Variable Expression of Notch1 and Pax5 in Classical Hodgkin Lymphoma and Infection with Epstein–Barr in Pediatric Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cases
2.2. Epstein–Barr Detection
2.3. Immunohistochemistry of Tissue Microarrays
2.4. Immunostaining
2.5. Variable Definitions
2.6. Statistical Analysis
3. Results
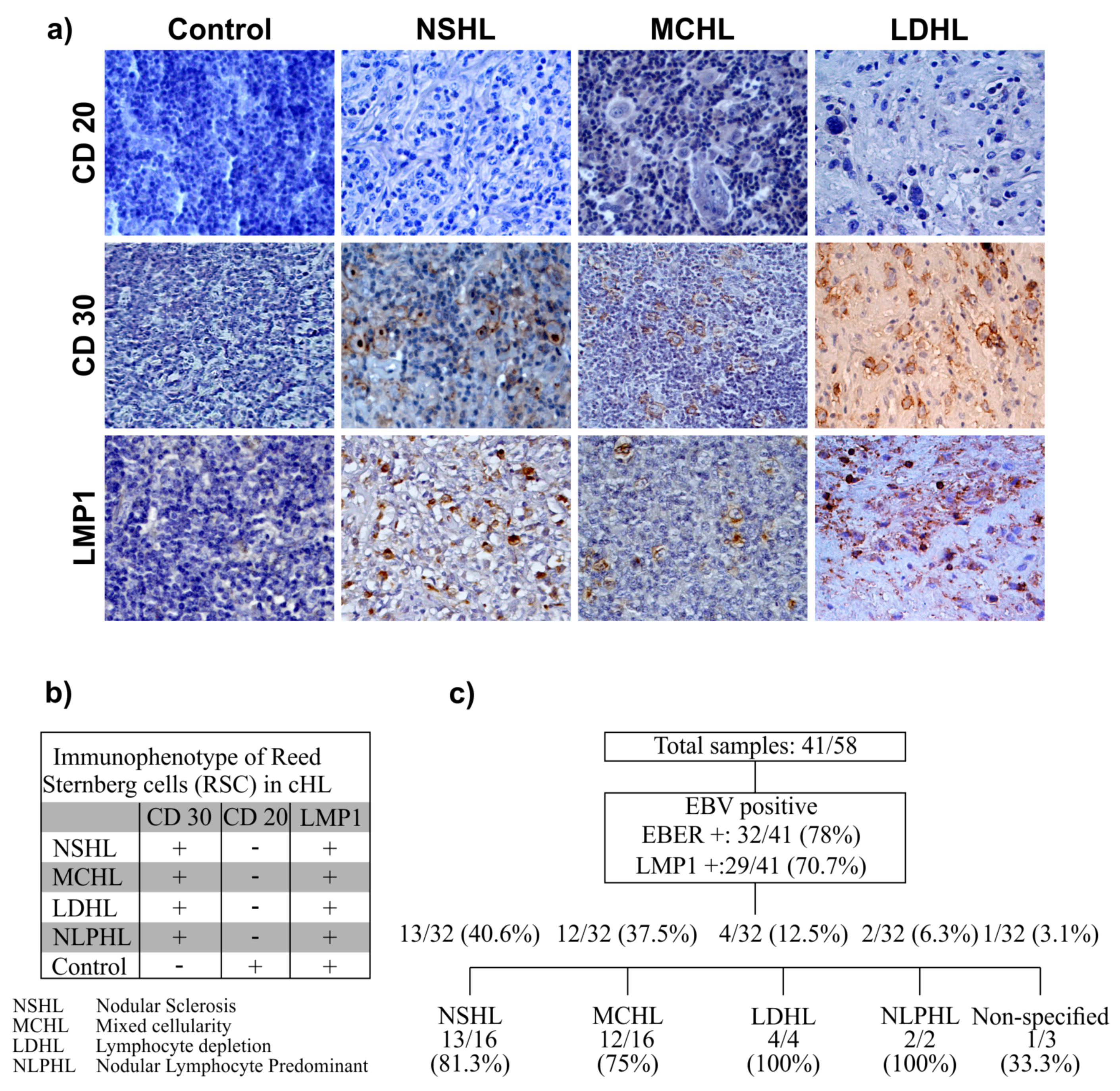
3.1. EBV Detection
3.2. Age at Presentation and Immunophenotype
3.3. NOTCH1 Expression
3.4. PAX5 Expression
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Jesus Loeza, J.; Montes, E.; Andrade, I.; Cortes, E.; Reyes, D.; Vazquez, J.; Pérez, C. Prevalence of Germ Cell Tumors in a Mexican Tertiary Hospital and the Creation of a Probabilistic Network of Risk Factors. Iran. J. Pediatr. 2020, 30, e96590. [Google Scholar]
- Swerdlow, S.; Harris, N.; Jaffe, E.; Pileri, S.; Stein, H.; Thiele, J.; Vierdiman, J. WHO Classification of Tumours; IARC: Lyon, France, 2008; p. 2. [Google Scholar]
- Kapatai, G.; Murray, P. Contribution of the Epstein Barr virus to the molecular pathogenesis of Hodgkin lymphoma. J. Clin. Pathol. 2007, 60, 1342–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarrett, A.; Alexander, A. Epidemiology of EBV and Hodgkin’s lymphoma. Ann. Oncol. 1996, 7, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Shaklawoon, K.; Altagazi, N.; Altorjman, F.; Alturki, A.; Eltaweel, M.; Alqawi, O. Molecular detection of Epstein–Barr virus in different types of lymphoma. Mol. Biol. Rep. 2020, 47, 1803–1807. [Google Scholar] [CrossRef] [PubMed]
- Carotta, S.; Nutt, S. Losing B cell identity. BioEssays 2008, 30, 203–207. [Google Scholar] [CrossRef]
- Carotta, S.; Holmes, M.; Pridans, C.; Nutt, S. Pax5 maintains cellular identity by repressing gene expression throughout B cell differentiation. Cell Cycle 2006, 5, 2452–2456. [Google Scholar] [CrossRef] [PubMed]
- Cobaleda, C.; Schebesta, A.; Delogu, A.; Busslinger, M. Pax5: The guardian of B cell identity and function. Nat. Immunol. 2007, 8, 463–470. [Google Scholar] [CrossRef]
- Roessler, S. Role of transcription factors in commitment and differentiation of early B lymphoid cells. Semin. Immunol. 2006, 18, 12–19. [Google Scholar] [CrossRef]
- Anderson, L.; Longnecker, R. Epstein-Barr virus latent membrane protein 2A exploits Notch1 to alter B cell identity in vivo. Lymphoid Neoplasia 2009, 113, 108–115. [Google Scholar] [CrossRef] [Green Version]
- Jundt, F.; Acikgöz, O.; Kwon, S.; Schwarzer, R.; Anagnostopoulos, I.; Wiesner Mathas, S.; Hummel, M.; Stein, H.; Reichardt, H.; Dörken, B. Aberrant expression of Notch1 interferes with the B-lymphoid phenotype of neoplastic B cells in classical Hodgkin lymphoma. Leukemia 2008, 22, 1587–1594. [Google Scholar] [CrossRef]
- Hansmann, M.; Willenbrock, K. Die WHO-Klassifikation des Hodgkin-Lymphoms und ihre molekularpathologische Relevanz [WHO classification of Hodgkin’s lymphoma and its molecular pathological relevance]. Pathologe 2002, 23, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Palma, I.; Sánchez, A.E.; Jiménez-Hernández, E.; Alvarez-Rodríguez, F.; Nava-Frias, M.; Valencia-Mayoral, P.; Salinas-Lara, C.; Velazquez-Guadarrama, N.; Portilla-Aguilar, J.; Pena, R.Y.; et al. Detection of Epstein-Barr Virus and Genotyping Based on EBNA2 Protein in Mexican Patients With Hodgkin Lymphoma: A Comparative Study in Children and Adults. Clin. Lymphoma Myeloma Leuk. 2013, 13, 266–272. [Google Scholar] [CrossRef]
- Piris, M.; Medeiros, L.; Chang, C. Hodgkin lymphoma: A review of pathological features and recent advances in pathogenesis. Pathology 2020, 52, 154–165. [Google Scholar] [CrossRef] [Green Version]
- Höflinger, S.; Kesavan, K.; Fuxa, M.; Hutter, C.; Heavey, B.; Radtke, F.; Busslinger, M. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J. Immunol. 2004, 173, 3935–3944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, G.; Wang, X.; Xiao, H.; Liu, X.; Fang, Y.; Zhai, B.; Xu, R.; Han, G.; Chen, G.; Hou, C.; et al. Both Notch1 and its ligands in B cells promote antibody production. Mol. Immunol. 2017, 91, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Arima, H.; Nishikori, M.; Otsuka, Y.; Kishimoto, W.; Izumi, K.; Yasuda, K.; Yoshimoto, T.; Takaori-Kondo, A. B cells with aberrant activation of Notch1 signaling promote Treg and Th2 cell-dominant T-cell responses via IL-33. Blood Adv. 2018, 2, 2282–2295. [Google Scholar] [CrossRef]
- Romero-Masters, J.; Ohashi, M.; Djavadian, R.; Eichelberg, M.; Hayes, M.; Bristol, J.; Shidong, M.; Ranheim, E.; Gumperz, J.; Johannsen, E.; et al. An EBNA3C-deleted Epstein-Barr virus (EBV) mutant causes B-cell lymphomas with delayed onset in a cord blood-humanized mouse model. PLoS Pathog. 2018, 14, e1007221. [Google Scholar] [CrossRef] [Green Version]
- Desouki, M.; Post, P.; Cherry, D.; Lazarchick, J. PAX-5: A Valuable Immunohistochemical Marker in the Differential Diagnosis of Lymphoid Neoplasms. Clin. Med. Res. 2010, 8, 84–88. [Google Scholar] [CrossRef] [Green Version]
- Kanteti, R.; Nallasura, V.; Loganathan, S.; Tretiakova, M.; Kroll, T.; Krishnaswamy, S.; Faoro, L.; Cagle, P.; Husain, A.; Vokes, E.; et al. PAX5 is expressed in small-cell lung cancer and positively regulates c-Met transcription. Lab. Investig. 2009, 89, 301–314. [Google Scholar] [CrossRef] [Green Version]
- Brzozowa-Zasada, M.; Piecuch, A.; Michalski, M.; Segiet, O.; Kurek, J.; Harabin-Słowińska, M.; Wojnicz, R. Notch and its oncogenic activity in human malignancies. Eur. Surg. 2017, 49, 199–209. [Google Scholar] [CrossRef]
- Jensen, K.; Montgomery, K.; Kaygusuz, G.; Natkunam, Y. The utility of PAX5 immunohistochemistry in the diagnosis of undifferentiated malignant neoplasms. Modern Pathol. 2007, 20, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Adams, H.; Liebisch, P.; Schmid, P.; Dirnhofer, S.; Tzankov, A. Diagnostic utility of the B-cell lineage markers CD20, CD79a, PAX5, and CD19 in paraffin-embedded tissues from lymphoid neoplasms. Appl. Immunohisto. Mol. Morphol. 2009, 17, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Ahmad, A.; Kayani, N.; Minhas, K. Expression of PAX-5 in B Cell Hodgkin and Non Hodgkin Lymphoma. Asian Pac. J. Cancer Prev. 2018, 19, 3463–3466. [Google Scholar] [CrossRef] [Green Version]
- Feldman, A.; Dogan, A. Diagnostic uses of Pax5 immunohistochemistry. Adv. Anat. Pathol. 2007, 14, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Sukswai, N.; Khoury, J. Immunohistochemistry Innovations for Diagnosis and Tissue-Based Biomarker Detection. Curr. Hematol. Malig. Rep. 2019, 14, 368–375. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, D.; Fedoriw, Y.; Weiss, L. Distinguishing classical Hodgkin lymphoma, gray zone lymphoma, and large B-cell lymphoma: A proposed scoring system. Appl. Immunohisto. Mol. Morphol. 2016, 24, 535–540. [Google Scholar]
- Cozzolino, I.; Vitagliano, G.; Caputo, A.; Montella, M.; Franco, R.; Ciaincia, G.; Selleri, C.; Zeppa, P. CD15, CD30, and PAX5 evaluation in Hodgkin’s lymphoma on fine-needle aspiration cytology samples. Diagn. Cytopathol. 2020, 48, 211–216. [Google Scholar] [CrossRef]
- Vali Betts, E.; Dwyre, D.; Wang, H.; Rashidi, H. PAX5-Negative Classical Hodgkin Lymphoma: A Case Report of a Rare Entity and Review of the Literature. Case Rep. Hematol. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Maillard, I.; Pear, W. Notch and the immune system. Immunity 2003, 19, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Adelusola, K.; Rotimi, O.; Durosinmi, M. Epstein Barr virus latent membrana protein-1 in Hodgkin’s lymphoma in Nigerians. Afr. Health Sci. 2009, 9, 174–178. [Google Scholar]
- Hofscheier, A.; Ponciano, A.; Bonzheim, I.; Adam, P.; Lome-Maldonado, C.; Vela, T.; Cortes, E.; Ortiz-Hidalgo, C.; Fend, F.; Quintanilla-Martínez, L. Geographic variation in the prevalence of Epsten-Barr virus-positive diffuse large B-cell lymphoma of the elderly: A comparative analysis of a mexican and german population. Mod. Pathol. 2011, 24, 1046–1054. [Google Scholar] [CrossRef] [Green Version]
- Nam-Cha, S.; Salcedo, M.; San juan, J.; Garcia, J.; Piris, M. Lymphocyte-rich classical Hodgkin’s lymphoma: Distinctive tumor and microenvironment markers. Modern Pathol. 2009, 22, 1006–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knecht, H.; Bachmann, E.; Joske, D.; Sahli, R.; Emery-Goodman, A.; Casanova, J.; Zilic, M.; Bachmann, F.; Odermatt, B. Molecular analysis of the LMP (latent membrane protein) oncogene in Hodgkin’s disease. Leukemia 1993, 7, 580–585. [Google Scholar] [PubMed]
- Chang, K.; Chen, P.; Chang, Y.; Wu, Y.; Chen, Y.; Lai, C.; Wang, H. Epstein–Barr virus latent membrane protein-1 up-regulates cytokines and correlates with older age and poorer prognosis in Hodgkin lymphoma. Histopathology 2017, 70, 442–455. [Google Scholar] [CrossRef] [PubMed]


| Subtype | Total cHL Cases | EBV Status | Reed–Sternberg Cell Counts | ||||
|---|---|---|---|---|---|---|---|
| Negative | Positive | ||||||
| n = 9 | n = 32 | <20 | 21–40 | 41–60 | >61 | ||
| NSHL | 16 | 3 | 13 | 7 | 0 | 0 | 6 |
| MCHL | 16 | 4 | 12 | 6 | 2 | 0 | 4 |
| LDHL | 4 | 0 | 4 | 0 | 0 | 1 | 3 |
| NLPHL | 2 | 0 | 2 | 0 | 0 | 0 | 2 |
| Not Specified | 3 | 2 | 1 | 1 | 0 | 0 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palma-Lara, I.; Sánchez-Aldana, A.E.; Jiménez-Hernández, E.; Martínez-Villegas, O.; Núñez-Enríquez, J.C.; Mejía-Aranguré, J.M.; Ochoa, S.A.; Xicohtencatl-Cortes, J.; Cruz-Córdova, A.; Zavala-Vega, S.; et al. Variable Expression of Notch1 and Pax5 in Classical Hodgkin Lymphoma and Infection with Epstein–Barr in Pediatric Patients. Microorganisms 2020, 8, 958. https://doi.org/10.3390/microorganisms8060958
Palma-Lara I, Sánchez-Aldana AE, Jiménez-Hernández E, Martínez-Villegas O, Núñez-Enríquez JC, Mejía-Aranguré JM, Ochoa SA, Xicohtencatl-Cortes J, Cruz-Córdova A, Zavala-Vega S, et al. Variable Expression of Notch1 and Pax5 in Classical Hodgkin Lymphoma and Infection with Epstein–Barr in Pediatric Patients. Microorganisms. 2020; 8(6):958. https://doi.org/10.3390/microorganisms8060958
Chicago/Turabian StylePalma-Lara, Icela, Ana Elena Sánchez-Aldana, Elva Jiménez-Hernández, Octavio Martínez-Villegas, Juan Carlos Núñez-Enríquez, Juan Manuel Mejía-Aranguré, Sara A. Ochoa, Juan Xicohtencatl-Cortes, Ariadnna Cruz-Córdova, Sergio Zavala-Vega, and et al. 2020. "Variable Expression of Notch1 and Pax5 in Classical Hodgkin Lymphoma and Infection with Epstein–Barr in Pediatric Patients" Microorganisms 8, no. 6: 958. https://doi.org/10.3390/microorganisms8060958
APA StylePalma-Lara, I., Sánchez-Aldana, A. E., Jiménez-Hernández, E., Martínez-Villegas, O., Núñez-Enríquez, J. C., Mejía-Aranguré, J. M., Ochoa, S. A., Xicohtencatl-Cortes, J., Cruz-Córdova, A., Zavala-Vega, S., García-Jiménez, M., Contreras-Ramos, A., Torres-Nava, J. R., Mora-Ramiro, G., & Arellano-Galindo, J. (2020). Variable Expression of Notch1 and Pax5 in Classical Hodgkin Lymphoma and Infection with Epstein–Barr in Pediatric Patients. Microorganisms, 8(6), 958. https://doi.org/10.3390/microorganisms8060958








