13, 190613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikaido, E.; Giraud, E.; Baucheron, S.; Yamasaki, S.; Wiedemann, A.; Okamoto, K.; Takagi, T.; Yamaguchi, A.; Cloeckaert, A.; Nishino, K. Effects of indole on drug resistance and virulence of Salmonella enterica serovar Typhimurium revealed by genome-wide analyses. Gut Pathog. 2012, 4, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domiciano, T.P.; Wakita, D.; Jones, H.D.; Crother, T.R.; Verri, W.A., Jr.; Arditi, M.; Shimada, K. Quercetin Inhibits Inflammasome Activation by Interfering with ASC Oligomerization and Prevents Interleukin-1 Mediated Mouse Vasculitis. Sci. Rep. 2017, 7, 41539. [Google Scholar] [CrossRef] [PubMed]
- Lamkanfi, M.; Mueller, J.L.; Vitari, A.C.; Misaghi, S.; Fedorova, A.; Deshayes, K.; Lee, W.P.; Hoffman, H.M.; Dixit, V.M. Glyburide inhibits the Cryopyrin/Nalp3 inflammasome. J. Cell Biol. 2009, 187, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Broz, P.; Newton, K.; Lamkanfi, M.; Mariathasan, S.; Dixit, V.M.; Monack, D.M. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 2010, 207, 1745–1755. [Google Scholar] [CrossRef]
| Internal Number | Specs-ID | Chemical Structure |
|---|---|---|
| G5 193 | AJ-292/43278258 | 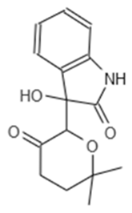 |
| E7 344 | AK-968/11036034 | 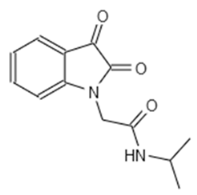 |
| A2 663 | AQ-911/40696225 | 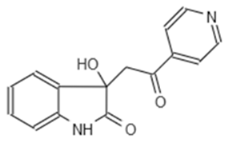 |
| B2 442 | AK-918/42028178 | 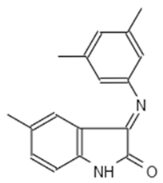 |
| B2 621 | AQ-900/41921933 |  |
| D5 682 | AG-219/3696225 | 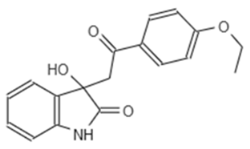 |
| Internal Number | Specs-ID | Chemical Structure |
|---|---|---|
| E9 423 | AN-584/43416482 | 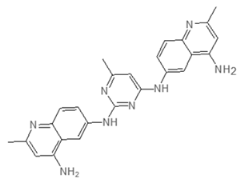 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
von Ambüren, J.; Schreiber, F.; Fischer, J.; Winter, S.; van Gumpel, E.; Simonis, A.; Rybniker, J. Comprehensive Host Cell-Based Screening Assays for Identification of Anti-Virulence Drugs Targeting Pseudomonas aeruginosa and Salmonella Typhimurium. Microorganisms 2020, 8, 1096. https://doi.org/10.3390/microorganisms8081096
von Ambüren J, Schreiber F, Fischer J, Winter S, van Gumpel E, Simonis A, Rybniker J. Comprehensive Host Cell-Based Screening Assays for Identification of Anti-Virulence Drugs Targeting Pseudomonas aeruginosa and Salmonella Typhimurium. Microorganisms. 2020; 8(8):1096. https://doi.org/10.3390/microorganisms8081096
Chicago/Turabian Stylevon Ambüren, Julia, Fynn Schreiber, Julia Fischer, Sandra Winter, Edeltraud van Gumpel, Alexander Simonis, and Jan Rybniker. 2020. "Comprehensive Host Cell-Based Screening Assays for Identification of Anti-Virulence Drugs Targeting Pseudomonas aeruginosa and Salmonella Typhimurium" Microorganisms 8, no. 8: 1096. https://doi.org/10.3390/microorganisms8081096
APA Stylevon Ambüren, J., Schreiber, F., Fischer, J., Winter, S., van Gumpel, E., Simonis, A., & Rybniker, J. (2020). Comprehensive Host Cell-Based Screening Assays for Identification of Anti-Virulence Drugs Targeting Pseudomonas aeruginosa and Salmonella Typhimurium. Microorganisms, 8(8), 1096. https://doi.org/10.3390/microorganisms8081096





